Abstract
Target cell overexpression of the integrase binding domain (IBD) of LEDGF/p75 (LEDGF) inhibits HIV-1 replication. The mechanism and protein structure requirements for this dominant interference are unclear. More generally, how and when HIV-1 uncoating occurs postentry is poorly defined, and it is unknown whether integrase within the evolving viral core becomes accessible to cellular proteins prior to nuclear entry. We used LEDGF dominant interference to address the latter question while characterizing determinants of IBD antiviral activity. Fusions of green fluorescent protein (GFP) with multiple C-terminal segments of LEDGF inhibited HIV-1 replication substantially, but minimal chimeras of either polarity (GFP-IBD or IBD-GFP) were most effective. Combining GFP-IBD expression with LEDGF depletion was profoundly antiviral. CD4+ T cell lines were rendered virtually uninfectable, with single-cycle HIV-1 infectivity reduced 4 logs and high-input (multiplicity of infection = 5.0) replication completely blocked. We restricted GFP-IBD to specific intracellular locations and found that antiviral activity was preserved when the protein was confined to the cytoplasm or directed to the nuclear envelope. The life cycle block triggered by the cytoplasm-restricted protein manifested after nuclear entry, at the level of integration. We conclude that integrase within the viral core becomes accessible to host cell protein interaction in the cytoplasm. LEDGF dominant interference and depletion impair HIV-1 integration at distinct postentry stages. GFP-IBD may trigger premature or improper integrase oligomerization.
LEDGF, a nuclear, chromatin-bound protein, was initially identified as an RNA polymerase II coactivator (17) and subsequently as an HIV-1 integrase (IN) binding protein (8) and integration cofactor (30, 36, 46, 52). LEDGF functions as an adaptor between chromatin and cellular proteins (2, 29, 35, 58). Current models posit that lentiviruses have exploited this tethering property to establish chromatin attachment (39, 40; for reviews, see references 13, 20, 39, 44, and 51). It is presently not clear whether the protein plays additional or sequential roles during the postentry path to integration (13, 44).
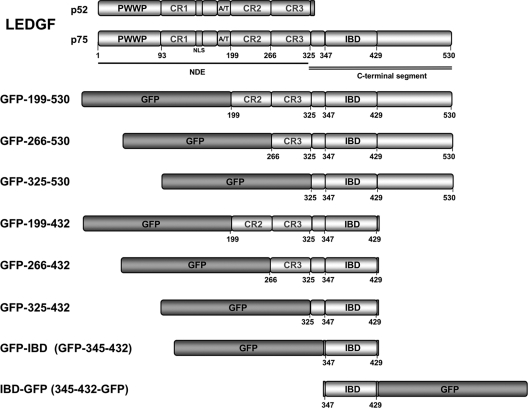
LEDGF has modular properties (Fig. 1). The chromatin linkage is mediated by an N-terminal domain ensemble (NDE), of which the key elements are the PWWP domain and a pair of A/T hooks (32, 50). Three other relatively charged regions (CR1 to CR3) are required for maximal chromatin affinity but lack autonomous chromatin-binding function (32). The integrase binding domain (IBD; residues 347 to 429) lies C-terminal to the NDE (7, 53). The IBD, which contains five alpha-helices (9), interacts with the integrase proteins of lentiviruses and with several cellular proteins (2-5, 7, 8, 31, 35, 53, 58).
FIG. 1.
GFP-LEDGF fusion proteins. The domains of LEDGF are indicated. The PWWP domain and the AT hooks are dominant chromatin binding elements (7, 30, 32, 50). The C-terminal domain (amino acids 325 to 530) contains the IBD (amino acids 347 to 429). In the present study “LEDGF” refers to the 75-kDa splice variant (i.e., LEDGF/p75) of the gene PSIP1. The gene also encodes an IBD-lacking splice variant LEDGF/p52, which has a different 8-amino-acid C-terminal domain and does not interact with integrase. CR, charged region; IBD, integrase binding domain; NDE, N-terminal domain ensemble involved in chromatin binding; NLS, nuclear localization signal.
Critical lentiviral IN contacts are made by two inter-helical IBD loops that engage a cleft formed by the IN catalytic core domain (CCD) dimer, with both hydrophobic and charged interactions participating at the interface (6). Additional charged residue interactions occur between the IBD and the N-terminal domain of IN (21). The function of the C terminus of LEDGF (amino acids 429 to 530) is unknown. This domain is also the least conserved across species (7).
Tethering of the lentiviral preintegration complex (PIC) to the chromatin fiber by LEDGF has not been demonstrated directly but has been inferred from a combination of protein interaction and virological experiments. If lentiviral IN proteins are artificially expressed outside the viral life cycle they are found tightly bound to chromatin, where they colocalize with LEDGF. Making cells LEDGF-deficient has two effects in such experiments. First, IN becomes untethered and relocates to the cytoplasm (31, 34). The second consequence is a much reduced steady-state level of IN at a given mRNA level, because LEDGF binding protects IN from degradation in the proteasome (28).
HIV-1 infection is inhibited by LEDGF knockdown in CD4+ human T cells when this is stringent enough to strip chromatin of detectable protein (30), or by LEDGF gene knockout in murine cells (46). Expression of the IBD in an untethered state, as a green fluorescent protein (GFP) fusion protein, can also inhibit HIV-1 replication to a similar extent (10, 25, 30). This is a dominant interference (DI) rather than dominant negative effect since GFP-IBD does not interact with LEDGF (9) or self-multimerize (40). Fusions of GFP to either the IBD (10, 25, 30, 40) or to a larger segment of LEDGF (residues 325 to 530 [10, 25]) has been reported to inhibit HIV-1 replication.
We previously found that introducing GFP-IBD on top of LEDGF-depletion caused multiplicative impairment, up to 560-fold for single-round HIV-1 reporter viruses (30). This observation suggested that the two inhibitory effects might impair the virus in different ways. In theory, any protein that contains the IBD but no chromatin binding module might inhibit HIV-1 integration. Conversely, recent evidence suggests that an IBD protein can function as a cofactor if it has a chromatin binding module and that highly diverse modules from unrelated proteins can suffice (16, 18, 40, 47). Importantly, GFP-IBD itself converts from a potent DI protein to an integration cofactor if a short core histone-binding peptide is attached to its N terminus (40).
Both GFP-IBD and GFP-325-530 distribute diffusely throughout cells but are more abundant in the nucleus, and mechanistically they have generally been presumed to act there since they interfere with the integration step (10, 25, 30, 40). However, although intranuclear PIC interdiction has appeared intuitively probable, it has not been excluded that GFP-IBD engages the PIC and/or exerts its decisive mechanistic effect earlier.
Indeed, whether retroviral IN enzymes within the reverse transcription complex become accessible to host cell protein binding prior to nuclear entry is not known. HIV-1 core uncoating is poorly understood, both in terms of the timing of disassembly and how and when core constituents become exposed to the cellular milieu. Here we investigated this question as we characterized LEDGF dominant interference in detail. LEDGF chimeras were analyzed with or without knockdown of the endogenous protein. We asked how incorporating different LEDGF segments affects the antiviral activity, extended testing to multiple cell types, determined the extent of antiviral effect that can be achieved, and devised experiments to test whether intracellular location of a DI protein matters.
MATERIALS AND METHODS
Plasmid construction.
The gfp cDNA used was the enhanced (eGFP) version in all cases. HIV-1-based lentiviral transfer vectors TSINGFP-IBD and TSINGFP-IBDD366N have been described (30). The additional GFP-LEDGF segment fusions were constructed with PCR primers indicated in Table 1; amplicons were digested with SacII and KpnI and ligated into the TSINGFP-IBD backbone. GFP-IBD comprises amino acids 345 to 432 of LEDGF. Changes in all plasmids were confirmed by sequencing. To make PK-GFP-IBD, the nucleotide sequence encoding amino acids 17 to 529 of pyruvate kinase cDNA3-myc-PK (53) were amplified with primers listed in Table 1 and ligated into a TSIN vector intermediate. PK-GFP-IBDD366N,5E has the D366N mutation and K401E, K402E, R405E, F406E, and V408E mutations. This mutant was constructed by overlap extension PCR in the PK-GFP-IBD vector using the primers indicated in Table 1. A lentiviral vector that expresses the LEDGF-targeting shRNA, as well as the DI protein, was constructed by cleaving TSINPK-GFP-IBD and TSINCherry 1340 (30) with SnaBI and AvrII and ligating the appropriate fragments.
TABLE 1.
PCR primers
| Construct | Junction | Primer sequence (5′-3′) |
|---|---|---|
| GFP-199-432 | VKQPCP | atatccgcggtaaaacagccctgtccttcagag |
| ttctcgaggtacctatccttcaccaaccaagaacatgttc | ||
| GFP-266-432 | KTGVTST | atatccgcggggaaaacaggggttacttcaacc |
| ttctcgaggtacctatccttcaccaaccaagaacatgttc | ||
| GFP-325-432 | EQQNKDEGKK | atatccgcggagcagcagaataaagatgaagg |
| ttctcgaggtacctatccttcaccaaccaagaacatgttc | ||
| GFP-199-530 | VKQPCP | atatccgcggtaaaacagccctgtccttcagag |
| atatggtaccctagttatctagtgtagaatc | ||
| GFP-266-530 | KTGVTST | atatccgcggggaaaacaggggttacttcaacc |
| atatggtaccctagttatctagtgtagaatc | ||
| GFP-325-530 | EQQNKDEGKK | atatccgcggagcagcagaataaagatgaagg |
| atatggtaccctagttatctagtgtagaatc | ||
| NETC-GFP-IBD | atatggatccaccatgtcaggagccggaggagtcggatctgcagatggtgagcaagggc | |
| atatctgcagaggggattccaacatggaaattgtcgacctatccttcaccaaccaa | ||
| NETC-GFP-IBDD366N | atatggatccaccatgtcaggagccggaggagtcggatctgcagatggtgagcaagggc | |
| atatctgcagaggggattccaacatggaaattgtcgacctatccttcaccaaccaa | ||
| PK-GFP-IBD | atatctcgaggccgccaccatgctgcacgctgccatggcagacacctttctg | |
| atatggatcccctggcacgggcaccacccgc | ||
| PK-GFP-IBDD366N,5E | atatggatccaccggtcgccaccatggtgagcaagg | |
| cttttccatgattacctgactctctttctcctcccgtatttcttccagtgtagtaatc | ||
| gattactacactggaagaaatacgggaggagaaagagagtcaggtaatcatggaaaag | ||
| atatactagtgctagagattttccacactgac |
TSIN.NETC-GFP-IBD was constructed by amplifying (i) the Nup153 nuclear envelope targeting cassette (NETC; amino acids 2 to 144 of Nup153) from a human Nup153 cDNA plasmid kindly provided by K. Ullman, University of Utah, and (ii) GFP-IBD using the primers indicated in Table 1. The amplicons were digested with BamHI and PstI (NETC) or PstI and SalI (GFP-IBD) and ligated into the polylinker of TSINpoly. The latter lentiviral transfer vector was constructed previously by cleaving TSINCherry (30) with BamHI and KpnI to remove mCherry and inserting a KpnI-BamHI fragment containing the multiple cloning site of pBluescript. TSINIBD-GFP-tubulin was constructed by amplification of the IBD from TSINGFP-IBD and GFP-alpha-tubulin from pAcGFP1-tubulin (Clontech). The amplicons were ligated into TSINpoly with BamHI and PstI flanking the IBD segment and PstI and KpnI flanking the coding segment for GFP-tubulin. The reverse polarity IBD-GFP protein vector TSINIBD-GFP was constructed as follows. TSINIBD-GFP-tubulin was digested with XhoI and BlpI to remove tubulin and the termini were blunted with Klenow polymerase and religated, adding six additional amino acids to the end of GFP (SGLRSR). MLV-luc was constructed by inserting firefly luc from pGL3 (Promega) into pJZ308 (43).
Human CD4+ T cell lines, PBMC, and primary human macrophages.
MT-4 and SupT1 cells were maintained in RPMI with 10% fetal calf serum and penicillin-streptomycin and l-glutamine. Normal donor peripheral blood mononuclear cells (PBMC) were obtained by Ficoll centrifugation of PBMC eluted from Mayo Clinic Blood Bank apheresis machine leukoreduction system chambers (11). PBMC were cultured in RPMI with 10% heat-inactivated filtered human AB serum (Irvine Scientific, catalog no. 5005) in T25 flasks at a density of 5 × 106 cells per ml and allowed to differentiate to macrophages by plastic adherence. At day 7 after isolation the cells formed a confluent adherent monolayer with characteristic fried-egg morphology. A 1:1 trypsin-EDTA solution was used to remove the macrophages, which were replated into 48-well plates at 105 cells per well and spinoculated (centrifuged at 2,800 rpm for 2 h) on alternate days three times with TSINScrGFP and TSINGFP-IBD or TSINGFP-IBDD366N at a multiplicity of infection (MOI) of 70 to 100. Eleven days after the first spinoculation the cells were challenged with NL4-3 luciferase at an MOI of 10. Cells were lysed in phosphate-buffered saline (PBS)-0.1% Tween and assayed for luciferase activity at 7 days postchallenge using SteadyGlo (Promega, catalog no. E2520) according to the manufacturer's instructions.
Retroviral vectors, RNA interference (RNAi), and stable cell lines.
Table 2 summarizes cell lines. The basic cell line generation method enables single-step de facto gene replacement in human CD4+ T cell lines. The same U3-deleted lentiviral transfer vector encodes the polymerase III (pol III)-promoted shRNA targeting the endogenous LEDGF mRNA transcript (target located at nucleotide 1340 of the protein-coding sequence) and the pol II-promoted DI protein cDNA. (Details of this and other retroviral vector methods are given in reference 30.) LEDGF re-expression in knockdown cells was done with a cDNA having 7 synonymous mutations at the shRNA target site (p75syn). MLV-based vectors coexpress neoR with 600 μg of G418/ml used for selection. ilvRNAi vectors were generated by three-plasmid calcium phosphate cotransfection in two-chamber (CF2) Cell Factories (33). The medium was changed at 12 to 16 h, and the supernatants were collected 48 h later, filtered through a 0.45-μm-pore-size filter, and concentrated by ultracentrifugation over a sucrose cushion in an SW28 swinging-bucket rotor at 25,000 rpm for 2 h. MLV-luc and MLV-LEDGFsyn7 vectors were produced in Phoenix A cells by calcium phosphate-mediated cotransfection of 10 μg of transfer vector and 3 μg of pMD.G. Luciferase reporters have been described previously (30).
TABLE 2.
Summary of stable cell lines
| Cell line | shRNA | Protein(s) expressed |
|---|---|---|
| Adherent cell lines | ||
| 293T | None | None |
| L1340 | p75 1340 | None |
| LH4 | p75 1340 | HIV-1 IN |
| CD4+ T cell lines | ||
| SupT1 | None | None |
| TC3 | Control | mCherry |
| TL3 | p75 1340 | mCherry |
| SupT1 GFP-IBD | None | GFP-IBD (345-432) |
| SupT1 GFP-IBDD366N | None | GFP-IBDD366N (345-432) |
| SupT1 GFP-IBD + p75shRNA | p75 1340 | GFP-IBD (345-432) |
| SupT1 GFP-IBD + p75shRNA + p75syn7 | p75 1340 | GFP-IBD (345-432) + LEDGF |
| SupT1 GFP-199-530 | None | GFP-199-530 |
| SupT1 GFP-199-530D366N | None | GFP-199-530D366N |
| MT4 | None | None |
| MT4 Scr GFP | Control | GFP |
| MT4 GFP-IBD | None | GFP-IBD (345-432) |
| MT4 IBD-GFP | None | IBD-GFP (345-432) |
| MT4 GFP-IBDD366N | None | GFP-IBDD366N (345-432) |
| MT4 GFP-IBD + p75shRNA | p75 1340 | GFP-IBD (345-432) |
| MT4 GFP-IBDD366N + p75shRNA | p75 1340 | GFP-IBDD366N (345-432) |
| MT4 GFP-199-530 | None | GFP-199-530 |
| MT4 GFP-199-530D366N | None | GFP-199-530D366N |
| MT4 199-432 | None | GFP-199-432 |
| MT4 266-432 | None | GFP-266-432 |
| MT4 266-432D366N | None | GFP-266-432D366N |
| MT4 325-432 | None | GFP-325-432 |
| MT4 325-432D366N | None | GFP-325-432D366N |
| MT4 266-530 | None | GFP-266-530 |
| MT4 325-530 | None | GFP-325-530 |
| MT4 NETC-GFP-IBD | None | NETC-GFP-IBD |
| MT4 NETC-GFP-IBDD366N | None | NETC-GFP-IBDD366N |
| MT4 PK-GFP-IBD | None | PK-GFP-IBD |
| MT4 PK-GFP-IBDD366N,5E | None | PK-GFP-IBDD366N,5E |
| MT4 PK-GFP-IBD + p75shRNA | p75 1340 | PK-GFP-IBD |
Indirect immunofluorescence, laser scanning confocal microscopy.
Cells were imaged as described previously (31, 53). The LH4 single cell clone was derived by sequential plasmid-based stable shRNA knockdown of LEDGF in 293T cells (which generated L cells), followed by stable expression of Myc-epitope-tagged HIV-1 IN (28). Antibodies used were primary mouse monoclonal anti-LEDGF (BD Transduction Laboratories) and a rabbit polyclonal anti-cMyc (A-14 sc-789; Santa Cruz Biotechnology). The secondary antibodies were Alexa-488-conjugated goat anti-mouse antibody and Alexa-594-conjugated goat anti-rabbit antibody (Molecular Probes). Rev-CFP was constructed by overlap extension PCR of CFP into phCMV rev, using the rev primers GACGCAAATGGGCGGTAG and CTTGCTCACCATTTCTTTAGCTCCTGACTCCAA and the CFP primers GGAGCTAAAGAAATGGTGAGCAAGGGCGAGGAG and CCTCTACAAATGTGGTATG. Leptomycin B (Sigma catalog no. L2913) at 10 ng/ml was added to either suspension cells or adherent cells for 4 h before fixing. Imaging of suspension cells was performed as previously described (40). The nuclear fluorescence intensity of MT4, GFP-IBD, GFP-IBDLOW, and PK-GFP-IBD cell lines (imaged at the same fluorescent gain) was obtained using LSM 510 software (Zeiss). Twenty randomly chosen cells were assessed in each cell line, and the average intensity was determined.
HIV-1 infections.
VSV-G pseudotyped HIV-1luc, FIV-luc, or MLV-luc was used for single-round infection (transduction) of 0.5 × 105 to 1.0 × 105 cells in 24-well plates at MOIs ranging from 0.2 to 5.0. Luciferase assays were performed with SteadyGlo (Promega) and measured in duplicate with a Packard TopCount NXT microplate luminescence counter. For real-time quantitative PCR analyses, half the sample was analyzed for luciferase, while DNA was collected from the other half with a tissue DNeasy kit (Qiagen). Full-length HIV-1 NL4-3 and LAI viruses were generated in 293T producer cells by calcium phosphate-mediated transfection. Virus particles were quantified in cell supernatants by HIV-1 p24 antigen capture enzyme-linked immunosorbent assay (Coulter). Titers were determined on GHOST cells. A total of 106 cells were infected with either full-length HIV-1 NL4-3 or LAI at MOIs of 5.0, 1.0, or 0.3 in 3 ml of RPMI. Cells were washed five times after 16 h. The cells were maintained in 6 ml of RPMI.
Subcellular fractionation and immunoblotting.
The chromatin binding assay has been described previously (32). S1 and S2 fractions and total cell lysates were separated by SDS-PAGE. Blots were probed for LEDGF by incubation with a mouse anti-LEDGF monoclonal antibody (BD Transduction Laboratories) at 1:500 overnight at 4°C or for eGFP using mouse anti-GFP MAb (Living Colors Aequorea victoria green fluorescent protein monoclonal antibody JL-8) at 1:1,000 for 1 h at room temperature. Mouse anti-α-tubulin MAb (clone B-5-1-2; Sigma) was used for a loading control of whole-cell lysates. Secondary goat anti-mouse conjugated to horseradish peroxidase (Calbiochem) was used at 1:1,000 for 1 h at room temperature. All antibodies were used in 5% milk-0.1% Tween. Blots were developed with the ECL chemiluminescence kit (Amersham Pharmacia).
Coimmunoprecipitation.
Aliquots (30 μl) of Dynal sheep anti-mouse IgG Dynabeads (product no. 110.31) were blocked in TBST-10% milk by rotating samples for 1.5 h at 4°C. A total of 2.8 × 106 LH4 cells were plated in T-75 flasks, and transfected the next day with 10 μg of the GFP-LEDGF fusion proteins. At 48 h after transfection, the cells were scraped off the flasks and lysed for 30 min in CSKII buffer (see “Subcellular fractionation and immunoblotting”) with DNase. One-third volume of 1 M Ammonium sulfate was added, followed by incubation at room temperature for an additional 30 min and then centrifugation at 1,300 × g for 6 min. A 500-μl portion of the cell lysate was incubated with 3 μg of monoclonal anti-GFP (BD Biosciences Living Colors, catalog no. 632381) or isotype control for 1 h on ice. The sample was then mixed by continuous rotation with the preblocked beads overnight at 4°C. The beads were washed three times with PBS, eluted in 50 μl of 2× Laemmli buffer, and boiled for 7 min at 95°C before immunoblotting.
2-LTR circle measurement.
Total DNA was extracted with a tissue DNeasy kit (Qiagen) and analyzed by real-time quantitative PCR in a Roche LightCycler using the FastStart DNA Master SYBR Green kit and Roche LCDA software as previously described (30, 31). 2-LTR circles were normalized to either mitochondrial DNA (mtDNA) or GAPDH (glyceraldehyde-3-phosphate dehydrogenase) signal (40).
Alu element-anchored PCR for integrated HIV-1 and genomic DNA integration standard, LEDGF mRNA PCR.
For Alu-PCR, a cellular integration standard was created as follows. A total of 106 293T cells were transduced at an MOI of 22 with an HIV-1 vector (TRIP-GFP), and single cell clones were derived. The number of integrants in each of five single-cell clones of this line was quantified by junction Southern blots, and the mean (12 integrants/cell clone) was used as the proviral copy number in the polyclonal cell standard. Genomic DNA from this polyclonal standard was then serially diluted in a fixed background of uninfected negative control DNA for Alu-PCR standard curves. Quantitative Alu-PCR for quantification of integrants was done at 6 days posttransduction in two phases. The first phase amplifies from Alu sequences to U3 sequences absent in self-inactivating (U3-deleted) HIV-1 vectors using 300 nM AluI (TCC CAG CTA CTG GGG AGG CTG AGG), 300 nM Alu2 (GCC TCC CAA AGT GCT GGG ATT ACA G), and 100 nM U3 AS (CTC CGG ATG CAG CTC TCG). Amplification conditions were as follows: 95°C for 10 s, 63°C for 10 s, and 72°C for 2 min 30 s for 12 cycles, with a temperature transition rate of 5°C/s. The second phase amplifies a nested product within U3 using 300 nM sense primer GAA CTA CAC ACC AGG GCC and 300 nM antisense primer CTC CGG ATG CAG CTC TCG. The PCR conditions were as follows: 95°C for 10 s, 65°C for 10 s, and 72°C for 9 s for 50 cycles, with a temperature transition rate of 5°C/s. To establish a loading control, human mtDNA was quantified by using 20 pmol of sense primer GAA TGT CTG CACA AGC CAC TT and 20 pmol of antisense primer TAG AAA GGC TAG GAC CAA AC. PCR amplification was performed as follows: 95°C for 10 s, 54°C for 20 s, and 72°C for 30 s for 40 cycles, with a temperature transition rate of 5°C/s. RNA isolation and reverse transcription-PCR (RT-PCR) for LEDGF mRNA was performed as described previously (30).
RESULTS
Characterization of DI proteins.
An initial series of GFP fusion proteins, illustrated in Fig. 1, were constructed based on described LEDGF domains (7, 32, 53). None contain the domains required for chromatin binding, and all contain the IBD. A protein with reversed polarity, IBD-GFP, was also constructed to determine whether the placement of GFP affects function. In addition, Asp366→Asn (D366N) control mutants were evaluated. This mutation disrupts an important hydrogen bond at the IBD-IN dimer interface (6).
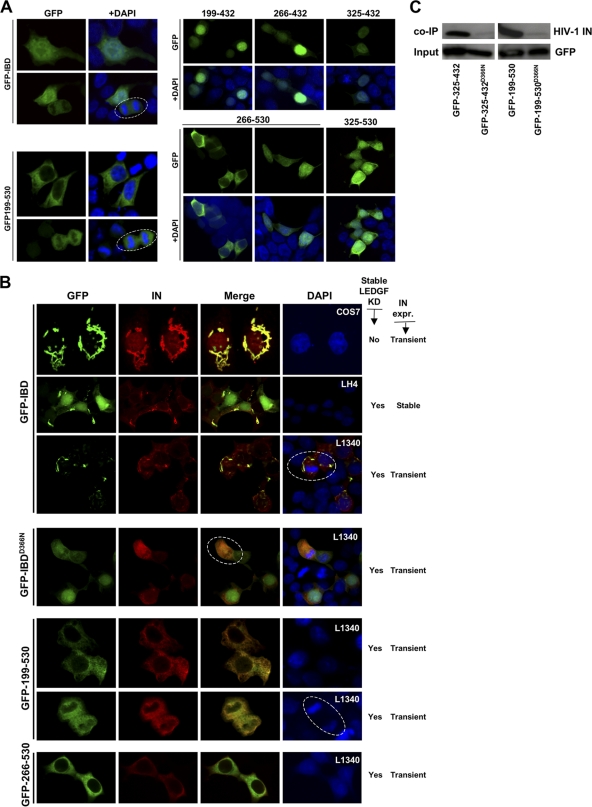
Subcellular localization was determined for each protein using confocal microscopy (Fig. 2 A and B). GFP-IBD was consistently found throughout the cell. In general, the addition of flanking portions of LEDGF tended to restrict location, with the largest protein, GFP-199-530, observed primarily in the cytoplasm. Proteins that contain the C-terminal 98 amino acids of LEDGF (residues 432 to 530) demonstrated greater cytoplasmic localization, while those that terminate with the IBD were predominantly nuclear. However, the terminal 98 amino acids did not confer cytoplasmic retention. For example, GFP-325-530 was found approximately equivalently in both the nucleus and the cytoplasm.
FIG. 2.
Subcellular localization of IBD proteins, with or without IN. (A) Cells were fixed at 48 h after transfection, stained with DAPI, and imaged with a confocal microscope. These transfections were done in LEDGF-deficient 293T cells (L1340 cells, Table 2), but similar results were seen in 293T, HeLa, and Cos7 cells (data not shown). Dashed line circles indicate cells in mitosis. Fusions terminating at LEDGF residue 432 are mainly nuclear, while those terminating at amino acid 530 have greater cytoplasmic localization. (B) GFP-LEDGF fusion proteins were coexpressed with HIV-1 IN in the presence or absence of endogenous LEDGF. Cells used are noted at right (white font). The morphologically distinctive inclusions seen in COS7 cells were also seen in HeLa cells (data not shown). As described in Table 2, L1340 (or “L”) cells have a stable LEDGF knockdown (31). LH4 cells are L1340 cells that contain a stably integrated HIV-1 IN expression plasmid (28, 40). IN is unstable in the absence of an IBD protein, so that very low levels are present at steady state in LH4 cells, too low to be detected at this confocal gain (28). Therefore, IN seen in these LH4 cells images is that which is stabilized by the GFP-IBD interaction (40). Wild-type LEDGF re-expression in LH4 cells (not shown here) results in nuclear (chromatin-bound) IN-LEDGF complexes (40). (C) Coimmunoprecipitation of HIV-1 IN with GFP-LEDGF fusion proteins. Immunoblotting for Myc epitope-tagged HIV-1 IN was performed after immunoprecipitation of indicated GFP-LEDGF fusion proteins from LH4 cells.
When imaged with coexpressed HIV-1 IN, all IBD proteins colocalized with it, but in two different ways. Each of the IBD-terminating proteins coalesced into discrete cytoplasmic structures with IN, generally with rod- or worm-like morphology (Fig. 2B). These structures were apparent even in the presence of endogenous LEDGF (Fig. 2B, COS7 cells). In contrast, those having the terminal 98 amino acids of LEDGF colocalized with IN in a diffusely cytoplasmic distribution (Fig. 2B). All of the fusion proteins lack the PWWP and A/T hook domains that mediate the chromatin association of full-length LEDGF (32), and none bound to condensed mitotic chromatin (Fig. 2, circled cells). In fact, IN-fusion protein complexes were seen only in the cytoplasm in all cases, even with proteins that were predominantly nuclear in the absence of IN. This was also the case when IN was underexpressed relative to GFP-IBD and considerable free nuclear GFP-IBD was present (Fig, 2B, second row). Coimmunoprecipitation experiments confirmed interaction with IN (Fig. 2C, and data not shown).
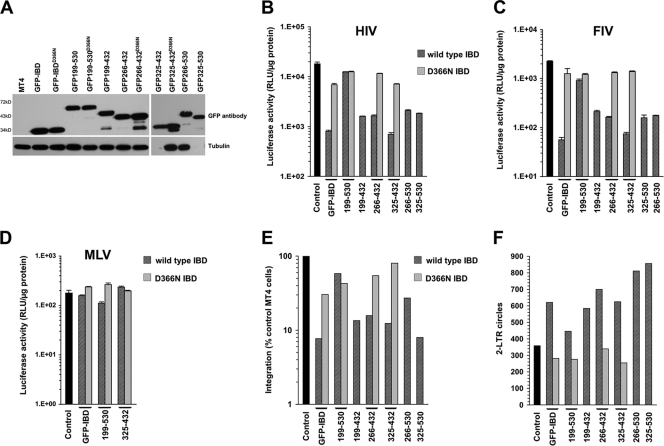
CD4+ human T cell lines were transduced with lentiviral vectors encoding fusion proteins and/or a LEDGF-targeted shRNA. Table 2 summarizes the generated cell lines. Expression of predicted proteins was confirmed by Western blotting (Fig. 3A). Stable cell lines were then challenged with HIV-1luc, FIV-luc, or MLV-luc (Fig. 3B, C, and D). As shown in Fig. 3B and C, all of the fusion proteins except GFP-199-530 inhibited HIV-1 and FIV infection, with excellent concordance between the two lentiviruses. The respective D366N mutants had minimal effect, indicating molecular specificity. MLV was not inhibited, demonstrating lentiviral specificity (Fig. 3D). The block to infection mapped to integration with a corresponding increase in 2-LTR circles (Fig. 3E and F). Note that the D366N mutants had reciprocal effects in the integration (Fig. 3E) and 2-LTR circle assays (Fig. 3F). The addition of C-terminal segment residues to the left or the right of the IBD tended to reduce activity (Fig. 3B to D). In three separate experiments directly comparing stable cell MT4 lines expressing GFP-IBD and GFP-325-432 (without RNAi), the average inhibition was 17-fold (range, 8- to 23-fold) for the GFP-IBD line versus the control and 18-fold (range, 9- to 26-fold) for the GFP-325-432 line versus the control. Thus, the minimal proteins GFP-IBD and GFP-325-432 consistently inhibited single-cycle infection the most, whereas the largest protein, GFP-199-530, had the least effect.
FIG. 3.
Fusion proteins inhibit lentiviral infection. (A) Western blotting of MT4 cell lysates with antibody to GFP. Tubulin was immunoblotted as a loading control. (B) HIV-1luc challenge of stable MT4 cells. WT/D366N pairs are indicated. Values were normalized to cell number or protein. Error bars are between duplicates in each experiment. All challenge experiments were performed at least twice with similar results. In panels B to D, cell lysates were collected 5 to 7 days after infection. (C) FIV-luc infection. (D) MLV-luc infection. (E) Alu-PCR assessment of integration. Values are normalized to GAPDH standard. (F) 2-LTR circles, analyzed at 22 h posttransduction with HIV-1luc. Values are normalized to MtDNA as a loading control.
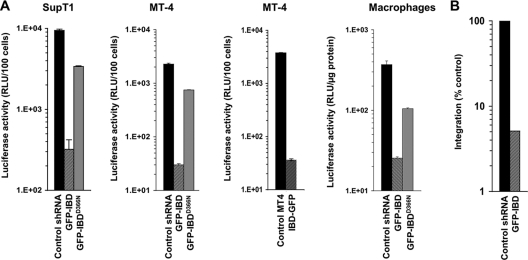
With this disparity in mind, GFP-IBD and GFP-199-530 were analyzed further in other cell lines (Fig. 4A). GFP-IBD expression inhibited single-round HIV-1 infectivity 57-fold in SupT1 cells, 73-fold in other challenges in MT-4 cells, and 8-fold in primary macrophages. The reversed polarity protein IBD-GFP was tested in MT4 cells, where it inhibited single-round infection 100-fold (Fig. 4A). In contrast, GFP-IBDD366N expression produced only a small (yet consistently detected) inhibition, between 3- and 5-fold. The latter effect is consistent with the slight residual interaction between the D366N-mutant IBD and IN that we have previously detected in coimmunoprecipitation experiments (40). The number of integrants measured by Alu element-anchored PCR was markedly decreased in SupT1 GFP-IBD cells compared to control cells (Fig. 4B). Thus, GFP-IBD inhibits HIV-1 infection in multiple T cell lines and primary macrophages, blocking integration and the dominant interfering phenotype is apparent whether GFP is situated on the IBD N or C terminus. The relatively lower antiviral effect in macrophages is consistent with the relative resistance of these cells to transduction by the DI protein-expressing vectors (data not shown).
FIG. 4.
GFP-IBD inhibits HIV-1 infection in multiple T cell lines and primary cells. (A) SupT1 and MT4 cell lines that stably express GFP-IBD or IBD-GFP were challenged with HIV-1luc, and luciferase was analyzed at 5 days. The macrophages were transduced as described in Materials and Methods and challenged with HIV-1luc, and expression was analyzed at 7 days. (B) Alu-PCR-based assay of integration in SupT1 GFP-IBD cell lines challenged with HIV-1luc.
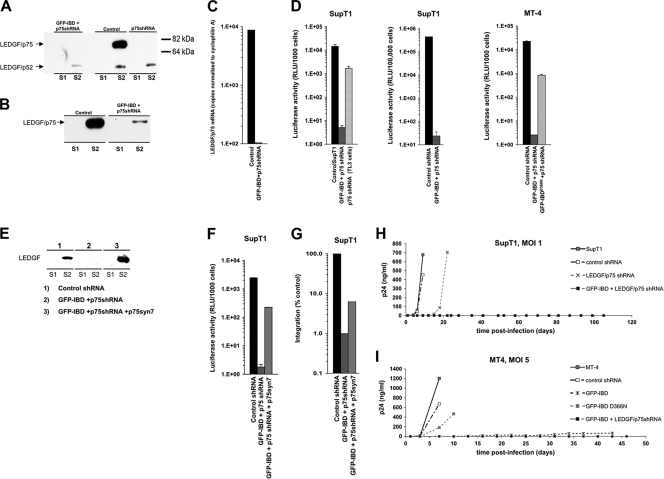
Profound synergy of DI and RNAi.
Llano et al. showed that combining LEDGF RNAi with GFP-IBD inhibited single cycle luciferase reporter virus infectivity up to 560-fold in SupT1 cells (30). The combination of RNAi with GFP-IBD was not tested for replicating HIV-1, but the knockdown alone did delay spreading viral replication approximately 2 weeks at an MOI of 0.001 to 0.1 (30). In the present study we extended these initial observations, performing detailed separate and combined testing of LEDGF-targeted DI and knockdown (KD), in both MT4 and SupT1 cell lines, and using 10- to 50-fold higher infectious HIV-1 challenges (MOI = 1.0 or 5.0). As shown in Fig. 5A, there was no detectable chromatin-associated LEDGF in the SupT1-derived GFP-IBD+KD cells. However, although much decreased, endogenous LEDGF could still be detected in the S2 fraction (32) of the equivalent MT4 cell line (Fig. 5B). LEDGF mRNA levels in SupT1-derived GFP-IBD+KD cells were decreased to approximately 1% of the controls, confirming knockdown stringency (Fig. 5C).
FIG. 5.
Combined dominant interference and knockdown. (A and B) Subcellular fractions of SupT1 (A) and MT4 cells (B) stably transduced with lentiviral vectors expressing LEDGF-targeted shRNA (p75shRNA) with or without GFP-IBD were analyzed for residual LEDGF expression using anti-LEDGF antibody. S1, Triton X-100-extractable, non-chromatin-bound fraction. S2, Triton X-100-resistant, chromatin-bound fraction releasable with salt and DNase treatment. See Fig. 3 in reference 32 for assay details and validation. (C) LEDGF mRNA levels in stable cell lines, assessed by real-time quantitative RT-PCR. The primers span the exons 14/15 junction and do not detect GFP-IBD. The results were normalized to copies of cyclophilin A mRNA. (D) SupT1 and MT4 cell lines stably expressing GFP-IBD and the LEDGF mRNA-targeting shRNA were challenged with HIV-1luc, and the luciferase activity was analyzed after 5 days. The fold inhibition values for the combined modalities in the three graphs from right to left are 2,860, 18,600, and 8,730, respectively. (E) SupT1 cells expressing both GFP-IBD and LEDGF shRNA were transduced with a retroviral vector encoding neoR and p75syn7, an LEDGF cDNA insensitive to the shRNA. Cells were selected in G418. Western blotting of subcellular fractions confirmed re-expression of LEDGF. (F and G) LEDGF re-expression partially restores HIV-1 infection in cells expressing GFP-IBD and LEDGF shRNA. A luciferase assay (F) and an Alu-PCR integration assay (G) were performed in SupT1 cell lines. (H and I) SupT1 (H) and MT4 (I) cells stably transduced with the indicated expression elements were challenged with HIV-1. Supernatant p24 antigen was determined at the various time points shown.
SupT1 knockdown cells had 10-fold-reduced susceptibility to single-round HIV-1 infection (Fig. 5D), a finding consistent with previous results (30). However, much greater and effectively lethal inhibition was obtained when GFP-IBD was cointroduced. In single cycle analyses infectivity was impaired by 3 to 4 logs (Fig. 5D). Re-expression of LEDGF produced partial rescue of the >3-log defect, presumably of the RNAi-induced component (Fig. 5E and F). Alu-PCR-detected integrants were also profoundly reduced and, similar to the results shown in Fig. 5F, this was rescued partially by LEDGF re-expression, while the DI-induced component of the block persisted (Fig. 5G).
The MT4 and SupT1 cell lines were then challenged with replication-competent HIV-1 at high inputs (MOIs of 1.0 and 5.0). HIV-1 NL4-3 and HIV-1 LAI each produced the same results, and the data for LAI are shown here. After infection at an MOI of 1.0 in SupT1 cells, peak p24 production occurred in the control cells at 4 days and in knockdown (TL3) cells at 22 days (Fig. 5H). In contrast, in the combined modality cells no p24 was detectable in supernatants out to 116 days postinfection, at which time the experiment was terminated. MT4 cell lines were challenged at MOIs of 1.0 and 5.0, with similar results (in Fig. 5I, the results for an MOI = 5.0 are shown). Again, no p24 was detectable in supernatants of the combined modality MT4 cells. Note also that low-level p24 antigen was detectable in the supernatants of GFP-IBD-expressing MT4 cells (without endogenous protein knockdown), but this was still markedly less than in control cells (8, 42, and 68 ng of p24 at 13, 31, and 43 days, respectively, versus 500 to 1,000 ng in the controls). Thus, combining LEDGF knockdown and GFP-IBD is multiplicative, with profound viral impairment consistently observed in assays that measure single cycle infectivity or resistance to the propagation of infectious HIV-1 after high MOI challenges that rapidly obliterate parental cells. The results in the MT4 cells also reveal that in the presence of GFP-IBD, the complete absence of detectable S2 fraction LEDGF is not a prerequisite for a profound antiviral effect.
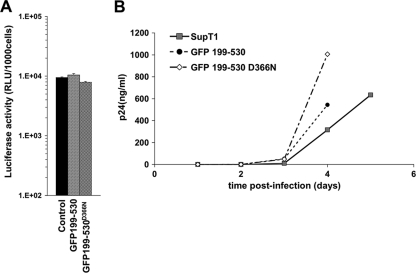
In clear contrast to the results with GFP-IBD, GFP-199-530 had minimal effect on HIV-1 infection (Fig. 6A and Fig. 3B) and FIV infection (Fig. 3C). The proteins were equivalently expressed (Fig. 3A and data not shown). We also confirmed transduction with the appropriate GFP-199-530 and GFP-199-530D366N vectors by isolating genomic DNA and amplifying and sequencing the 199-530 segment (data not shown). Challenge with replication-competent HIV-1 confirmed the lack of inhibition by GFP-199-530 (Fig. 6B).
FIG. 6.
GFP-199-530 does not inhibit HIV-1 infection. SupT1 cells stably expressing GFP-199-530 or GFP-199-530D366N were challenged with either HIV-1luc (A) or replication-competent HIV-1 (B).
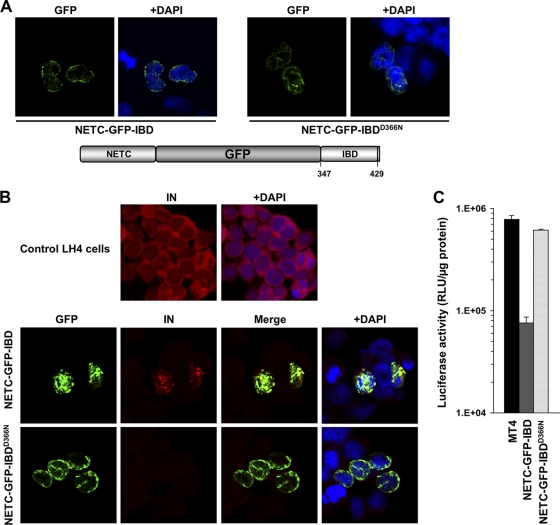
Nuclear envelope targeted GFP-IBD (NETC-GFP-IBD) inhibits HIV-1 infection.
Given that GFP-IBD distributes throughout the cell and we observed GFP-199-530 to be largely cytoplasmic (Fig. 2A), we considered it likely that PIC-associated IN is not accessible to a cytoplasmic DI protein, a hypothesis we tested further. Experimentally, we sought to restrict the IBD to earlier points along the viral infection pathway. IN (56) and Vpr (54) have been reported to interact with Nup153, a nuclear pore protein, suggesting that viral components may encounter this protein during the nucleopore traverse. To similarly localize GFP-IBD, we placed at its N terminus the Nup153 nuclear envelope targeting cassette (NETC), which comprises residues 2 to 144 of this 1,475-amino-acid nucleoporin (12). NETC-GFP-IBD localized to the nuclear envelope as intended (Fig. 7A), where it functioned to recruit ectopically expressed IN (Fig. 7B). As shown in Fig. 7C, HIV-1 reporter virus infection was inhibited 10-fold by NETC-GFP-IBD but not by NETC-GFP-IBDD366N. These results suggested that IN is accessible to IBD binding at least at the point of nuclear pore traverse.
FIG. 7.
NETC-GFP-IBD inhibits HIV-1 infection. (A) NETC-GFP-IBD and NETC-GFP-IBDD366N display a nuclear envelope localization pattern. (B) NETC-GFP-IBD colocalizes with HIV-1 IN at the nuclear envelope, in contrast to NETC-GFP-IBDD366N, which does not. Cells were transiently transfected with the indicated constructs, and immunofluorescence analyses were performed as described in Materials and Methods. Untransfected LH4 cells are shown in the top panel for comparison. Note that IN is poorly visualized in LH4 cells in the presence of NETC-GFP-IBDD366N because it is not protected from degradation (28); the confocal gain is much higher in the control LH4 panel at the top than in the images of the bottom two rows, enabling the low level of IN to be seen in these LEDGF-deficient cells. (C) NETC-GFP-IBD and NETC-GFP-IBDD366N were stably expressed in MT4 with lentiviral vectors. The lines were challenged with HIV-1luc, and the luciferase activity was analyzed at 5 days.
Cytoplasm-confined GFP-IBD inhibits HIV-1 infection.
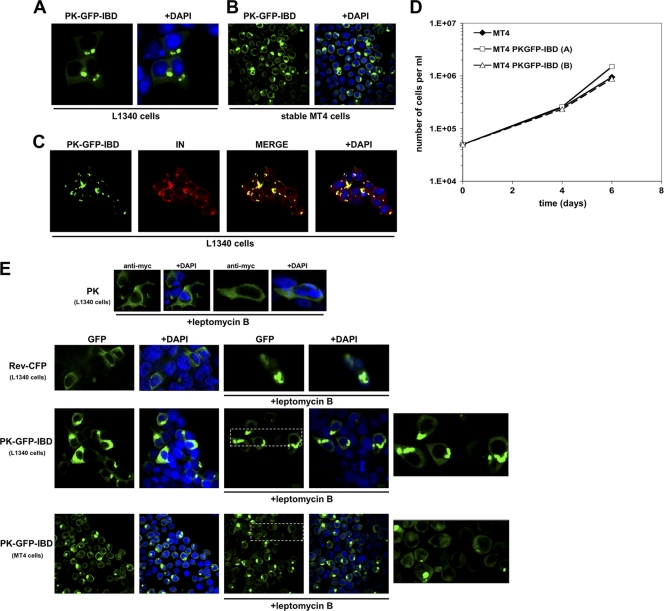
Additional approaches were undertaken to examine how intracellular location influences dominant interference. An initial one was to fuse the simian virus 40 (SV40) NLS or the HIV-1 Rev NES to the GFP-IBD and GFP-199-530 proteins. Confocal microscopy showed that the targeting domains functioned as predicted to restrict steady-state cellular localization to the nucleus or cytoplasm. However, when they were expressed in MT4 cells, reduced anti-HIV-1 effect was observed consistently with either protein (data not shown). We considered that our conferring of shuttle protein status to GFP-IBD might be responsible for this blunting of the antiviral effect. For example, binding of these proteins to Importin family members could interfere with concurrent binding to PIC-associated IN. A further limitation to drawing clear conclusions from this approach is that the continuous nucleocytoplasmic shuttling such proteins undergo may be expected to result in relative local abundance in the vicinity of both faces of the nuclear envelope. α-tubulin-GFP-IBD fusion proteins were constructed, with the rationale that lentiviruses have been reported to transit to the nucleus via molecular motors that traffic along microtubules (37). However, we found that tubulin fusions could either bind IN or assemble into microtubules, but not both (data not shown). Consistent with this, no tubulin fusion protein had antiviral properties (data not shown).
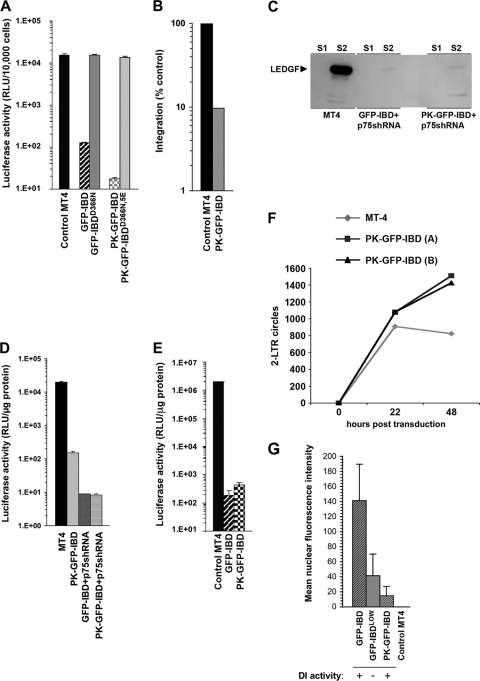
Since restricting the IBD to fixed cytoskeletal elements was not informative, we devised a cytoplasm-restricted GFP-IBD by fusing it to chicken pyruvate kinase (PK). This glycolytic pathway enzyme exceeds the size limit for diffusion through nucleopores and is exclusively cytoplasmic (48). A control version of the PK fusion, termed PK-GFP-IBDD366N,5E, was also constructed. PK-GFP-IBDD366N,5E has the previously described D366N mutation and five additional mutations predicted on the basis of recent structural evidence (23) to fully abrogate IBD interaction with IN (K401E, K402E, R405E, F406E, and V408E). K401, K402, and R405 amino acids engage in charged interactions with acidic residues in the IN N terminus (23), whereas F406 and V408 engage in hydrophobic interactions with IN W131 (9). Confocal microscopy showed that, as predicted, PK-GFP-IBD and PK-GFP-IBDD366N,5E are cytoplasmic proteins (Fig. 8A to C and data not shown). The fusion proteins also formed distinctive cytoplasmic inclusions, while PK did not. These cells were otherwise indistinguishable from the parental cells, with identical morphology, growth rates, and peak density (Fig. 8D). When HIV-1 IN was also expressed, it clearly colocalized with PK-GFP-IBD in similar structures, confirming that PK fusion did not disrupt the interaction of the IBD with IN (Fig. 8C). PK-GFP-IBDD366N,5E, in contrast, did not colocalize with IN (data not shown).
FIG. 8.
Characterization of a cytoplasmically restricted DI protein. (A) LEDGF-deficient 293T cells (L1340 cells [31]) were transfected with PK-GFP-IBD. Cells were fixed, and stained with DAPI at 48 h after transfection, and imaged with a confocal microscope. (B) MT4 cells stably expressing PK-GFP-IBD were centrifuged onto slides and imaged as described in Materials and Methods. (C) PK-GFP-IBD and Myc-epitope-tagged IN expression plasmids were transiently transfected into L1340 cells. Cells were fixed 48 h later and stained with anti-Myc. (D) Proliferative properties of MT4 cell lines derived by lentiviral vector transduction. (The “A” cell line is shown in the photomicrographs of this figure). (E) PK-GFP-IBD does not shuttle. PK-, Rev-CFP-, or PK-GFP-IBD-transfected L1340 cells or a stable CD4+ T cell line (MT4) expressing PK-GFP-IBD were each treated with leptomycin B to assess shuttling via the CRM1 pathway. The positive control in the experiment was Rev-CFP, which was imaged in the green channel to distinguish it from DAPI. In the presence of leptomycin B, Rev-CFP becomes completely nucleolar. In contrast, pyruvate kinase (PK; detected with an Alexa Green-conjugated antibody to the N-terminal Myc epitope tag) and PK-GFP-IBD remain completely cytoplasmic under the same conditions. The areas marked by dashed line boxes are enlarged at right.
HIV-1 challenges showed the PK-GFP-IBD protein had potent antiviral function. HIV-1luc infection was inhibited in the stable PK-GFP-IBD cell lines but not in the control PK-GFP-IBDD366N,5E (Fig. 9A). A concomitant decrease in integration was noted with the antiviral effect (Fig. 9B). When combined with LEDGF knockdown, the antiviral effect of PK-GFP-IBD was multiplicative to approximately the same extent as GFP-IBD, yielding between 3 and 4 logs of inhibition of single cycle reporter virus infection (Fig. 9C and D). Moreover, an alternative fusion protein having a fragment of PK (amino acids 21 to 418) also displayed leptomycin B-resistant nuclear exclusion, nontoxicity allowing long-term stable human CD4+ T cell line generation, and the same potent antiviral activity (data not shown).
FIG. 9.
Inhibition of HIV-1 infection by PK-GFP-IBD. (A) MT4 cell lines stably expressing GFP-IBD, GFP-IBDD366N, PK-GFP-IBD, and PK-GFP-IBDD366N,5E were challenged with HIV-1luc. (B) Alu-PCR based integration assay of HIV-1luc in MT4 PK-GFP-IBD cells. (C) Western blot of subcellular fractions of MT4 cells expressing either GFP-IBD+LEDGFshRNA or PK-GFP-IBD+LEDGFshRNA. LEDGF is markedly decreased in both cell lines. See the Fig. 5A legend for assay details. (D) MT4 cells stably expressing PK-GFP-IBD, a LEDGF-targeted shRNA and GFP-IBD, or a LEDGF-targeted shRNA and PK-GFP-IBD were challenged with HIV-1luc. (E) LAI luciferase challenge of indicated cell lines. Luciferase was analyzed 4 days after infection. (F) Time course of HIV-1luc 2-LTR circle formation in the indicated cell lines. (G) Mean nuclear fluorescence intensity versus DI activity. The MFI was obtained for 10 control MT4 cells, and 20 each of GFP-IBD, GFP-IBDLOW, and PK-GFP-IBD cells (see Fig. S1A to C in the supplemental material). The legend at the bottom indicates whether the cell line produces antiviral dominant interference versus MT4 control cells.
Further studies confirmed that PK-GFP-IBD acts in the cytoplasm. First, leptomycin B experiments established that the protein does not undergo nucleocytoplasmic shuttling (Fig. 8E). Therefore, steady-state absence of the protein from the nucleus (Fig. 8A to C) does not reflect continuous export but rather exclusion. We also quantified fluorescence intensity in individual nuclei and correlated this with antiviral effects (see Fig. S1A, B, and C in the supplemental material). For this purpose, an additional GFP-IBDLOW cell line was generated. In contrast to PK-GFP-IBD cells, GFP-IBDLOW cells have visibly green nuclei (see Fig. S1A). To analyze this disparity quantitatively, green fluorescence at the midpoint of the DAPI (4′,6′-diamidino-2-phenylindole) signal was determined with Zeiss LSM 510 software in 20 randomly selected cells in each cell line, all imaged under identical microscopic conditions (Fig. S1B and Fig. 9G). In agreement with the optical data, mean nuclear fluorescence intensity in GFP-IBDLOW cells exceeded fluorescence in PK-GFP-IBD cells. No antiviral effect was seen in the former cells while profound effects were seen in the latter cells (Fig. S1 and Fig. 9). The aggregate results are presented in Fig. 9G. These experiments showed unequivocally that nuclear levels of PK-GFP-IBD are well below levels that are antiviral. A threshold level of DI protein is required for the antiviral effect, and this is not reached in either GFP-IBDLOW cells or in the nuclei of the PK-GFP-IBD cells.
To determine whether entry via the normal receptors rather than via the VSV-G (endosomal) pathway influenced PK-GFP-IBD antiviral activity, we challenged these cells with an env-intact HIV-1 reporter virus. Marked inhibition was seen in both GFP-IBD and PK-GFP-IBD cells, confirming that virus entering by both routes is blocked (Fig. 9E).
Because the antiviral effect is triggered in the cytoplasm, we considered that PK-GFP-IBD might function by blocking PIC nuclear entry. However, determination of the time course of 2-LTR circle formation did not support this hypothesis (Fig. 9F). 2-LTR circles were increased at both 22 and 48 h with PK-GFP-IBD compared to parental MT4 cells. These results are intriguing since they suggest that PK-GFP-IBD does not derail RTC/PIC nuclear import. We conclude that the chimeric protein either transits into the nucleus with the PIC or, more likely, its binding causes loss of PIC integration capacity, perhaps by triggering IN multimerization prematurely. In summary, the block induced by this cytoplasm-confined protein becomes manifest not at the nuclear entry step, but after nuclear entry and at integration.
DISCUSSION
Lentiviruses are in general thought to exploit LEDGF as a chromatin docking receptor, but a complete picture of this dependency factor's role in the viral life cycle is lacking. Cofactor function requires intact NDE and IBD attachment functions (30, 46). DI protein experiments have also shown that uncoupling these functions can inhibit HIV-1 replication (10, 25, 30, 40) and that basic cofactor activity is sustained when the NDE is replaced with several unrelated chromatin-binding modules (16, 18, 40). Nevertheless, only some proteins that uncouple chromatin and integrase binding have DI function. For example, a LEDGF mutant lacking the key NDE elements (PWWP and AT hook domains) neither rescued nor intensified the impairment to lentiviral infection in LEDGF-deficient cells (30).
We determined how DI activities correlate with cellular context, protein compositional features, and intracellular location. Combining LEDGF depletion with GFP-IBD rendered highly permissive human CD4+ T cells virtually uninfectable, with single-round assays decreased by up to 4 logs, high-MOI replicating challenges unable to establish infection, and integration levels reduced to <1% of controls. We previously found that in the human CD4+ T cell line SupT1, stripping of detectable chromatin-bound (S2 fraction) LEDGF by intensified RNAi correlated with the antiviral effect (30). Here, in MT4 cells, a several log multiplicative effect was produced even when an S2 fraction residuum of endogenous LEDGF was detectable. The consistent multiplicative synergy with knockdown and the persistence of the effect with a cytoplasm-confined protein leads us to conclude that depletion of endogenous LEDGF and the DI proteins exert their effects at temporally different stages. Experimentally, the single-step knockdown/re-expression lentiviral vectors we are able to use allow de facto gene replacement with a single manipulation of cells. The extent of inhibition achievable is experimentally useful in providing a high dynamic range for testing variables and suggests scope for therapeutic applications.
Minimal proteins (GFP-IBD, IBD-GFP, or GFP-325-432) consistently displayed the strongest antiviral phenotypes, alone and in combination with endogenous LEDGF depletion. Additional domains of LEDGF did not augment dominant interfering capacity, and indeed, with the exception of the GFP-325-432 protein, they somewhat diminished it.
Adding the C-terminal 98 amino acids of LEDGF rendered more of the protein cytoplasmic, irrespective of size. However, complexes with independently expressed IN were always cytoplasmic for any of the DI proteins. With the proviso that overexpression of IN does not recapitulate the normal viral situation where the enzyme lies within an organized subviral complex, this observation may be significant virologically. Proteins that lack these terminal 98 amino acids formed elongated, often worm-shaped inclusions with IN, while those containing the terminal 98 amino acids tended to be more diffusely distributed (Fig. 2A and B). Proteins that formed the worm-shaped structures with IN did tend to have greater antiviral effects (GFP-IBD, GFP-325-432) perhaps correlating with higher IN affinity and/or a particular structural orientation. De Rijck et al. also compared GFP-325-530 and GFP-IBD proteins in MT4 cells and found they were roughly equivalent in single round viral inhibition assays, with a slight trend toward better activity of GFP-IBD in some but not all HIV-1 replication assays (10). Although we found GFP-325-530 to be active (Fig. 3), it was consistently less so than either GFP-325-432 or GFP-IBD (i.e., GFP-345-432), again reinforcing our overall conclusion that including the portion of LEDGF that lies C-terminal to the IBD subtracts from maximal DI activity.
That GFP-IBD antiviral function can be initiated in the cytoplasm is informative in two main ways. The results demonstrate first of all that prenuclear uncoating of the viral core proceeds to the point where functional IN molecules become exposed to the cytoplasmic milieu and are accessible to interaction with cellular proteins. We initially found that GFP-199-530, which in the absence of IN localized predominantly to the cytoplasm of transfected cells, was not effective, suggesting that cytoplasmic DI proteins may in fact not access the viral core-associated IN. In contrast, targeting GFP-IBD to the nuclear envelope with the nuclear envelope targeting cassette of Nup153 produced dominant interference, suggesting that IN is accessible in the vicinity of the nucleopore. To further examine this issue, we excluded GFP-IBD from the nucleus by fusing it to pyruvate kinase, a cytoplasmic protein. The resulting fusion protein is cytoplasmic and was also documented to not shuttle and yet it retained the antiviral activity of GFP-IBD.
These clear functional data complement prior biochemical experiments. LEDGF as well as some cellular proteins that are thought to associate with the viral cDNA have been identified as PIC components (14, 27, 31). Nevertheless, protein detection experiments of this nature have inherent limitations, among them being that PICs cannot be purified to homogeneity with current methods and that many if not most such complexes are likely to be defective. In addition, HIV-1 CA appears to be involved in nuclear import (57). Most CA is shed by the time of nuclear entry (41), but the time course of this process, and the access it provides to key viral components has been unclear. A recent study suggested, for example, that APOBEC3 proteins that can interdict and edit exogenous nonviral (plasmid) DNA in the cytoplasm cannot similarly access the HIV-1 cDNA (49).
The second point is that the results shed light on the DI mechanism. Interestingly, 2-LTR circle measurements indicated that viral nuclear import was not impaired, which is contrary to a simple scenario in which engagement of the uncoating particle by PK-GFP-IBD blocks nuclear import. In addition, directing GFP-IBD to a steady-state nuclear location with the SV40 NLS even reduced its activity. It cannot be excluded that PK-GFP-IBD is imported with the PIC into the nucleus, where it simply interferes with binding of endogenous LEDGF and hence with PIC attachment to the chromatin fiber. Alternatively and more likely in our view, we propose that dominant interference produced by PK-GFP-IBD and other DI proteins involves a triggering effect in which premature or improper IN multimerization, some other dysfunctional intasome state, or even dysregulated uncoating ensues from prenuclear engagement of IN by an IBD. Previous studies have suggested three general mechanistic scenarios for the role of LEDGF in HIV-1 integration: tethering of the PIC to the chromosomal DNA target (LEDGF as a chromosome receptor for the intranuclear PIC), allosteric regulation of catalysis, and enhancement of virologically relevant IN multimerization (5, 30, 38, 40, 44, 45). In regard to the latter possibility, much evidence suggests properly concerted integration requires orderly assembly of enzyme, cofactor, and DNA substrates. The functional form of IN is likely a tetramer (1, 15, 19, 21, 22, 26, 55). In vitro, a dimer enables 3′ end processing but a tetramer appears needed for DNA strand transfer activity (15, 19, 26). The IBD has been demonstrated in vitro to stabilize IN subunit-subunit interactions and promote tetramer formation, which may impact concerted insertion of both LTR ends at the retroviral integration site (21, 24, 38). The in vitro order of addition experiments with purified proteins and model DNA substrates suggest that LEDGF-IN interaction before proper assembly of the IN-cDNA synaptic complex can impair concerted integration (42, 45). Furthermore, PICs isolated from the cytoplasmic fraction of LEDGF-deficient mouse cells had undiminished competence for integrating into a DNA target (46). Analyses of PICs purified from cells with differentially localized DI proteins may help establish whether these proteins trigger a dysfunctional intasome.
Supplementary Material
Acknowledgments
The study was funded by NIH AI77344 to E.M.P.
We thank K. Ullman (University of Utah) for providing a Nup153 expression plasmid and Robert Sikkink for technical assistance.
Footnotes
Published ahead of print on 26 January 2011.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bao, K. K., et al. 2003. Functional oligomeric state of avian sarcoma virus integrase. J. Biol. Chem. 278:1323-1327. [DOI] [PubMed] [Google Scholar]
- 2.Bartholomeeusen, K., et al. 2007. Differential interaction of HIV-1 integrase and JPO2 with the C terminus of LEDGF/p75. J. Mol. Biol. 372:407-421. [DOI] [PubMed] [Google Scholar]
- 3.Bartholomeeusen, K., et al. 2009. Lens epithelium-derived growth factor/p75 interacts with the transposase-derived DDE domain of pogZ. J. Biol. Chem. 284:11467-11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Busschots, K., et al. 2005. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 280:17841-17847. [DOI] [PubMed] [Google Scholar]
- 5.Cherepanov, P. 2007. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 35:113-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherepanov, P., A. L. Ambrosio, S. Rahman, T. Ellenberger, and A. Engelman. 2005. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl. Acad. Sci. U. S. A. 102:17308-17313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherepanov, P., E. Devroe, P. A. Silver, and A. Engelman. 2004. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional coactivator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 279:48883-48892. [DOI] [PubMed] [Google Scholar]
- 8.Cherepanov, P., et al. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278:372-381. [DOI] [PubMed] [Google Scholar]
- 9.Cherepanov, P., et al. 2005. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat. Struct. Mol. Biol. 12:526-532. [DOI] [PubMed] [Google Scholar]
- 10.De Rijck, J., et al. 2006. Overexpression of the lens epithelium-derived growth factor/p75 integrase binding domain inhibits human immunodeficiency virus replication. J. Virol. 80:11498-11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dietz, A. B., et al. 2006. A novel source of viable peripheral blood mononuclear cells from leukoreduction system chambers. Transfusion 46:2083-2089. [DOI] [PubMed] [Google Scholar]
- 12.Enarson, P., M. Enarson, R. Bastos, and B. Burke. 1998. Amino-terminal sequences that direct nucleoporin nup153 to the inner surface of the nuclear envelope. Chromosoma 107:228-236. [DOI] [PubMed] [Google Scholar]
- 13.Engelman, A., and P. Cherepanov. 2008. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 4:e1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farnet, C. M., and F. D. Bushman. 1997. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell 88:483-492. [DOI] [PubMed] [Google Scholar]
- 15.Faure, A., et al. 2005. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 33:977-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferris, A. L., et al. 2010. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc. Natl. Acad. Sci. U. S. A. 107:3135-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge, H., Y. Si, and R. G. Roeder. 1998. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 17:6723-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gijsbers, R., et al. 2010. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol. Ther. 18:552-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guiot, E., et al. 2006. Relationship between the oligomeric status of HIV-1 integrase on DNA and enzymatic activity. J. Biol. Chem. 281:22707-22719. [DOI] [PubMed] [Google Scholar]
- 20.Hare, S., and P. Cherepanov. 2009. Review: the interaction between lentiviral integrase and LEDGF: structural and functional insights. Viruses 1:780-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hare, S., et al. 2009. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog. 5:e1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hare, S., S. S. Gupta, E. Valkov, A. Engelman, and P. Cherepanov. 2010. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464:232-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hare, S., et al. 2009. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 5:e1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayouka, Z., et al. 2007. Inhibiting HIV-1 integrase by shifting its oligomerization equilibrium. Proc. Natl. Acad. Sci. U. S. A. 104:8316-8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hombrouck, A., et al. 2007. Virus evolution reveals an exclusive role for LEDGF/p75 in chromosomal tethering of HIV. PLoS Pathog. 3:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, M., M. Mizuuchi, T. R. Burke, Jr., and R. Craigie. 2006. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 25:1295-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, C. W., and A. Engelman. 2003. The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J. Virol. 77:5030-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Llano, M., S. Delgado, M. Vanegas, and E. M. Poeschla. 2004. LEDGF/p75 prevents proteasomal degradation of HIV-1 integrase. J. Biol. Chem. 279:55570-55577. [DOI] [PubMed] [Google Scholar]
- 29.Llano, M., J. Morrison, and E. Poeschla. 2010. Virological and cellular roles of the transcriptional coactivator LEDGF/p75. Curr. Top. Microbiol. Immunol. 339:125-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Llano, M., et al. 2006. An essential role for LEDGF/p75 in HIV integration. Science 314:461-464. [DOI] [PubMed] [Google Scholar]
- 31.Llano, M., et al. 2004. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral pre-integration complexes. J. Virol. 78:9524-9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Llano, M., et al. 2006. Identification and characterization of the chromatin binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 360:760-773. [DOI] [PubMed] [Google Scholar]
- 33.Loewen, N., et al. 2003. FIV vectors. Methods Mol. Biol. 229:251-271. [DOI] [PubMed] [Google Scholar]
- 34.Maertens, G., et al. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278:33528-33539. [DOI] [PubMed] [Google Scholar]
- 35.Maertens, G. N., P. Cherepanov, and A. Engelman. 2006. Transcriptional coactivator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. J. Cell Sci. 119:2563-2571. [DOI] [PubMed] [Google Scholar]
- 36.Marshall, H. M., et al. 2007. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One 2:e1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonald, D., et al. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKee, C. J., et al. 2008. Dynamic modulation of HIV-1 integrase structure and function by cellular lens epithelium-derived growth factor (LEDGF) protein. J. Biol. Chem. 283:31802-31812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meehan, A. M., and E. M. Poeschla. 2010. Chromatin tethering and retroviral integration: recent discoveries and parallels with DNA viruses. Biochim. Biophys. Acta 1799:182-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meehan, A. M., et al. 2009. LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors. PLoS Pathog. 5:e1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pandey, K. K., S. Sinha, and D. P. Grandgenett. 2007. Transcriptional coactivator LEDGF/p75 modulates human immunodeficiency virus type 1 integrase-mediated concerted integration. J. Virol. 81:3969-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poeschla, E., F. Wong-Staal, and D. Looney. 1998. Efficient transduction of nondividing cells by feline immunodeficiency virus lentiviral vectors. Nat. Med. 4:354-357. [DOI] [PubMed] [Google Scholar]
- 44.Poeschla, E. M. 2008. Integrase, LEDGF/p75 and HIV replication. Cell. Mol. Life Sci. 65:1403-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raghavendra, N. K., and A. Engelman. 2007. LEDGF/p75 interferes with the formation of synaptic nucleoprotein complexes that catalyze full-site HIV-1 DNA integration in vitro: implications for the mechanism of viral cDNA integration. Virology 360:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shun, M. C., et al. 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 21:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silvers, R. M., et al. 2009. Modification of integration site preferences of an HIV-1-based vector by expression of a novel synthetic protein. Hum. Gene Ther. 21:337-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siomi, H., and G. Dreyfuss. 1995. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stenglein, M. D., M. B. Burns, M. Li, J. Lengyel, and R. S. Harris. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat. Struct. Mol. Biol. 17:222-229. [DOI] [PMC free article] [PubMed]
- 50.Turlure, F., G. Maertens, S. Rahman, P. Cherepanov, and A. Engelman. 2006. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 34:1663-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Maele, B., K. Busschots, L. Vandekerckhove, F. Christ, and Z. Debyser. 2006. Cellular cofactors of HIV-1 integration. Trends Biochem. Sci. 31:98-105. [DOI] [PubMed] [Google Scholar]
- 52.Vandekerckhove, L., et al. 2006. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 80:1886-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanegas, M., et al. 2005. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 118:1733-1743. [DOI] [PubMed] [Google Scholar]
- 54.Varadarajan, P., et al. 2005. The functionally conserved nucleoporins Nup124p from fission yeast and the human Nup153 mediate nuclear import and activity of the Tf1 retrotransposon and HIV-1 Vpr. Mol. Biol. Cell 16:1823-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, J. Y., H. Ling, W. Yang, and R. Craigie. 2001. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 20:7333-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woodward, C. L., S. Prakobwanakit, S. Mosessian, and S. A. Chow. 2009. Integrase interacts with nucleoporin NUP153 to mediate the nuclear import of human immunodeficiency virus type 1. J. Virol. 83:6522-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamashita, M., and M. Emerman. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 78:5670-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yokoyama, A., and M. L. Cleary. 2008. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell 14:36-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.