Abstract
Major histocompatibility complex class I (MHC I) molecules present antigenic peptides for CD8+ T-cell recognition. Prior to cell surface expression, proper MHC I loading is conducted by the peptide-loading complex (PLC), composed of the MHC I heavy chain (HC) and β2-microglobulin (β2m), the peptide transporter TAP, and several chaperones, including tapasin. Tapasin connects peptide-receptive MHC I molecules to the PLC, thereby facilitating loading of high-affinity peptides onto MHC I. To cope with CD8+ T-cell responses, human cytomegalovirus (HCMV) encodes several posttranslational strategies inhibiting peptide transport and MHC I biogenesis which have been studied extensively in transfected cells. Here we analyzed assembly of the PLC in naturally HCMV-infected fibroblasts throughout the protracted replication cycle. MHC I incorporation into the PLC was absent early in HCMV infection. Subsequently, tapasin neosynthesis became strongly reduced, while tapasin steady-state levels diminished only slowly in infected cells, revealing a blocked synthesis rather than degradation. Tapasin mRNA levels were continuously downregulated during infection, while tapasin transcripts remained stable and long-lived. Taking advantage of a novel method by which de novo transcribed RNA is selectively labeled and analyzed, an immediate decline of tapasin transcription was seen, followed by downregulation of TAP2 and TAP1 gene expression. However, upon forced expression of tapasin in HCMV-infected cells, repair of MHC I incorporation into the PLC was relatively inefficient, suggesting an additional level of HCMV interference. The data presented here document a two-pronged coordinated attack on tapasin function by HCMV.
Human cytomegalovirus (HCMV) belongs to the β-subgroup of herpesviruses, which are a family of viruses with a large double-stranded DNA genome. The HCMV genome carries approximately 200 genes, which are transcribed in a cascading fashion of immediate-early (IE), early (E), and late (L) genes. Completion of the protracted HCMV replication cycle takes 72 to 96 h. Like all herpesviruses, HCMV persists in the infected host for life, with alternating episodes of latent infection and recurrent replication. While being clinically symptomless in immunocompetent individuals, HCMV can cause severe disease in immunocompromised individuals, such as transplant and AIDS patients, reflecting the delicate balance between the immune system responding to the infection and viral evasion of immune control.
Antigen presentation to CD8+ T cells is a major defense mechanism against virally infected cells. The major histocompatibility complex class I (MHC I) antigen presentation pathway exposes peptide antigens on the cell surface to surveilling CD8+ T cells. Upon a fitting contact between the T cell receptor and an MHC I-peptide complex, the CD8+ T cell becomes activated to induce lytic destruction of the recognized target cell.
The antigenic peptides displayed by MHC I molecules on the cell surface are degradation products of proteins that have been targeted to the proteasome. To reach the luminal side of the endoplasmic reticulum (ER), the peptides are bound to the transporter associated with antigen processing (TAP) before being translocated across the ER membrane (43), where MHC I molecules encounter their peptide ligands. TAP is a heterodimeric ABC (ATP binding cassette) transporter consisting of the subunits TAP1 and TAP2. The efficient loading of peptides onto MHC I molecules requires participation of several additional chaperones. The peptide-receptive heterodimeric MHC I, comprising the MHC I heavy chain (HC) and β2-microglobulin (β2m), is recruited to the peptide-loading complex (PLC). In the PLC, MHC I is bridged to TAP via the chaperone tapasin (48). Tapasin connects PLC constituents but also exerts a critical quality control on the MHC I loading process that determines the release of MHC I molecules and cell surface expression (23, 57). In addition, calreticulin and the oxidoreductase ERp57 are found as further constituents of the PLC (34, 44). Within the PLC, tapasin and ERp57 form a stable interaction through a disulfide bond (41, 42). Recently, it was shown that this specific interaction promotes loading of high-affinity peptides even in the presence of an excess of low-affinity peptides (54).
Most of the constituents of the MHC I antigen presentation pathway are extensively controlled on the transcriptional level. Expression of the genes encoding MHC I, β2m, TAP1, TAP2, and tapasin is strongly induced by gamma interferon (IFN-γ) (1, 26, 35). In contrast, ERp57 is not elevated by IFN-γ stimulation, but under these conditions, the majority of the existing molecules are recruited to the PLC (42). Analysis of the mouse tapasin promoter showed that several gamma activated sequence (GAS) elements located shortly upstream of the transcription start site are functional. Enhancement of gene transcription by IFN-γ was found to be dependent on secondary transcription factors such as interferon regulatory factor 1 (IRF-1) (1, 22). Conversely, a transcriptional repressor belonging to the Krüppel family of zinc finger proteins, PRDM-1 (positive regulatory domain I; also called BLIMP1 [B-lymphocyte-induced maturation protein 1]), is able to inhibit IFN-γ-induced transcription of tapasin (10).
Elaborate strategies by which HCMV inhibits the MHC I antigen presentation pathway have been characterized extensively, highlighting the inherent necessity of this virus to escape CD8+ T-cell immune control. Multiple glycoproteins encoded by the unique short region of the HCMV genome block defined checkpoints of the class I pathway. Specifically, US2 and US11 degrade the MHC I HC (28, 55), whereas US3 inhibits the export of MHC I out of the ER (2, 29). Ahn and colleagues also found that US3 is able to inhibit the function of tapasin (37) and that of protein disulfide isomerase (PDI) (38). US3 is a short-lived protein which is expressed only during the IE phase of HCMV gene expression (13, 49), implying that the inhibition of tapasin through US3 is restricted to a few hours of HCMV infection. US6 binds to TAP and blocks the peptide supply, preventing the formation of ternary MHC I (3, 19, 32). US6 interacts with both TAP subunits to inhibit peptide transport, thereby freezing the PLC in a complex with TAP and US6 (15). Reports on as yet undefined mechanisms suggest that US10, US8, and UL82/pp71 might also be involved in evasion of antigen presentation by MHC I molecules (11, 50, 51).
Here we analyzed the formation of the PLC throughout the HCMV replication cycle in detail. Deficient association of MHC class I heterodimers with the PLC was observed early during the course of infection. In addition, HCMV blocked nascent tapasin gene transcription and protein neosynthesis. Forced tapasin transcription in HCMV-infected cells only partly reconstituted the incorporation of MHC I into the PLC and the density of MHC I molecules on the cell surface, pointing at possible unidentified mechanisms by which HCMV prevents proper PLC assembly and further impedes peptide loading of MHC I.
MATERIALS AND METHODS
Cell lines and antibodies.
MRC-5 (ATCC CCL171) and HEK 293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (FCS), penicillin, streptomycin, and glutamine. The STC anti-tapasin rabbit antiserum recognizing the C terminus of tapasin was used for immunoprecipitation (48). The Stressgen CSA-630 anti-tapasin rabbit antibody or N-17 (Santa Cruz Biotechnology) was applied for immunoblot detection. For ERp57 immunoprecipitation and immunoblot detection, mouse ascitic fluid (Upstate) was used, and the mouse monoclonal antibodies (MAbs) 148.3 and 439.3 were used as human TAP1- and TAP2-specific reagents, respectively (36, 52). Anti-calreticulin was obtained from Stressgen (SPA-600). For precipitation and fluorescence-activated cell sorter (FACS) analysis of MHC I HC-β2m complexes, MAb W6/32 (ATCC HB-95) was used. In addition, we used fluorescein isothiocyanate (FITC)-conjugated IgG Fcγ fragments (Rockland) for FACS staining of viral Fcγ receptors (vFcγRs).
Viruses.
In this study, the HCMV wild-type (wt) strain AD169 (17), the ΔUS6-US11 deletion mutant RV35 (27), and the AD169-derived HCMV ΔUS2-US6 deletion mutant HB5 (6) and ΔUS2-US11 mutant (14) were employed. The viruses were propagated on MRC-5 cells, and purified HCMV stocks were prepared as described previously (31). In addition, a vFcγR-deficient HB5 mutant lacking both IRL11- and TRL11-encoded gp34 and UL119-118-encoded gp68 (4) was generated using PCR-directed bacmid-based mutagenesis as described previously (4, 6, 53). The bacterial artificial chromosome (BAC) plasmid pHB5-ΔIRL-frt (kindly provided by Eva Borst, Hannover, Germany), lacking nucleotides 179150 to 192329 of pHB5 (6), was used to generate the BAC plasmid pΔIRL/ΔTRL11/ΔUL118. For deletion of UL118 from pHB5-ΔIRL-frt, the following primers were used: 5′-GGACCAGTCCCGCCGGCGCGCCAGCATGTAGGTCACGTACAAAAGAATAATCACCATGAACGATTTATTCAACAAAGCCACG-3′ (forward) and 5′-CCTTTTCCTCCCCCGCCAACCTGCCAACAAAATAAACAACTTATGAATTTTATCCACAGGGCCAGTGTTACAACCAATTAACC-3′ (reverse). The primer pair was used to amplify the kanamycin resistance (Knr) gene from plasmid pSLFRTKn (4) for homologous recombination into pΔIRL-frt, thereby disrupting the UL118 locus. The primer pair 5′-ACAGACGACGAAGAGGACGAGGACGACAACGTCTGATAAGGAAGGCGAGAACGTGTTTTGTCCAGTGAATTCGAGCTCGGTAC-3′ (forward) and 5′-TGTATACGCCGTATGCCTGTACGTGAGATGGTGAGGTCTTCGGCAGGCGACACGCATCTTGACCATGATTACGCCAAGCTCC-3′ (reverse) was used to amplify the tetracycline resistance gene (Tetr) from pCP16 (8) for insertion into pΔIRL/ΔUL118, resulting in the BAC plasmid pΔIRL/ΔTRL11/ΔUL118 (referred to here as ΔΔΔvFcγR). Recombinant HCMV was reconstituted from BAC plasmids by SuperFect transfection as described by Borst et al. (6).
Lentiviral vectors.
Tapasin was amplified from cDNA prepared from MRC-5 cells by applying the primers 5′-CGGCTAGCATGAAGTCCCTGTCTCTGC-3′ (forward) and 5′-GCGGATCCTCACTCTGCTTTCTTCTTTGAATCCTTGC-3′ (reverse). The amplified product was cloned into the puc2CL6Ipwo lentiviral vector by using the NheI and BamHI sites and was subsequently sequenced. Lentiviruses were produced in HEK 293T cells by transfection of 6 μg pCDNL-BH, 6 μg of vesicular stomatitis virus G, and 6 μg of puc2CL6IP-tpn or empty vector, using SuperFect transfection reagent (Qiagen). At 48 h posttransfection, the supernatant was collected and filtered through a 0.45-μm filter prior to transduction. MRC-5 cells were transduced at a low passage number and kept in normal medium for 4 to 5 days before transduced cells were selected with 5 μg/ml puromycin for 4 to 5 days.
siRNA nucleofection.
A small interfering RNA (siRNA) targeting tapasin was purchased from Dharmacon (L-017387). For nucleofection of siRNA, kit R from Lonza (Cologne, Germany) was applied. A total of 106 cells were resuspended in 100 μl solution R, together with 20 to 80 nM siRNA, prior to nucleofection with a Lonza Nucleofector device.
Immunoprecipitation and immunoblotting.
Immunoprecipitations were performed as follows. Cells cultured in 6-well plates were washed with phosphate-buffered saline (PBS) and metabolically labeled (Easytag Express [35S]Met-Cys protein labeling mix; Perkin Elmer) with 100 μCi/ml for various times prior to lysis of cells. About 0.5 × 106 cells were lysed in 1 ml of digitonin lysis buffer (140 mM NaCl, 20 mM Tris [pH 7.6], 5 mM MgCl2, and 1% digitonin). Lysates were cleared from membrane debris at 13,000 rpm for 30 min at 4°C. For analysis of lysates with several antibodies, identical lysates were pooled after centrifugation and then split up into equal aliquots. Lysates were incubated with antibodies for 1 h at 4°C in an overhead tumbler before immune complexes were retrieved by protein A-Sepharose (GE Healthcare). Sepharose pellets were washed five times with increasing NaCl concentrations (0.15 to 0.5 M in lysis buffer containing 0.2% detergent). Immune complexes were dissociated at 95°C for 5 min in a dithiothreitol (DTT) (40 mM)-containing sample buffer, samples were cooled on ice, and subsequently, iodoacetamide (80 mM) was added to the samples before gel loading. Proteins were separated by SDS-PAGE. For quantification, dried gels were exposed to a phosphor screen for 2 to 16 h, depending on the strengths of the protein bands of interest, and band intensities were quantified using Aida Image Analyzer software by Raytest (Straubenhardt, Germany).
For immunoblotting, adjusted protein amounts were separated by SDS-PAGE. Proteins were blotted onto a Protran membrane and incubated with specific antibodies, followed by peroxidase-conjugated secondary antibodies and detection using the enhanced chemiluminescence substrate ECL Plus (Amersham Biosciences).
Northern blotting and semiquantitative reverse transcription-PCR (RT-PCR).
RNA was isolated using a QIAshredder and RNeasy kit (Qiagen). Equal amounts of total RNA (2 to 5 μg) were subjected to gel electrophoresis and cross-linked to a nylon membrane. Hybridization, washing, and detection were performed using standard techniques. Equal loading of RNA was confirmed by ethidium bromide staining of agarose gels and reblotting of nylon membranes with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe. Detection of specific transcripts was performed with digoxigenin (DIG)-labeled probes amplified using the primers listed in Table 1.
TABLE 1.
Primers for Northern blot probes
| Primer | Sequence |
|---|---|
| TPN_Frwd | CAAAGTGTCCCTGATGCCAGC |
| TPN_Rev | GGTGAATTCGACAGGCATAGCG |
| TAP1_Frwd | TCTCCTCTCTTGGGGAGATG |
| TAP1_Rev | GAGACATGATGTTACCTGTCTG |
| TAP2_Frwd | GTGCTGTTCTCCGGTTCTGT |
| TAP2_Rev | TATCATGAACTTCCAAAGGG |
| 2m_Frwd | GGGCATTCCTGAAGCTGACA |
| 2m_Rev | TGCGGCATCTTCAAACCTCC |
| MHC I HC_Frwd | CCCGCTTCATCTCAGTGGGCTA |
| MHC I HC_Rev | GCGATGTAATCCTTGCCGTCGT |
| GAPDH_Frwd | ACCACAGTCCATGCCATCAC |
| GAPDH_Rev | TCCACCACCCTGTTGCTGTA |
| TRL10_Frwd | TATGTATCCGCGTGTAATGCAC |
| TRL10_Rev | GACGTTGTCGTCCTCGTCC |
For OneStep RT-PCR (Qiagen), 20 ng of total RNA was applied, and intron-overlapping primers were used when possible (Table 2). For semiquantitative analysis, the number of cycles was adjusted to 30 cycles. PCR products were run in a 3% Metaphor gel. For actinomycin D (actD) treatment of cells, a final concentration of 5 μg/ml was used.
TABLE 2.
Primers for RT-PCR
| Primer | Sequence | Exon | Amplicon size (bp) |
|---|---|---|---|
| TPN_Frwd | CAAAGTGTCCCTGATGCCAGC | 1 | 280 |
| TPN_Rev | GGTGAATTCGACAGGCATAGCG | 2 | |
| SOCS3_Frwd | GGAGTTCCTGGACCAGTACG | 2 | 115 |
| SOCS3_Rev | TTCTTGTGCTTGTGCCATGT | 2 | |
| β2m _Frwd | GGGCATTCCTGAAGCTGACA | 1 | 424 |
| β2m_Rev | TGCGGCATCTTCAAACCTCC | 4 |
Analysis of nascent RNA.
Labeling of nascent RNA by 4-thiouridine (4sU) was performed as described previously (9). In brief, 4sU (Sigma) was added to the cells at a final concentration of 200 μM in prewarmed, CO2-equilibrated medium for 60 min. Total cellular RNA was isolated using Trizol reagent (Invitrogen). The -SH groups were biotinylated by EZ-Link N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (biotin-HPDP; Pierce) in 10 mM Tris (pH 7.4), 1 mM EDTA, and 0.2 mg/ml biotin-HPDP at a final RNA concentration of 100 ng/μl. Unbound biotin-HPDP was removed by chloroform-isoamyl alcohol (24:1) extraction in Phase Lock Gel Heavy tubes (5 Prime). Subsequently, the RNA was precipitated in isopropanol. RNA pellets were resuspended in 40 μl RNase-free water. Following denaturation at 65°C for 10 min, biotinylated RNA was isolated using μMACS streptavidin beads and columns (Miltenyi). For cDNA synthesis, 2 to 5 μl of RNA (equivalent to 1 to 2.5 μg total or 100 to 250 ng labeled RNA) was reverse transcribed with SuperScript III First-Strand reverse transcriptase (Invitrogen), using random hexamer primers. Quantitative RT-PCR (qRT-PCR) was performed using LightCycler DNA master SYBR green I mix (Roche). Transcripts were amplified using the primers listed in Table 3, and relative quantification was carried out by normalization against 18S rRNA. Reactions were performed in duplicate, and water controls were included for all primers. A melting curve analysis was performed to demonstrate amplification of the distinct amplicon.
TABLE 3.
Primers for qRT-PCR
| Transcript | Primer direction | Primer sequence (5′-3′) | Primer Tm (°C) | Exon junction | Amplicon size (bp) | Amplicon Tm (°C) |
|---|---|---|---|---|---|---|
| Tapasin | Forward | TGAAAGTGAAAGGAGGAAGAGG | 54 | 1-2 | 147 | 90 |
| Reverse | ATCCTCCACGAACCAACACT | |||||
| TAP1 | Forward | ATGTCTCTTGGGGATCATGC | 53 | 4-5 | 144 | 86 |
| Reverse | TGGACTTTGCCAGAGATTCC | |||||
| TAP2 | Forward | TTCACAATAGCAGCGGAGAA | 53 | 5-6 | 158 | 90 |
| Reverse | GCGACAGACTTCATGCTCCT | |||||
| β2m | Forward | GTGCTCGCGCTACTCTCTCT | 52 | 1-2 | 143 | 84 |
| Reverse | TTCAATGTCGGATGGATGAA | |||||
| HLA-B | Forward | TCCGCAGATACCTGGAGAAC | 55 | 3-4 | 140 | 89 |
| Reverse | GTGTGATCTCCGCAGGGTAG | |||||
| ERp57 | Forward | CCTAAAAGCAGCCAGCAACT | 52 | 5-6 | 150 | 81 |
| Reverse | TGCCACAGTCTTGTCCTCAA | |||||
| GAPDH | Forward | TGCACCACCAACTGCTTA | 54 | 7-8 | 177 | 88 |
| Reverse | GGATGCAGGGATGATGTTC |
FACS analysis.
MRC-5 cells were detached with trypsin and washed with PBS containing 3% FCS. The first staining was performed with W6/32 before incubation with allophycocyanin (APC)-coupled anti-mouse antibodies. Cells were washed thoroughly, fixed in 3% paraformaldehyde (PFA), and permeabilized using 0.1% saponin. Subsequently, cells were stained with FITC-labeled human IgG Fc fragments to identify HCMV-infected cells expressing viral FcγRs gp34 and gp68. The data were analyzed with FloJo software.
RESULTS
Altered composition of the PLC in HCMV-infected cells.
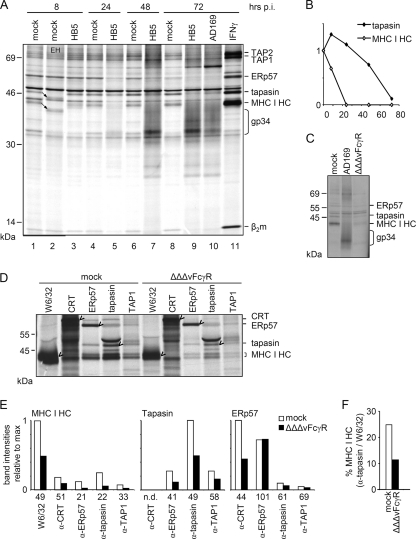
HCMV inhibitors interfering with the MHC I pathway of antigen presentation have been studied extensively in transfected cells overexpressing the inhibitors, whereas no comprehensive approaches to analyze the PLC in the context of HCMV infection have been reported so far. In a previous study, we noted that MHC I and tapasin might be absent from TAP1/2 immunoprecipitates of HCMV-infected cells at 72 h postinfection (p.i.) (18). This hypothesis prompted us to monitor the composition of the PLC throughout HCMV infection. MRC-5 fibroblasts were infected with the HCMV AD169-derived mutant HB5 (6), lacking the genes US2 to US6, to exclude the effects of previously described inhibitors. The cells were radiolabeled for 6 h. Lysis was performed using the mild detergent digitonin, which preserves the PLC for precipitation by antibodies directed against various components of the PLC. PLC assembly was analyzed at 8, 24, 48, and 72 h p.i. IFN-γ treatment of mock-infected cells enhanced the bands corresponding to tapasin, MHC I HC and β2m, ERp57, and TAP1/2 (Fig. 1 A, lane 11), and the expected band shift resulting from endo-β-N-acetylglucosaminidase H (endo H) treatment confirmed the identities of tapasin and MHC I HC (indicated by arrows in Fig. 1A, lane 2). The experiment revealed two interesting phenotypes. First, as the replication cycle proceeded, the tapasin band weakened in comparison to that for mock-infected cells. The first indication was observed at 24 h p.i. (Fig. 1A, lane 5), but it was most pronounced at 72 h p.i. both in wild-type AD169-infected cells and in ΗΒ5-infected cells (Fig. 1A, lanes 9 and 10). Second, MHC I HC and β2m had already disappeared from the PLC in ΗΒ5-infected cells at 24 h p.i. (Fig. 1A, lane 5), which was a surprising observation since the gene for US2 is deleted from HB5 and, consequently, fewer MHC I molecules are degraded. Interestingly, the two observed phenotypes followed different kinetics. Whereas the band for tapasin disappeared slowly, MHC I coprecipitation was reduced already at 8 h p.i. and was completely abolished at 24 h p.i. (Fig. 1B).
FIG. 1.
HCMV impairs PLC assembly. MRC-5 cells were mock or IFN-γ (500 U/ml, 72 h) treated or infected with HB5, AD169, or ΔΔΔFcγR virus at a multiplicity of infection (MOI) of 5. (A) Cells were labeled for 6 h and then lysed at the indicated time points of infection. Lysates were snap-frozen in liquid N2 and stored at −80°C. After thawing, proteins were recovered by immunoprecipitation using anti-tapasin antibodies. Arrows indicate endo H (EH)-deglycosylated proteins in lane 2. The smear produced by the HCMV-encoded vFcγR gp34 is indicated. (B) Quantification of tapasin and MHC I HC precipitation from HB5-infected cells. The band intensities are displayed relative to those for the mock control at 8 h (lane 1 in panel A; depicted as time point 0). (C) HCMV-encoded FcγRs gp34 and gp68 are precipitated by rabbit antiserum. Cells were labeled for 3 h and lysed at 48 h p.i. Using rabbit anti-tapasin antiserum, precipitation of viral FcγRs was demonstrated. The vFcγR deletion mutant (ΔΔΔvFcγR) lacks the bands caused by the highly glycosylated vFcγR proteins gp34 and gp68 seen in the AD169 lane. (D) Analysis of PLC composition. Cells were labeled for 3 h and lysed at 48 h p.i. Immunoprecipitations were performed with the indicated antibodies. Immune complexes were separated by 10 to 11.5% SDS-PAGE. Marked bands indicate primary targets of the antibodies. (E) Quantification of PLC components. Diagrams show intensities of bands corresponding to the PLC components indicated above, relative to the maximum intensity. The antibody used for immunoprecipitation is indicated on the x axis. Numbers beneath show the percentages of bands from ΔΔΔvFcγR-infected cells in comparison to bands from mock cells. n.d., not determined. (F) Quantification of the amount of MHC I HC coprecipitated by anti-tapasin antibodies in comparison to the amount of MHC I HC precipitated by W6/32.
To further examine the changes taking place in the formation of the PLC during HCMV infection, we made use of specific antibodies directed against distinct PLC components for analysis of MHC I incorporation into the PLC. Since HCMV produces highly glycosylated viral FcγRs, i.e., TRL11- and IRL11-encoded gp34 and UL119-118-encoded gp68, which have high affinities for rabbit IgG (4, 33), we constructed an HB5 (ΔUS2-US6)-derived mutant lacking gp34 and gp68 (ΔΔΔvFcγR) to avoid precipitation of vFcγRs (causing background bands as demonstrated in Fig. 1C). The cells were infected with HCMV ΔΔΔvFcγR for 48 h, a time point at which a reduced level of tapasin incorporation into the PLC was observed and no MHC I HC-β2m complexes were coprecipitated by anti-tapasin antibodies (Fig. 1A). Using MAb W6/32, recognizing only heterodimeric MHC I HC-β2m complexes and not free HC, the levels of radiolabeled MHC I in mock- and HCMV-infected cells were determined. Due to the deletion of US2 to US6, there was a substantial amount (50% in comparison to that for mock-infected cells) of newly synthesized MHC I assembled in infected cells (Fig. 1D). A reduction of about 50% was observed for newly synthesized tapasin molecules in HCMV-infected cells compared to mock-infected cells, which was confirmed by the equally reduced coprecipitation of tapasin by antibodies directed to other PLC components, such as anti-ERp57 (Fig. 1F). In contrast, however, MHC I was coprecipitated to an even lesser extent by antibodies to PLC components. From HCMV-infected cells, anti-tapasin recovered only 22% of MHC I in comparison to MHC I from mock-infected cells, anti-ERp57 recovered 21%, anti-TAP1 recovered 33%, and anti-calreticulin recovered 51% (Fig. 1E). The relative amount of MHC I recovered by anti-tapasin compared to W6/32-immunoprecipitated MHC I heterodimers was determined to be 25% in the case of mock-infected cells, whereas for infected cells it was 10% (Fig. 1F). This implies that whereas the calreticulin-MHC I interaction seems to be maintained in infected cells, the recruitment of MHC I heterodimers to the PLC is disturbed independent of the downregulation of tapasin molecules.
Reduced biosynthesis of tapasin.
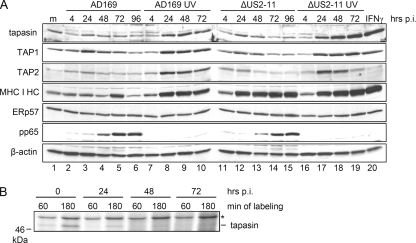
Tapasin is known to be a long-lived protein (5). To assess the time point at which the reduction of newly synthesized tapasin results in a loss of overall protein abundance, the steady-state levels of tapasin were examined in comparison with other PLC components during the progression of HCMV replication. Cell lysates from AD169- and ΔUS2-US11 mutant-infected cells were analyzed by immunoblotting. No drastic effect on tapasin amounts was observed in infected cells, but a moderate reduction was eventually recognizable (Fig. 2 A, compare lane 1 with lanes 4 to 6 and 12 to 15), which contrasted with the clearly increased amounts in cells treated with UV-inactivated HCMV particles (Fig. 2A, lanes 8 to 10 and 17 to 19). Hence, HCMV gene expression controls the induced tapasin expression triggered by HCMV virions. This was also the case for TAP1 and TAP2 but not for ERp57. The reduction occurred independently of the US2 to US11 genes, as revealed by cells infected with the HCMV ΔUS2-US11 deletion mutant. We also detected MHC I HC by immunoblotting and found only moderate changes in the steady-state levels during HCMV AD169 wt infection, whereas in ΔUS2-US11 mutant-infected cells, MHC I HC was abundantly detected, reaching a level close to that seen after treatment of cells with IFN-γ or inactivated HCMV particles (Fig. 2A). Taken together, the data from the immunoblot analysis demonstrated only a slow diminishment of tapasin steady-state levels in infected cells, suggesting a blocked synthesis rather than enhanced protein degradation.
FIG. 2.
De novo synthesis of tapasin is delayed in HCMV-infected cells. MRC-5 cells were mock (m) or IFN-γ (500 U/ml, 72 h) treated or infected with AD169 or the ΔUS2-US11 HCMV deletion mutant at an MOI of 5. (A) Immunoblot analysis of PLC components. At the indicated time points, cell lysates were prepared and protein concentrations of the samples were adjusted. Immunoblot detection was performed as indicated. Using stripping solutions, several proteins were detected consecutively on one blot. Blot 1, tapasin and TAP1; blot 2, TAP2, HC, ERp57, pp65, and β-actin. (B) HCMV inhibition of tapasin biosynthesis. Cells were metabolically labeled for 60 or 180 min prior to lysis at various time points p.i. Tapasin-specific antibodies (STC) were used for immunoprecipitation.
To illustrate this hypothesis, tapasin biosynthesis was investigated in more detail by shortened metabolic pulse labeling of cells for 60 and 180 min. Under these conditions, the viral effect on tapasin neosynthesis was much more pronounced (Fig. 2B). Reduction of tapasin biosynthesis could be observed at 24 h p.i., and at 48 and 72 h p.i., tapasin was barely detectable (equal translation rates of samples were verified [data not shown]). In pulse-chase experiments using metabolically labeled cells, we observed no change in the half-life of tapasin upon HCMV infection (data not shown). Taken together, the data set presented in Fig. 2 strongly argues that the loss of tapasin precipitation from radiolabeled HCMV-infected cells was not due to protein degradation but rather to blocked de novo synthesis.
Reduced levels of tapasin mRNA in HCMV-infected cells.
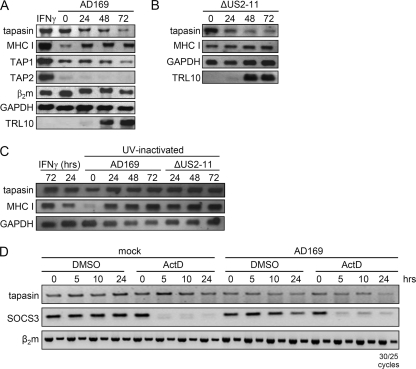
Since existing tapasin molecules are stable in HCMV-infected cells and the rate of newly synthesized molecules is downregulated, we set out to analyze whether this regulation occurs at the transcriptional or translational level. Northern blot analysis was performed at different time points along the HCMV replication cycle. Similar to tapasin protein levels, tapasin transcript levels were found to be reduced continuously in comparison to the mock control level, eventually reaching very low levels (Fig. 3 A). This effect was seen in cells infected with the HCMV wt strain AD169 and was reproduced with the ΔUS2-US11 mutant virus (Fig. 3B). In contrast to the results of immunoblot analysis (Fig. 2A), there was no difference in MHC I transcripts between AD169- and ΔUS2-US11 mutant-infected cells, and the overall level of MHC I mRNA was induced throughout HCMV infection. Interestingly, TAP1 and TAP2 RNAs were also reduced in HCMV-infected cells, but the effect appeared later in infection and was clearly visible at 72 h p.i. At 24 h p.i., the TAP1 RNA level was even slightly upregulated. The β2m RNA level was only marginally affected by HCMV infection at 72 h p.i. By analyzing transcripts in cells treated with UV-inactivated HCMV particles, the strength of the transcriptional upregulation in the absence of viral gene expression was assessed (Fig. 3C). Transcription of MHC I was found to be induced in cells treated with inactivated HCMV particles, reaching levels seen in IFN-γ-stimulated cells. Similarly, the steady-state level of tapasin transcripts was upregulated in cells infected with UV-inactivated HCMV, strongly contrasting with the progressive decline of tapasin mRNA induced by replicating HCMV.
FIG. 3.
HCMV infection downregulates tapasin mRNA. (A and B) Reduced tapasin mRNA in HCMV-infected cells, as determined by Northern blotting. MRC-5 cells were left untreated or incubated with IFN-γ (500 U/ml, 72 h). Cells were infected with AD169 or the ΔUS2-US11 deletion mutant at an MOI of 5. Total RNA was collected at various time points and analyzed by Northern blotting, using the indicated hybridization probes. (C) Inactivated virus particles induce transcription of PLC components. MRC-5 cells were IFN-γ (500 U/ml) treated for 24 or 72 h or treated with UV-inactivated AD169 or ΔUS2-US11 HCMV particles (5 × 106 particles per 106 cells) for the indicated times. Northern blot analysis was performed as described for panel A. (D) Tapasin mRNA half-life is not altered by HCMV infection. MRC-5 cells were mock infected or infected with AD169 at an MOI of 5. At 36 h p.i., cells were treated with solvent (DMSO) or actD for 0, 5, 10, and 24 h. RNAs were prepared, and semiquantitative RT-PCR analysis was performed for tapasin, SOCS3, and β2m. Amplicons from 25 and 30 rounds of PCR cycles are shown to demonstrate equal levels of the β2m mRNA standard.
HCMV inhibits tapasin gene expression at a pretranscriptional level.
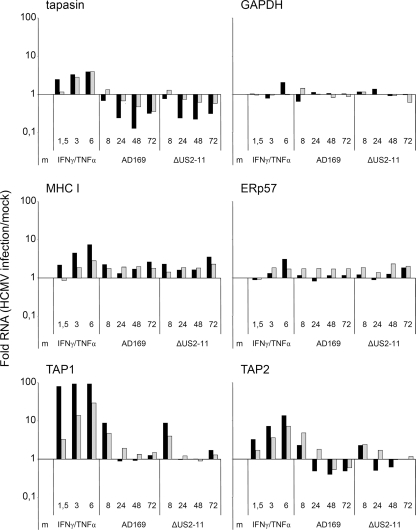
In order to test whether the tapasin mRNA is destabilized, e.g., by a degrading microRNA (miRNA), or if HCMV interferes with tapasin gene expression on a pretranscriptional level, the stability of tapasin RNA was measured. Cells were treated with solvent (dimethyl sulfoxide [DMSO]) only or with actD for 0, 5, 10, and 24 h, starting at 36 h p.i. Tapasin mRNA was analyzed by semiquantitative RT-PCR (Fig. 3D). Tapasin mRNA is relatively stable, and the half-life exceeded the time frame of the experiment. Successful actD blocking of the RNA polymerase was confirmed by the disappearance of SOCS3 mRNA, which has been shown to have a half-life of only 30 min (59). In line with the previous results obtained by Northern blotting, tapasin mRNA was already reduced at 36 h p.i. in infected cells without actD treatment (Fig. 3). Interestingly, actD treatment of infected cells did not reveal a decrease in tapasin mRNA stability. This finding strongly argues against a destabilization of tapasin transcripts. Rather, we considered it more likely that the reduced mRNA level was caused by events taking place at a pretranscriptional level. However, conclusions concerning transcriptional activity based on levels of total RNA may be deceptive in cases of long-lived transcripts, as for tapasin. Hence, we took advantage of a novel method in which nascent RNA is isolated from the total pool of cellular RNA subsequent to RNA labeling by 4sU (9). This method allows for comparison of transcriptional regulation of different genes. To test the power of the applied method, the kinetics of transcriptional regulation of individual target genes in IFN-γ- and tumor necrosis factor alpha (TNF-α)-treated cells were included in this experimental setting (Fig. 4). In parallel, transcriptional induction was measured for TAP1, TAP2, and MHC I at 8 h post infection, whereas the tapasin transcripts were downregulated at this time point, implying different regulatory mechanisms of their respective promoters. The same pattern of gene regulation was observed in cells infected with the ΔUS2-US11 mutant virus. Interestingly, a strong downregulation of TAP1 and TAP2 could be observed between 8 and 24 h p.i., while transcription of tapasin was only weakly affected at this time point. Tapasin transcription continued to decrease throughout the HCMV replication cycle. In contrast, MHC I transcription accelerated toward E/L times of HCMV infection preceding the increased abundance at the protein level (Fig. 2A). After an initial induction, TAP1 transcripts remained approximately at control levels. TAP2 transcription, on the other hand, was repressed, although the effect was not as pronounced as that for tapasin. Taken together, the data show that the initial peak of transcription was inhibited at later time points for both TAP1 and TAP2, whereas tapasin gene expression, followed by that of TAP2, became repressed irreversibly early during HCMV infection.
FIG. 4.
Newly synthesized tapasin RNA is downregulated throughout the HCMV replication cycle. MRC-5 cells were mock (m) or IFN-γ/TNF-α treated (500 U/ml and 25 ng/ml, respectively) or infected with AD169 or the ΔUS2-US11 HCMV deletion mutant at an MOI of 5. At the indicated time points, cells were incubated for 1 h with 4sU prior to RNA preparation and isolation of 4sU-incorporated RNA. qRT-PCR was performed, and relative quantification was carried out by normalization against 18S rRNA. Reactions were performed in duplicate. Data for one of two independent experiments with very similar outcomes are shown. Black bars indicate pulse-labeled newly synthesized RNA, and gray bars indicate total RNA.
Tapasin association of MHC I HC-β2m heterodimers is impaired in HCMV-infected cells.
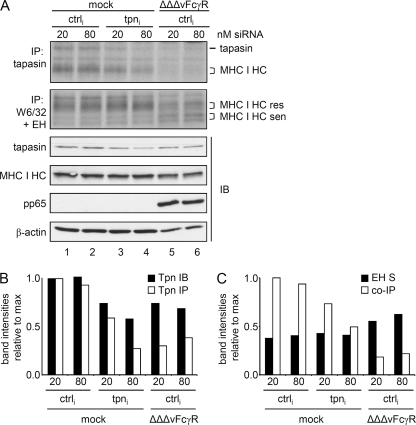
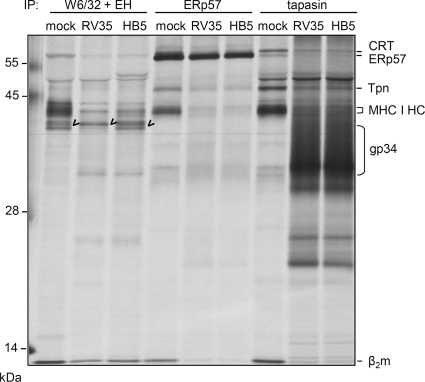
The transcriptional block explains the absence of newly synthesized tapasin in HCMV-infected cells. As demonstrated above, the lack of MHC I HC-β2m incorporation into the PLC was observed earlier in infection (Fig. 1A, lane 5), when tapasin protein was still abundantly detectable in Western blots (Fig. 2A). We therefore wondered whether newly synthesized tapasin rather than aged molecules is necessary for MHC I recruitment and incorporation into the PLC. To approach this issue, we nucleofected of siRNA directed against tapasin transcripts into MRC-5 cells and assessed the PLC formation by immunoprecipitation. Cells infected with the ΔΔΔvFcγR HCMV mutant were analyzed in parallel. Knockdown of tapasin was successful, as tapasin biosynthesis was strongly reduced with increasing concentrations of siRNA (Fig. 5 A). At 48 h p.i., the HCMV-mediated reduction of tapasin biosynthesis was as efficient as inhibition in mock-infected cells by siRNA treatment (Fig. 5A and B). Comparing the level of MHC I coprecipitation by tapasin-specific antibodies and the levels of tapasin (Fig. 5A, IP [biosynthesis] and IB [total]), we noticed that in mock-infected cells the efficiency of MHC I coprecipitation was rather proportional to the total level of tapasin, and not to the amount of newly synthesized molecules (compare white bars in Fig. 5C with black and white bars in Fig. 5B), excluding the possibility that newly synthesized tapasin molecules preferentially bind to MHC I HC-β2m complexes. To obtain a more precise estimation of the amounts of MHC I HC-β2m molecules available for PLC recruitment, an endo H treatment was performed subsequent to precipitation by MAb W6/32 (Fig. 5A). Whereas endo H-resistant forms of MHC I have been transported beyond the cis-Golgi compartment, the endo H-sensitive forms localize to earlier compartments, where formation of the PLC is taking place. Remarkably, although the overall MHC I level was reduced in ΔΔΔvFcγR HCMV-infected cells, an increased amount of endo H-sensitive forms was found (Fig. 5A, lanes 5 and 6, and C, black bars). Despite the 1.5-fold increase of endo H-sensitive MHC I molecules in infected cells, the coprecipitation of MHC I by tapasin was by far not as efficient as that in the control cells (Fig. 5A and C, white bars), demonstrating that the lack of MHC I recruitment to tapasin cells is not a consequence of reduced numbers of MHC I molecules. In addition to the ΔΔΔvFcγR mutant, lacking the US2 to US6 genes, this phenotype was also observed in RV35 (ΔUS6-US11) (27)-infected cells (Fig. 6), suggesting that the US2 to US11 genes are not responsible for the disturbed tapasin interaction with MHC I HC-β2m heterodimers. Based upon the findings that (i) MHC I molecules are not limited for PLC incorporation into HCMV-infected cells and (ii) MHC I association with tapasin is not directly linked to tapasin neosynthesis, we concluded that the transcriptional block of tapasin cannot be the cause of the lack of MHC I recruitment to the PLC in HCMV-infected cells.
FIG. 5.
Blocked interaction between MHC I and tapasin in HCMV-infected cells. (A) MRC-5 cells were subjected to nucleofection with siRNA targeting tapasin transcripts or with a random sequence (ctrl) and then infected as indicated. Cells were metabolically labeled for 3 h prior to lysis at 48 h postnucleofection/postinfection. Lysates were analyzed by anti-tapasin (STC) or anti-MHC I (W6/32) immunoprecipitation (IP) or by immunoblotting (IB) with the indicated antibodies. (B) Diagram showing relative intensities of bands corresponding to tapasin (Tpn) IB (black bars) and IP (white bars). (C) Relative intensities of endo H (EH)-sensitive MHC I molecules (black bars) and tapasin-coprecipitated MHC I (white bars).
FIG. 6.
Blocked interaction between MHC I and tapasin is independent of the US2 to US11 genes. MRC-5 cells were infected with the ΔΔΔvFcγR mutant, lacking the US2 to US6 genes, and the RV35 mutant (ΔUS6-US11), as indicated, at an MOI of 5. Cells were metabolically labeled for 3 h prior to lysis at 48 h p.i. Lysates were analyzed by anti-tapasin (STC), anti-ERp57, or anti-MHC I (W6/32) immunoprecipitation. Subsequent to the W6/32 precipitation, the samples were treated by endo H. Marked bands indicate endo H-sensitive MHC I.
Restoration of MHC I cell surface expression after forced expression of tapasin in HCMV-infected cells.
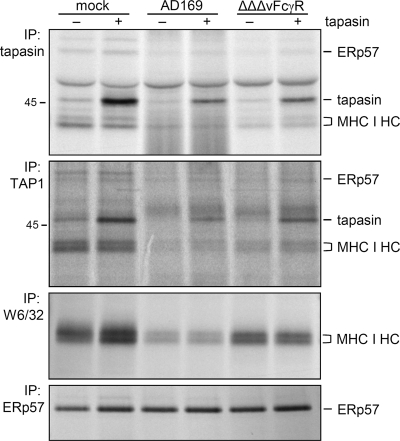
To examine the relative impact of inhibited tapasin transcription in the context of HCMV infection, we reconstituted newly synthesized tapasin molecules in HCMV-infected cells. MRC-5 cells were transduced with either a tapasin-expressing or control lentiviral vector before PLC assembly was assessed by tapasin-specific antibodies under mock infection conditions or HCMV infection at 48 h p.i. Biosynthesis of tapasin was restored to levels above those observed in mock-treated cells, although HCMV reduced the lentiviral tapasin overexpression compared to that in mock-infected cells (Fig. 7, upper panel). Remarkably, there was no further increase of MHC I-tapasin association in mock-infected fibroblasts as a result of tapasin overexpression. Likewise, tapasin synthesis in AD169-infected cells largely failed to restore MHC I incorporation into the PLC, while a marginal gain of MHC I coprecipitation was seen in ΔΔΔvFcγR-infected cells (Fig. 7). Clearly, MHC I association in HCMV-infected cells did not reach the levels observed in mock-infected cells. As an independent tool, anti-TAP1 antibodies were applied to analyze PLC assembly (Fig. 7, second panel from top), basically confirming the PLC composition revealed by anti-tapasin antibodies. Again, MHC I association by forced tapasin synthesis was marginally induced in ΔΔΔvFcγR- and AD169-infected cells and did not change the levels of MHC I as revealed by MAb W6/32 (Fig. 7, third panel from top). Taken together, the data showing the incomplete restoration of MHC I incorporation by overexpressed tapasin confirm the previous finding that an independent HCMV-induced factor prevents MHC I association with tapasin molecules. Since overexpression of tapasin had no or only a marginal effect on MHC I recruitment to the PLC, it is assumable that HCMV targets a molecule other than tapasin.
FIG. 7.
Forced expression of tapasin is not sufficient to rescue MHC I incorporation into the PLC in HCMV-infected cells. Tapasin-transfected or control MRC-5 cells were mock infected or infected with AD169 or with the HB5 (ΔUS2-US6)-derived ΔΔΔFcγR HCMV deletion mutant, as indicated, at an MOI of 5. Cells were labeled for 3 h prior to lysis at 48 h p.i. Immunoprecipitations were performed as indicated. Proteins were separated by 10 to 11.5% SDS-PAGE.
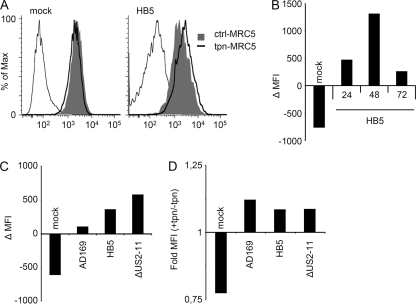
In the experiments described above, conditions of forced tapasin expression were used to study early events of PLC assembly restricted to a time interval of 3 h. To study the outcome of retained tapasin neosynthesis on steady-state levels on the cell surface, FACS analysis was performed. First, lentivirus-transduced control or tapasin-expressing MRC-5 cells were mock treated or infected with HB5. Cell surface expression of MHC I was determined by MAb W6/32 throughout the HB5 replication cycle. Interestingly, mock-treated tapasin-expressing MRC-5 cells had smaller amounts of MHC I than control MRC-5 cells (Fig. 8 A). Tapasin transduction of MRC-5 cells was repeated three times with the same outcome, convincing us that selectively overexpressed tapasin indeed retards MHC I trafficking to the cell surface. This was confirmed in a pulse-chase experiment (data not shown), consistent with earlier reports (45, 46). In contrast, in HCMV-infected cells, the trend was reversed and MHC I was consistently induced in the tapasin-expressing MRC-5 cells. Already at 24 h p.i., the reduced MHC I cell surface expression observed in mock-infected tapasin-expressing MRC-5 cells became reversed (Fig. 8B). At 48 h p.i., the impact of restored tapasin synthesis was even stronger (Fig. 8A and B), before it dropped at 72 h p.i. In addition, the effect was followed up with viruses allowing different degrees of MHC I expression: AD169, HB5 (ΔUS2-6), and the ΔUS2-11 mutant. The more MHC I expression the different viruses allowed for, the larger was the increase of MHC I due to forced tapasin expression (Fig. 8C). However, the relative gains of MHC I cell surface expression were similar for all viruses (Fig. 8D). In conclusion, the data provided proof of principle that restoration of tapasin expression is able to convey a higher level of MHC I cell surface expression in HCMV-infected cells, albeit to a rather limited extent, possibly due to the inhibitory effect of HCMV targeting MHC I HC-β2m association with tapasin.
FIG. 8.
Tapasin overexpression induces MHC I cell surface expression on HCMV-infected cells. Tapasin-expressing or control MRC-5 cells were mock treated or infected with AD169, HB5, or the ΔUS2-US11 deletion mutant at an MOI of 5. MHC I cell surface expression was determined by FACS analysis using W6/32. (A) MHC I detection at 48 h p.i. for mock and HB5 infections. (B) Mean changes in fluorescence intensity (ΔMFI) between tapasin-expressing and control MRC-5 cells at various time points of HB5 infection. (C) ΔMFI values between tapasin-expressing and control MRC-5 cells at 72 h p.i. for the indicated infections. (D) MFI values from the experiment shown in panel C, depicted as fold MFI values measured for tapasin-expressing MRC-5 cells in comparison to control MRC-5 cells.
DISCUSSION
In this study, we analyzed the fate of the PLC in the context of all phases of the protracted HCMV replication cycle. A defective incorporation of MHC I heterodimers into the PLC was observed, and specifically, the presence of tapasin was strikingly reduced. While the stability of tapasin remained unaffected, an immediate effect on its biosynthesis was found. In accordance, tapasin mRNA was constantly diminishing in HCMV-infected cells, but only the selective analysis of newly synthesized RNAs revealed the instantly occurring transcriptional inhibition of the tapasin gene by HCMV. Unexpectedly, forced tapasin overexpression in HCMV-infected cells only marginally rescued PLC recruitment of MHC I, while at least augmenting surface MHC I. This relatively inefficient rescue suggests that additional unidentified factors may be involved in the distraction of MHC I heterodimers from the PLC. Taken together, our data establish for the first time that HCMV targets the transcription of selected genes of the MHC I pathway and prohibits MHC I association with the PLC.
Transcriptional control of tapasin gene expression.
The lack of tapasin biosynthesis in HCMV-infected cells is a result of viral inhibition of tapasin transcription. The overall levels of tapasin transcripts gave the impression that the RNA was downregulated at a slower pace than the observed reduction of protein neosynthesis. Only the selective measurement of newly transcribed mRNAs revealed the immediate repression of tapasin followed by TAP2 transcription. Based upon these findings, it is tempting to speculate that newly synthesized tapasin transcripts preferentially become translated into protein, while another, long-lived pool of mRNAs still exists without being translated. Alternatively, an HCMV-encoded miRNA could interfere with the translation of tapasin, similar to the case described for the NKG2D ligand MICB (47). Indeed, in a collaboration with O. Mandelboim, who performed bioinformatic screens for targets of HCMV-encoded miRNAs, a high-score target of the HCMV-encoded miRNA miRUS25.2 was identified at the tapasin 3′-untranslated region (3′-UTR). However, cells overexpressing miRUS25.2 did not show a reduced biosynthesis of tapasin (data not shown), suggesting that this miRNA is unlikely to cause the effect.
Treatment of cells with UV-inactivated virus particles showed that these trigger a strong induction of MHC I and tapasin transcription. While the massive induction of MHC I was essentially counterregulated on the protein level by the US2 and US11 glycoproteins (Fig. 2A), the reduction of tapasin, TAP1, and TAP2 transcription was mediated by HCMV genes outside the US2-US11 region. It has been suggested that the HCMV tegument protein pp71 (UL82) delays the maturation of MHC I molecules (51). pp71 also relieves HCMV replication from Daxx-mediated repression (25), raising the possibility that pp71 could be involved in the regulation of tapasin transcription. However, we have not been able to find any influence of pp71 expression on tapasin transcription and neosynthesis (data not shown). Moreover, tapasin is located in the extended MHC class II locus, and interestingly, the gene for the transcriptional corepressor Daxx is located next to tapasin. We wondered whether the regulation of tapasin transcription by HCMV might be part of a broader effect and analyzed the transcriptional activities of neighboring genes of tapasin, including Daxx, RGL2, and 18S(RP) (21). However, the transcription of none of these was affected by HCMV infection (data not shown), indicating that the early repression of tapasin transcription is astonishingly gene selective.
This might suggest that tapasin is regulated in HCMV-infected cells either by inhibitory viral factors or by mobilization of cellular factors that have the propensity to repress the transcription machinery directly. Indeed, it was recently described that the transcriptional repressor PRDM-1 (positive regulatory domain I) targets the tapasin promoter (10). PRDM-1 was found to be able to inhibit IFN-γ-induced transcription of tapasin, qualifying this factor to exert a possible function in tapasin gene repression during HCMV infection.
Phenotypic disruption of the PLC.
Most studies analyzing the molecular function of HCMV-encoded MHC inhibitors (i.e., US2, US3, US6, US10, and US11) have been performed with experimental systems of isolated gene expression, highlighting the sophisticated and redundant control mechanisms of this virus over the MHC I pathway of antigen presentation (3, 19, 28, 29, 32, 37-39, 55, 56). These established inhibitors interact with defined target molecules, i.e., MHC I, TAP1/2, and tapasin. Our finding that the disruption of PLC assembly was not only evident in cells infected with HCMV wt strains but also seen with ΔUS gene deletion mutants indicates that the effects must be caused by other factors, thus excluding the previously described MHC I inhibitors. The following two dominant phenotypic changes of the PLC could be defined by immunoprecipitation analysis of radiolabeled cells: (i) a lack of tapasin synthesis and (ii) disruption of molecular interactions between PLC components.
Multiple effects target tapasin in HCMV-infected cells.
Previous work by Park et al. (37) revealed that the US3 glycoprotein acts as an inhibitor of tapasin function, resulting in impaired peptide loading onto MHC I. However, the immediate-early kinetics of US3 transcription (49) and the short half-life of the US3 glycoprotein (13) exclude a sustained effect of US3 on tapasin function during the long course of HCMV replication. The early onset and sustained nature of blocked tapasin gene expression by HCMV, as documented here, could complement the transient US3-mediated effect in an optimal way by depleting this factor critical for the selection of MHC I epitopes. We found it remarkable that HCMV limits its effect to the replenishment of tapasin and does not downregulate overall levels of this PLC component. However, the restoration of the HCMV-induced effect on the PLC by ectopic expression of tapasin was grossly incomplete. This implies that, in fibroblasts, the impact of retarded tapasin transcription is diminished due to a redundant and dominant interference of HCMV with MHC I maturation, as illustrated by the inhibited association of MHC I HC-β2m heterodimers with tapasin. Indeed, augmentation of tapasin levels did not correct the deficient recruitment of MHC I HC-β2m heterodimers to tapasin (Fig. 7). Moreover, the level of MHC I is also not the limiting factor, since high levels of endo H-sensitive MHC I heterodimers were found in ΔUS2-6- and ΔUS6-11-infected cells. The obvious conclusion is that HCMV is able to keep these molecules from interacting. Such a posttranslational mechanism would perfectly complement the time-limited effect of US3 (37) and the successive suppression of tapasin transcription.
Remarkably, whereas the disruption of MHC I interaction could be documented for tapasin, ERp57, and TAP, the MHC I-calreticulin interaction was not reduced in infected cells (Fig. 1D). This drew our attention to a recent publication by Elliott, Springer, et al. (24) in which an additional function outside the PLC was described for calreticulin. Not by affecting the rate of MHC I molecules leaving the ER but by redirecting suboptimally loaded MHC I molecules from post-ER compartments back to the ER, calreticulin mediated a further point of quality control for MHC I. Accordingly, defects in peptide loading (e.g., TAP deficiency) induced the colocalization of MHC I and calreticulin in the ER-Golgi intermediate compartment (ERGIC) and the cis-Golgi compartment. It is tempting to hypothesize that the MHC I-calreticulin interaction we observed in HCMV-infected cells might reflect an accumulation of calreticulin and MHC I in the ERGIC and the cis-Golgi compartment. The increased portion of endo H-sensitive MHC I molecules in HCMV-infected cells supports the assumption that MHC I is trapped in the ER, ERGIC, and/or cis-Golgi compartment. Besides calreticulin, tapasin, TAP, and peptide-free MHC I have also been reported to cycle between the ER, ERGIC, and cis-Golgi compartment (12, 24, 40). In such a dynamic scenario, HCMV is apparently confronted with many opportunities to take control over checkpoints of PLC assembly and disassembly.
While the primary goal of established HCMV stealth features is the quantitative downregulation of the bulk of MHC class I molecules from the cell surface (16, 20), the targeting of tapasin is supposed to exert a qualitative influence. Tapasin promotes loading of immunodominant peptides (7), which allowed us to assume that deficiency of tapasin function in HCMV-infected cells could alter the quality of HCMV-specific CD8+ T-cell responses. Since HCMV infects many cell types and cycles through productive as well as latent phases of infection, the precise consequences of tapasin downregulation on HCMV infection biology remain to be established. Another conundrum of HCMV infection is the extreme convergence of CD8+ T-cell responses to a few epitopes, despite the very large number of HCMV antigens produced in infected cells (30, 58). In view of this peculiarity of the cytotoxic T-lymphocyte response to HCMV, we must consider the possibility that it is the sustained tapasin deficiency in HCMV-infected cells that contributes to the apparently ineffective presentation of a large majority of HCMV epitopes.
Acknowledgments
We are grateful to Alexandra Niepel and Bernd Rädle for expert technical assistance. We also acknowledge Robert Tampé for valuable reagents and Sebastian Springer for helpful discussions.
This work was supported by the Deutsche Forschungsgemeinschaft (He 2526/7-1 and SFB455 A7) and by the Forschungskommission of the Medical Faculty of the Heinrich Heine University (grant 9772359).
Footnotes
Published ahead of print on 19 January 2011.
REFERENCES
- 1.Abarca-Heidemann, K., et al. 2002. Regulation of the expression of mouse TAP-associated glycoprotein (tapasin) by cytokines. Immunol. Lett. 83:197-207. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, K., et al. 1996. Human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc. Natl. Acad. Sci. U. S. A. 93:10990-10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn, K., et al. 1997. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity 6:613-621. [DOI] [PubMed] [Google Scholar]
- 4.Atalay, R., et al. 2002. Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcgamma receptor homologs. J. Virol. 76:8596-8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bangia, N., and P. Cresswell. 2005. Stoichiometric tapasin interactions in the catalysis of major histocompatibility complex class I molecule assembly. Immunology 114:346-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulanger, D. S., et al. 2010. Absence of tapasin alters immunodominance against a lymphocytic choriomeningitis virus polytope. J. Immunol. 184:73-83. [DOI] [PubMed] [Google Scholar]
- 8.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 9.Dolken, L., et al. 2008. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14:1959-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doody, G. M., S. Stephenson, C. McManamy, and R. M. Tooze. 2007. PRDM1/BLIMP-1 modulates IFN-gamma-dependent control of the MHC class I antigen-processing and peptide-loading pathway. J. Immunol. 179:7614-7623. [DOI] [PubMed] [Google Scholar]
- 11.Furman, M. H., N. Dey, D. Tortorella, and H. L. Ploegh. 2002. The human cytomegalovirus US10 gene product delays trafficking of major histocompatibility complex class I molecules. J. Virol. 76:11753-11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghanem, E., et al. 2010. The transporter associated with antigen processing (TAP) is active in a post-ER compartment. J. Cell Sci. 123:4271-4279. [DOI] [PubMed] [Google Scholar]
- 13.Gruhler, A., P. A. Peterson, and K. Fruh. 2000. Human cytomegalovirus immediate early glycoprotein US3 retains MHC class I molecules by transient association. Traffic 1:318-325. [DOI] [PubMed] [Google Scholar]
- 14.Guma, M., et al. 2006. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood 107:3624-3631. [DOI] [PubMed] [Google Scholar]
- 15.Halenius, A., et al. 2006. Physical and functional interactions of the cytomegalovirus US6 glycoprotein with the transporter associated with antigen processing. J. Biol. Chem. 281:5383-5390. [DOI] [PubMed] [Google Scholar]
- 16.Hansen, T. H., and M. Bouvier. 2009. MHC class I antigen presentation: learning from viral evasion strategies. Nat. Rev. Immunol. 9:503-513. [DOI] [PubMed] [Google Scholar]
- 17.Hengel, H., C. Esslinger, J. Pool, E. Goulmy, and U. H. Koszinowski. 1995. Cytokines restore MHC class I complex formation and control antigen presentation in human cytomegalovirus-infected cells. J. Gen. Virol. 76:2987-2997. [DOI] [PubMed] [Google Scholar]
- 18.Hengel, H., T. Flohr, G. J. Hammerling, U. H. Koszinowski, and F. Momburg. 1996. Human cytomegalovirus inhibits peptide translocation into the endoplasmic reticulum for MHC class I assembly. J. Gen. Virol. 77:2287-2296. [DOI] [PubMed] [Google Scholar]
- 19.Hengel, H., et al. 1997. A viral ER-resident glycoprotein inactivates the MHC-encoded peptide transporter. Immunity 6:623-632. [DOI] [PubMed] [Google Scholar]
- 20.Hengel, H., and U. H. Koszinowski. 1997. Interference with antigen processing by viruses. Curr. Opin. Immunol. 9:470-476. [DOI] [PubMed] [Google Scholar]
- 21.Herberg, J. A., S. Beck, and J. Trowsdale. 1998. TAPASIN, DAXX, RGL2, HKE2 and four new genes (BING 1, 3 to 5) form a dense cluster at the centromeric end of the MHC. J. Mol. Biol. 277:839-857. [DOI] [PubMed] [Google Scholar]
- 22.Herrmann, F., J. Trowsdale, C. Huber, and B. Seliger. 2003. Cloning and functional analyses of the mouse tapasin promoter. Immunogenetics 55:379-388. [DOI] [PubMed] [Google Scholar]
- 23.Howarth, M., A. Williams, A. B. Tolstrup, and T. Elliott. 2004. Tapasin enhances MHC class I peptide presentation according to peptide half-life. Proc. Natl. Acad. Sci. U. S. A. 101:11737-11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howe, C., et al. 2009. Calreticulin-dependent recycling in the early secretory pathway mediates optimal peptide loading of MHC class I molecules. EMBO J. 28:3730-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang, J., and R. F. Kalejta. 2007. Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology 367:334-338. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, D. R., and J. S. Pober. 1990. Tumor necrosis factor and immune interferon synergistically increase transcription of HLA class I heavy- and light-chain genes in vascular endothelium. Proc. Natl. Acad. Sci. U. S. A. 87:5183-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, T. R., et al. 1995. Multiple independent loci within the human cytomegalovirus unique short region down-regulate expression of major histocompatibility complex class I heavy chains. J. Virol. 69:4830-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, T. R., and L. Sun. 1997. Human cytomegalovirus US2 destabilizes major histocompatibility complex class I heavy chains. J. Virol. 71:2970-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones, T. R., et al. 1996. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc. Natl. Acad. Sci. U. S. A. 93:11327-11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kern, F., et al. 1999. Target structures of the CD8+-T-cell response to human cytomegalovirus: the 72-kilodalton major immediate-early protein revisited. J. Virol. 73:8179-8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le, V. T., M. Trilling, M. Wilborn, H. Hengel, and A. Zimmermann. 2008. Human cytomegalovirus interferes with signal transducer and activator of transcription (STAT) 2 protein stability and tyrosine phosphorylation. J. Gen. Virol. 89:2416-2426. [DOI] [PubMed] [Google Scholar]
- 32.Lehner, P. J., J. T. Karttunen, G. W. Wilkinson, and P. Cresswell. 1997. The human cytomegalovirus US6 glycoprotein inhibits transporter associated with antigen processing-dependent peptide translocation. Proc. Natl. Acad. Sci. U. S. A. 94:6904-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lilley, B. N., H. L. Ploegh, and R. S. Tirabassi. 2001. Human cytomegalovirus open reading frame TRL11/IRL11 encodes an immunoglobulin G Fc-binding protein. J. Virol. 75:11218-11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindquist, J. A., O. N. Jensen, M. Mann, and G. J. Hammerling. 1998. ER-60, a chaperone with thiol-dependent reductase activity involved in MHC class I assembly. EMBO J. 17:2186-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma, W., P. J. Lehner, P. Cresswell, J. S. Pober, and D. R. Johnson. 1997. Interferon-gamma rapidly increases peptide transporter (TAP) subunit expression and peptide transport capacity in endothelial cells. J. Biol. Chem. 272:16585-16590. [DOI] [PubMed] [Google Scholar]
- 36.Meyer, T. H., P. M. van Endert, S. Uebel, B. Ehring, and R. Tampe. 1994. Functional expression and purification of the ABC transporter complex associated with antigen processing (TAP) in insect cells. FEBS Lett. 351:443-447. [DOI] [PubMed] [Google Scholar]
- 37.Park, B., et al. 2004. Human cytomegalovirus inhibits tapasin-dependent peptide loading and optimization of the MHC class I peptide cargo for immune evasion. Immunity 20:71-85. [DOI] [PubMed] [Google Scholar]
- 38.Park, B., et al. 2006. Redox regulation facilitates optimal peptide selection by MHC class I during antigen processing. Cell 127:369-382. [DOI] [PubMed] [Google Scholar]
- 39.Park, B., E. Spooner, B. L. Houser, J. L. Strominger, and H. L. Ploegh. 2010. The HCMV membrane glycoprotein US10 selectively targets HLA-G for degradation. J. Exp. Med. 207:2033-2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paulsson, K. M., et al. 2002. Association of tapasin and COPI provides a mechanism for the retrograde transport of major histocompatibility complex (MHC) class I molecules from the Golgi complex to the endoplasmic reticulum. J. Biol. Chem. 277:18266-18271. [DOI] [PubMed] [Google Scholar]
- 41.Peaper, D. R., and P. Cresswell. 2008. The redox activity of ERp57 is not essential for its functions in MHC class I peptide loading. Proc. Natl. Acad. Sci. U. S. A. 105:10477-10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peaper, D. R., P. A. Wearsch, and P. Cresswell. 2005. Tapasin and ERp57 form a stable disulfide-linked dimer within the MHC class I peptide-loading complex. EMBO J. 24:3613-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Powis, S. J., et al. 1991. Restoration of antigen presentation to the mutant cell line RMA-S by an MHC-linked transporter. Nature 354:528-531. [DOI] [PubMed] [Google Scholar]
- 44.Sadasivan, B., P. J. Lehner, B. Ortmann, T. Spies, and P. Cresswell. 1996. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity 5:103-114. [DOI] [PubMed] [Google Scholar]
- 45.Schneeweiss, C., M. Garstka, J. Smith, M. T. Hutt, and S. Springer. 2009. The mechanism of action of tapasin in the peptide exchange on MHC class I molecules determined from kinetics simulation studies. Mol. Immunol. 46:2054-2063. [DOI] [PubMed] [Google Scholar]
- 46.Schoenhals, G. J., et al. 1999. Retention of empty MHC class I molecules by tapasin is essential to reconstitute antigen presentation in invertebrate cells. EMBO J. 18:743-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stern-Ginossar, N., et al. 2007. Host immune system gene targeting by a viral miRNA. Science 317:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan, P., et al. 2002. Recruitment of MHC class I molecules by tapasin into the transporter associated with antigen processing-associated complex is essential for optimal peptide loading. J. Immunol. 168:1950-1960. [DOI] [PubMed] [Google Scholar]
- 49.Tenney, D. J., and A. M. Colberg-Poley. 1991. Human cytomegalovirus UL36-38 and US3 immediate-early genes: temporally regulated expression of nuclear, cytoplasmic, and polysome-associated transcripts during infection. J. Virol. 65:6724-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tirabassi, R. S., and H. L. Ploegh. 2002. The human cytomegalovirus US8 glycoprotein binds to major histocompatibility complex class I products. J. Virol. 76:6832-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trgovcich, J., C. Cebulla, P. Zimmerman, and D. D. Sedmak. 2006. Human cytomegalovirus protein pp71 disrupts major histocompatibility complex class I cell surface expression. J. Virol. 80:951-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Endert, P. M., et al. 1994. A sequential model for peptide binding and transport by the transporters associated with antigen processing. Immunity 1:491-500. [DOI] [PubMed] [Google Scholar]
- 53.Wagner, M., A. Gutermann, J. Podlech, M. J. Reddehase, and U. H. Koszinowski. 2002. Major histocompatibility complex class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J. Exp. Med. 196:805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wearsch, P. A., and P. Cresswell. 2007. Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin-ERp57 heterodimer. Nat. Immunol. 8:873-881. [DOI] [PubMed] [Google Scholar]
- 55.Wiertz, E. J., et al. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:769-779. [DOI] [PubMed] [Google Scholar]
- 56.Wiertz, E. J., et al. 1996. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384:432-438. [DOI] [PubMed] [Google Scholar]
- 57.Williams, A. P., C. A. Peh, A. W. Purcell, J. McCluskey, and T. Elliott. 2002. Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity 16:509-520. [DOI] [PubMed] [Google Scholar]
- 58.Wills, M. R., et al. 1996. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J. Virol. 70:7569-7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wormald, S., et al. 2006. The comparative roles of suppressor of cytokine signaling-1 and -3 in the inhibition and desensitization of cytokine signaling. J. Biol. Chem. 281:11135-11143. [DOI] [PubMed] [Google Scholar]