Abstract
Murine polyomavirus middle T-antigen (MT) induces tumors by mimicking an activated growth factor receptor. An essential component of this action is a 22-amino-acid hydrophobic region close to the C terminus which locates MT to cell membranes. Here, we demonstrate that this sequence is a transmembrane domain (TMD) by showing that a hemagglutinin (HA) tag added to the MT C terminus is exposed on the outside of the cells, with the N terminus inside. To determine whether this MT TMD is inserted into the endoplasmic reticulum (ER) membrane, we added the ER retention signal KDEL to the MT C terminus (MTKDEL). This mutant protein locates only in the ER, demonstrating that MT does insert into membranes solely at this location. In addition, this ER-located MT failed to transform. Examination of the binding proteins associated with the MTKDEL protein demonstrated that it associates with PP2A and c-Src but fails to interact with ShcA, phosphatidylinositol 3-kinase (PI3K), and phospholipase C-γ1 (PLC-γ1), despite being tyrosine phosphorylated. Additional mutant and antibody studies show that MT binding to PP2A is probably required for MT to efficiently exit the ER and migrate to the plasma membrane though the TMD also plays a role in this relocation. Overall, these data, together with previous publications, illustrate that MT associates with signaling proteins at different sites in its maturation pathway. MT binds to PP2A in the cytoplasm, to c-Src at the endoplasmic reticulum, and to ShcA, PI3K, and PLC-γ1 at subsequent locations en route to the plasma membrane.
The polyomaviruses feature a small, double-stranded DNA (dsDNA) genome coated with cell histones and surrounded by three virally encoded virion proteins (29). The simple genome consists of approximately 5 kb of closed circular dsDNA that is transcribed in two units, either early or late after infection. After RNA splicing to produce mRNA, the early region encodes the T-antigens that alter the infected cell in order to provide a suitable environment for replication of the viral genome. The large T-antigen (LT) also promotes initiation of viral DNA synthesis and a concomitant switch to transcription of the viral late region, which encodes the viral capsid proteins. As the T-antigens exert a mitogenic effect, their expression in the absence of the rest of the lytic cycle has the potential to convert a normal cell into a tumorigenic one; hence, many of the polyomaviruses are considered tumor viruses. The ability of these viruses to readily induce tumors in animals and to transform cells in culture and their simplicity have meant they have been used extensively as model systems to investigate the molecular mechanisms underlying tumor formation (1, 35). Consequently, they have been instrumental in unraveling many of the important factors involved in carcinogenesis. This includes the discovery of tyrosine kinases (15) and phosphatidylinositol 3 kinases (PI3Ks) (47) from work on the middle T-antigen (MT) encoded by the murine polyomavirus (10).
In addition to the two main T-antigens, LT and small T (ST), the rodent polyomaviruses uniquely express a third T-antigen, MT. MT is potently mitogenic and has the ability to convert normal fibroblasts in culture into a transformed tumorigenic state (43). Murine polyomavirus MT is a 421-residue polypeptide with an apparent molecular mass of 55 kDa (19). It is synthesized early in the viral lytic cycle and is produced by alternative splicing from the early synthesized RNA. It has the same N-terminal amino acid sequence as LT and ST, followed by a region in common only with ST, and then a unique C terminus. Like all T-antigens, MT exerts its effects on cells not by possessing an enzymatic property of its own but by interacting with cellular proteins and changing the way they are regulated. In an ordered sequence of interactions, MT binds to the core dimer of PP2A (MT-PP2A) (32, 45) and then to a member of the src family of tyrosine kinases, usually pp60c-src (8) or pp62c-yes (21). This activates the kinase activity of Src, which phosphorylates a number of tyrosines within MT. Three of these phosphotyrosines act as binding sites for the SH2 or PTB domains of PI3K (MT Y315) (41), ShcA (Y250) (5, 11) and phospholipase C-γ1 ([PLC-γ1] Y322) (40). As a consequence of their interaction with MT, each of these polypeptides is, in turn, tyrosine phosphorylated, which activates PI3K- and PLC-γ1-dependent signaling pathways and creates a binding site on ShcA for Grb2 (11). The guanine nucleotide exchange factor Sos1 and the adapter molecule Gab1 (30) are brought into the MT complex through their interactions with Grb2, thereby activating Ras and the extracellular signal-regulated kinase (ERK) kinase cascade (23, 33). These interactions and phosphorylation events closely resemble the reactions occurring during the activation of tyrosine kinase-associated growth factor receptors. Consequently, MT is now considered a permanently active analogue of such a receptor (10, 17) and therefore is a useful model that is providing important insights into the function of both normal and oncogenic receptors.
To transform cells, MT has to associate with cell membranes. This requires a 22-residue stretch of hydrophobic amino acids located near the MT C terminus (37). Deletion of this region abolishes transforming activity and prevents binding to all the signaling molecules except PP2A (6, 31). Despite a number of attempts, MT has been undetectable on the outside of the cells, so it was proposed that it is probably situated on the cytoplasmic face of the membrane (34). Previously, we have shown that MT is located on most cell membranes (13), so the hydrophobic region presumably does not target one specific membrane site. However, this region must do more than specify general membrane association as hydrophobic domain mutants have been isolated that still bind to membranes but fail to transform (25). In addition, mutants where the hydrophobic sequence of MT was replaced with the transmembrane domain (TMD) from vesicular stomatitis virus glycoprotein G (42) or cytochrome b5 (20) still targeted MT to membranes but, again, were transformation negative. However, the lipid-modified CAAX sequences at the C terminus of H-Ras can functionally replace the C terminus of MT (16), suggesting a common specificity to MT and H-Ras membrane binding. Consequently, it is likely that the hydrophobic domain in MT does more than target generalized membrane location, but what this extra activity consists of is unknown.
To examine how MT is inserted into membranes, we have isolated and studied a series of proteins with mutations in the C-terminal region of MT. We report here that an epitope tag inserted at the C terminus of MT is exposed on the exterior of transformed cells, demonstrating that MT is a tail-anchored (TA), integral membrane protein. Inserting the endoplasmic reticulum (ER) retention sequence KDEL at the C-terminal end of MT (yielding MTKDEL) located it just to the ER and cis-Golgi, showing that MT is inserted into membranes at the ER. MTKDEL failed to bind ShcA, PI3K, and PLC-γ1 and did not transform, demonstrating that MT has to migrate out of the ER to generate a mitogenic signal. Exit of MT from the ER and migration to the cell periphery were shown to be dependent upon both the TMD and PP2A binding by replacement of the MT TMD.
MATERIALS AND METHODS
Cells and plasmids.
All cell work was undertaken in a recloned Rat2 rat fibroblast cell line grown in Dulbecco's Modified Eagles Medium High Glucose (Invitrogen) supplemented with glutamine and 10% fetal bovine serum and incubated at 37°C and 10% CO2. The MT plasmid (pUCMT) used for all the work consisted of a BamHI-EcoRI fragment of polyomavirus DNA lacking the MT intron cloned into pUC19. Escherichia coli DH5α bacteria carrying the appropriate plasmid were grown in LB broth containing ampicillin, and then the plasmid DNA was purified using a Qiagen Hi Speed Midi Plasmid purification kit according to the manufacturer's instructions.
Mutation generation.
MT tagged with the HA epitope (MTHA+) was isolated by a PCR from the MT plasmid. A forward primer, CCGCACATACTGCTGGAAGAAGACG, which is an exact match to the polyomavirus sequence, was used with a reverse mutation primer that contained an EcoRI site, the sequence encoding the HA tag (HA+), and finally, a priming sequence that matched the appropriate polyomavirus sequence. The primers used were wtMTHA+reverse, Δ419-421MTHA+reverse, and Δ416-421MTHA+reverse (all oligonucleotide sequences are shown in Table 1). PCR was performed using Phusion polymerase (New England BioLabs) under the manufacturer's conditions. PCR conditions were 94°C for 180 s, and then 25 cycles of 94°C for 45 s, 57°C for 60 s, and 72°C for 120 s. The reaction was finished off by incubation at 72°C for 600 s. Following PCR, the DNA product was digested with restriction enzymes BlpI and EcoRI (NEB) and cloned into pUCMT cleaved with the same enzymes. MTKDEL and MT with the addition of the control sequence GGDV that lacks amino acids recognized by the KDEL receptor (MTGGDV) were made in the same fashion using the reverse oligonucleotides MTKDELrev and MTGGDVrev (Table 1).
TABLE 1.
Oligonucleotide sequences
| Oligonucleotide | Sequence |
|---|---|
| wtMTHA+reverse | CGCGAATTCCTAAAGAGAAGCGTAATCTGGAACATCGTATGGGTAAGCGAAATGCCGGGAACGTTTTATTAG |
| Δ419-421MTHA+reverse | GCGAATTCCTAAAGAGAAGCGTAATCTGGAACATCGTATGGGTAAGCGGAACGTTTTATTAGAATAAATAGCATGAGAC |
| Δ416-421MTHA+reverse | GCGAATTCCTAAAGAGAAGCGTAATCTGGAACATCGTATGGGTAAGCTATTAGAATAAATAGCATGAGACAAATACCCAG |
| MTKDELrev | CGCGAATTCTTACAACTCGTCCTTGAAATGCCGGGAACGTTTTATTAG |
| MTGGDVrev | CGCGAATTCTTAAACGTCTCCTCCGAAATGCCGGGAACGTTTTATTAG |
| DelMT-F | GCAACGCCACCTAAGAAGGCTCGGCCGATATCAGATCTCGGCATTTCTAGTATACTCCACC |
| DelMT-R | GGTGGAGTATACTAGAAATGCCGAGATCTGATATCGGCCGAGCCTTCTTAGGTGGCGTTGC |
| −40F | GTTTTCCCAGTCACGACGTTGTA |
| mtN1-F | TGTATCCAGAAAGCGACCAAGAC |
| Sec61β-F | CGCCGGCCGGGTGGGCCCTGTCCCAGTGCTGGTGATGAGTCTTCTGTTCATCGCTGCTGTATTTATGCTGCACATTTGGGGCAAAAGATCTCGC |
| Sec61β-R | GCGAGATCTTTTGCCCCAAATGTGCAGCATAAATACAGCAGCGATGAACAGAAGACTCATCACCAGCACTGGGACAGGGCCCACCCGGCCGGCG |
To construct a mutant with the hydrophobic domain of MT replaced with other sequences, we first isolated a deletion mutant that removed the hydrophobic domain and mutated the surrounding MT sequence to generate unique EagI and BglII restriction enzyme sites. This was achieved by performing two PCRs using the oligonucleotide pair DelMT-F and −40F and the pair mtN1-F and DelMT-R. The DelMT oligonucleotides are complementary matches and hybridize to the MT sequence either side of the hydrophobic domain-encoding region, and they contain the deletion and mutations required. Primers −40F and mtN1-F are exact matches to the polyomavirus sequence on either side of these oligonucleotides. The PCR products were purified using a Qiagen Gel Extraction kit, mixed, and combined with the outside primers mtN1-F and −40F and subjected to a further PCR. This created a fragment that contained the appropriate DelMT sequence and extended into the MT sequence. This fragment was cleaved with restriction enzymes NcoI and EcoRI and joined in a three-way ligation with an ApaI to NcoI MT fragment of MT cDNA and a pUCMT cleaved with ApaI and EcoRI fragments to create mutant MTdel. MT replacement mutants were then created by making complementary oligonucleotides that contained the required sequences plus appropriate overhanging EagI and BglII sites. These were mixed, treated with polynucleotide kinase and ATP, heated to 90°C for 10 min, and then cooled slowly to allow hybridization. The resulting double-stranded fragment was then ligated into EagI-BglII-cleaved MTdel. The oligonucleotides used were Sec61β-F and Sec61β-R to create MTSec61β. Sequences are shown in Table 1.
All mutants were verified as correct by DNA sequencing before use.
Transformation assay and cell line isolation.
Subconfluent Rat2 cells were transfected with plasmids containing MT and mutant DNA using a calcium phosphate technique. For focus assays, the cells were then fed every 3 days for 14 days, and the foci were stained by incubation with Leishmann's staining solution (BDH Chemicals) for 5 min at room temperature, followed by washing with water. For cell line isolation, cells were transfected with MT plasmid DNA plus 1/10 quantity of plasmid pSV2neo, which contains the neomycin resistance gene under the control of the simian virus 40 (SV40) early promoter. Following transfection, the cells were selected by addition of 750 μg/ml G418 (Calbiochem) for 14 days. Individual colonies were picked and subcloned a number of times.
Immunofluorescence assays.
Cells were seeded onto 13-mm glass coverslips in a 24-well plate and allowed to grow for 24 h. For fixed-cell staining, the medium was then removed, and the cells were washed twice with complete Dulbecco's phosphate-buffered saline (DPBS) at 37°C and then fixed by incubation with 4% formaldehyde solution (Agar Laboratories) in DPBS for 10 min at room temperature. The cells were then washed three times with DPBS and permeabilized by incubation with 1% NP-40 for 5 min at room temperature. The cells were washed with DPBS a further three times and incubated for 30 min with Signal IT Image Enhancer (Invitrogen). After a further three washes, the cells were incubated with a 1:200 dilution of primary monoclonal antibody for 60 min at room temperature. After the cells were washed three times with DPBS, the antibody was detected by incubation with a 1:1,000 dilution of Cy3-labeled anti-mouse antibody (Jackson Immunoresearch). The cells were washed a further three time in DPBS and then mounted in ProLong Gold mounting medium (Invitrogen). Immunofluorescent images were viewed on a Nikon inverted fluorescence microscope and captured using a Princeton Instruments MicroMax charge-coupled device (CCD) camera using Metamorph software. Live-cell labeling was achieved by washing the cells three times with DPBS at 37°C, followed by incubation for 20 min at 37°C with a 1:200 dilution of antibody. The cells were then washed three times with DPBS and fixed as above with formaldehyde solution. After treatment with Image Enhancer, the antibodies were detected with Cy3-labeled anti-mouse antibody as above. Antibodies used were PAb762, a mouse monoclonal directed against the N-terminal 72 amino acids of MT (11), PAb702, a rat monoclonal antibody directed against an N-terminal region of MT required for PP2A binding (12), PAb754, a mouse monoclonal antibody directed against amino acids 282 to 304 of MT (14), and antibody HA11, an anti-HA epitope monoclonal antibody from Covance.
Coimmunoprecipitation and in vitro protein kinase assays.
Confluent cells grown in a 100-mm petri dish were washed twice with cold phosphate-buffered saline (PBS) and then lysed by addition of 1 ml of ice-cold lysis buffer (100 mM Tris-Cl, pH 8.3, 100 mM NaCl, 0.5% Nonidet P-40 [Roche] plus complete protease inhibitor cocktail-EDTA [Roche]) and incubation on ice for 20 min. The supernatant was removed and centrifuged at 15,000 × g before removal of the final supernatant and storage at −80°C. For immunoprecipitation, a volume of lysate corrected for equal amounts of MT was thawed and incubated for 60 min on ice with 4 μg of PAb762-purified antibody. Thirty microliters of a 1:1 suspension of protein A-Sepharose 4B (GE Healthcare) in TBS-N (150 mM NaCl, 50 mM Tris-Cl, pH 8.3, 0.05% Nonidet P-40) buffer was added and incubated at 4°C for 30 min with constant mixing. The Sepharose was collected by very brief centrifugation and then washed three times by resuspension in TBS-N buffer. Following removal of the last wash supernatant, proteins were eluted from the protein A-Sepharose by incubation with 30 μl of Laemmli SDS sample buffer. After removal from the Sepharose, the samples were heated to 90°C for 5 min and then separated by electrophoresis on a 10% SDS-containing polyacrylamide gel run as described by Laemmli (22). After separation, the proteins were transferred onto nitrocellulose membranes (0.2-μm pore size; GE Healthcare) by Western blotting, and the filters were stored in PBS at 4°C.
Blots were blocked by incubation with 5% powdered skim milk in PBS for 60 min at room temperature. After the blots were washed with TBS-T (150 mM NaCl, 50 mM Tris-Cl, pH 7.5, 0.1% Tween 20), they were incubated for 60 min at room temperature with the appropriate dilution of primary antibody in 5% milk in TBS-T. After the blots were washed again for 30 min with frequent changes, antibody binding was detected by incubation with a 1:5,000 dilution of either anti-mouse or anti-rabbit IgG coupled to horseradish peroxidase (HRP; Dako) for 60 min at room temperature. After a further 30 min of washes, the proteins were revealed by incubation with Immobilon Western Chemiluminescent substrate (Millipore) and exposure to X-ray film. Antibodies used included the following: biotinylated anti-MT monoclonal PAb762 detected with HRP-streptavidin; F2.5G4, an anti-PP2A PR35 antibody, and G3 8F11, an anti-PP2A PR65 monoclonal antibody (our own unpublished clones); biotinylated clone 327 anti-pp60c-src; polyclonal anti-ShcA and -PI3K 85-kDa; and monoclonal anti-PLC-γ1 (BD Biosciences).
In vitro protein kinase assays were performed by immunoprecipitating MT as above until the final collection of the protein A-Sepharose. This was incubated with 50 μl of 25 mM Tris-Cl, pH 7.4, 5 mM manganese acetate, and 2μ Ci of [γ-32P]ATP for 30 min at 30°C. The Sepharose was then washed twice with TBS-N, eluted, and subjected to SDS-PAGE as above. After separation, the gel was fixed by incubation for 60 min with 40% methanol-10% acetic acid, dried, and autoradiographed.
RESULTS
MT is a transmembrane tail-anchored protein.
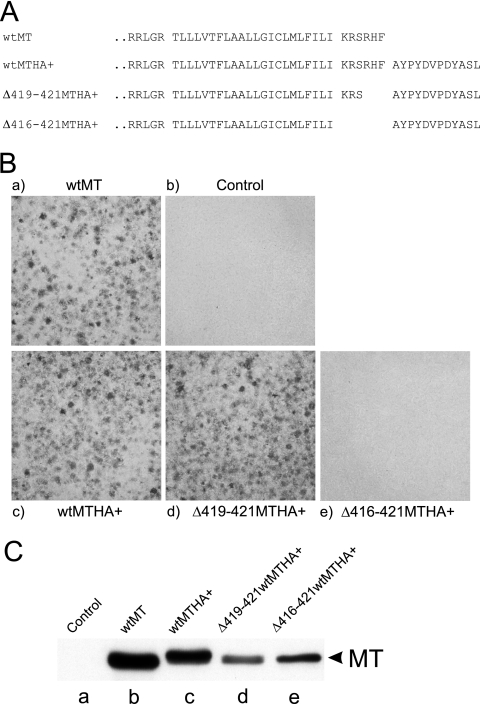
The location of the hydrophobic domain only six amino acids from the MT C terminus suggests strongly that MT belongs to the class of tail-anchored (TA) proteins inserted posttranslationally into membranes (3). However, a principle characteristic of TA proteins is that the hydrophobic sequence spans the membrane, and this has not been proven for MT. To address this, we inserted a slightly extended HA tag, AYPYDVPDYASL, at the C terminus of MT, with and without removal of all or part of the six amino acids C-terminal to the TMD (Fig. 1A), and then assayed for the ability to form foci when cells were transfected into rat fibroblasts. The HA tag had no inhibitory effect on transformation efficiency compared to wild-type MT (Fig. 1B). Removal of the C-terminal three amino acids of MT also had no effect on the ability of the HA-tagged MT to transform. However, deletion of all six amino acids C-terminal to the MT hydrophobic domain abolished transformation completely.
FIG. 1.
An epitope tag can be added to the C terminus of MT without disrupting transformation. (A) Amino acid sequences of the MT C terminus and three mutants tagged with the HA epitope. The amino acid sequence of wild-type MT is shown at the top, together with the sequence of three mutants where the HA epitope (shown on the right) is attached to MT. The designation of each MT species is listed on the left. The hydrophobic region of MT is identified with a space on either side, and the HA tag is also marked by a space between it and MT. (B) Transformation assays using the mutants shown in panel A. Plasmid DNA encoding each of the mutants shown in panel A was transfected into Rat2 fibroblasts and stained for focus formation after 14 days. The MT species used is indicated above or below each image. (C) MT polypeptides expressed by the mutants shown in panel A. Cell lines expressing each of the mutant MTs shown in panel A were isolated, and lysates were prepared. An equal volume of cell lysate from each line was separated by SDS-gel electrophoresis, Western blotted, and then probed with anti-MT monoclonal antibody PAb762. The migration position of MT is shown on the right, and the mutant used is indicated above each lane.
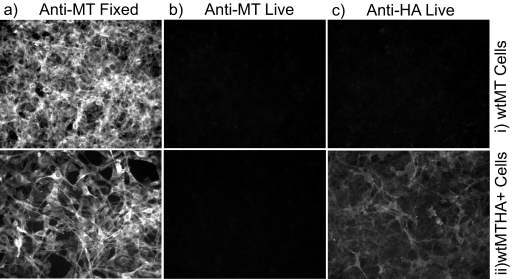
Cell lines expressing wild-type MT and MT plus the HA tag (wtMTHA+) were established and then examined by immunofluorescence, with the primary antibody added before or after fixation. As shown in Fig. 2, an antibody directed against the N terminus of MT (PAb762) reacted strongly with both lines when it was added after the cells were fixed with formaldehyde and then permeabilized. This antibody did not react with either cell line when it was added to live cells before fixation, further confirming that the N terminus of MT is not exposed on the outside of the plasma membrane. However, an anti-HA antibody added to living cells did bind to the surface of the wtMTHA+ line though not to wild-type MT-expressing cells (Fig. 2, wtMTHA+). Together, these data show clearly that some, if not all, of the C-terminal tail of MT is exposed on the outer surface of the cell, whereas the N terminus is not, confirming that MT is an integral transmembrane TA protein and that the hydrophobic region must act as a TMD.
FIG. 2.
Surface expression of the HA tag. A cell line expressing wtMTHA+ was established, and immunofluorescence was performed with the primary antibody added either after formaldehyde fixation (Fixed) or before (Live), as described in Materials and Methods. Cells were then photographed at low power on an epifluorescence microscope. The antibody reactivity used and the time of antibody addition are shown above each column. The cell line used, normal wt MT or wt MT plus the HA tag (wtMTHA+), is shown to the right of each row.
Endoplasmic reticulum-located MT fails to transform.
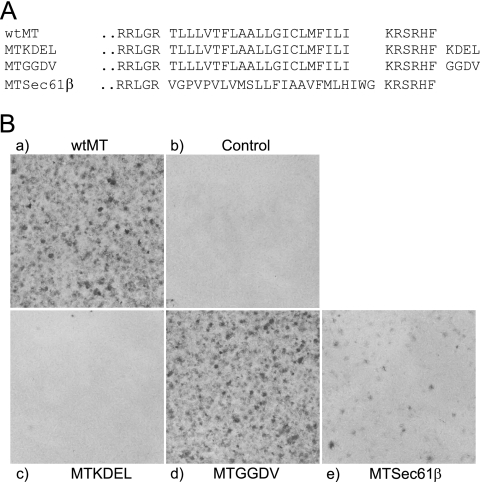
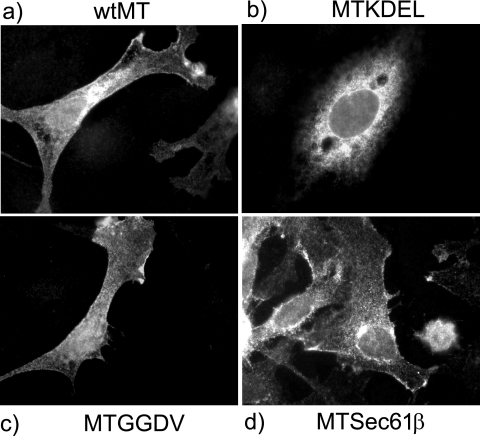
The finding above that small sequence segments can be added to the C terminus of MT without affecting its ability to induce cell transformation suggested that it may be possible to use this approach to examine the role of subcellular location in MT function. MT is found in most internal cell membranes (13), but where the transforming signal is generated is unknown. To determine whether MT located solely in the ER could produce a transforming signal, we added the ER retention signal KDEL (MTKDEL) and the control sequence GGDV (MTGGDV), which lacks the significant amino acids recognized by the KDEL receptor, to the C terminus of MT (Fig. 3A) and assayed for transforming ability. These are the same additions used to demonstrate that bovine papillomavirus E5 must locate to the Golgi apparatus to transform (38). MTKDEL failed to form foci in fibroblasts, but MTGGDV transformed at the same level as wt MT (Fig. 3B). Immunofluorescence analysis of the MT in transiently expressing cells demonstrated that the MTKDEL protein was located in the cytoplasm in structures that resemble those detected by an antibody to calreticulin, an ER marker (Fig. 4 and data not shown). Retention of the MTKDEL polypeptide in the ER indicates that the KDEL sequence must be exposed to the lumen and, hence, to the KDEL receptor, thereby showing that the C terminus of MT must be translocated across the membrane at the ER as well as in the plasma membrane. ER retention also suggests strongly that the MTKDEL protein is initially inserted into membranes at the ER as integration at any other membrane organelle would bypass the KDEL receptor and thus generate a different distribution.
FIG. 3.
Addition of a KDEL sequence to MT keeps it localized it in the endoplasmic reticulum and abolishes transformation. (A) The amino acid sequence of the C terminus of MT is shown together with that of three mutants containing either KDEL (MTKDEL), a nonfunctional mutated version of KDEL sequence (MTGGDV), or an MT mutant where the hydrophobic domain has been replaced with the ER-targeting transmembrane sequence from the Sec61β polypeptide (MTSec61β). (B) MTKDEL fails to form foci. Plasmids containing each of the mutants shown in panel A were transfected into Rat2 fibroblasts and assayed for focus formation. The MT mutant used is shown either above or below each panel.
FIG. 4.
MTKDEL is retained in the ER, whereas MTGGDV and MTSec61β are not. Immunofluorescence analysis was performed on cells transiently expressing the mutants shown in Fig. 3. Cells were transfected with each of the mutants, and, after 48 h, fixed, and stained with PAb762 anti-MT monoclonal antibody. Images were then taken at high magnification. The mutant expressed is shown either above or below each micrograph.
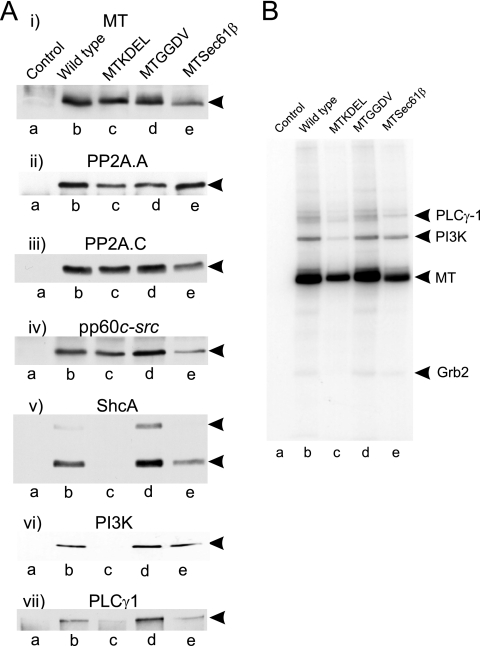
The lack of transforming ability shown by MTKDEL was intriguing as all the required protein binding sites were present in the polypeptide. To determine whether these sites were occupied, we established cell lines expressing each mutant and then performed Western blotting on the immunoprecipitated MT. Figure 5A shows that the MTKDEL polypeptide binds the core dimer of PP2A and pp60c-src at levels similar to the level of the wild-type MT. However, the signaling molecules ShcA, PI3K, and PLC-γ1 were absent in the MTKDEL immunoprecipitates. These polypeptides bind to MT through tyrosine sites phosphorylated by pp60c-src, so to determine whether the lack of interaction was a consequence of an inactive c-Src, we next performed an in vitro kinase assay on these immunoprecipitates (Fig. 5B). The result demonstrated that the MTKDEL protein-associated pp60c-src was active and able to phosphorylate MT though at a reduced level. A small amount of phosphorylation of an 85-kDa polypeptide, which is probably the regulatory subunit of PI3K, is also observed in these MTKDEL reaction products, possibly because the kinase reaction has greater sensitivity than Western blotting. These data suggest that the absence of ShcA, PI3K, and PLC-γ1 in the MT precipitate was not due to the lack of a phosphorylated binding site but was probably a consequence of these signaling molecules not having access to MT located in ER membranes. Therefore, MT is inserted into the ER membrane, where it can bind to PP2A and pp60c-src, but probably must migrate to other membranes before it becomes fully accessible to additional signaling molecules.
FIG. 5.
MTKDEL fails to bind the signaling proteins ShcA, PI3K, and PLC-γ1. (A) The protein binding properties of each of the mutants shown in Fig. 3 were determined by immunoprecipitating the MT present in equal volumes of lysate from cells expressing each of the mutants, separating it by SDS-PAGE, and Western blotting. The antibody reactivity used to probe each blot is shown above each panel, and the mutant used is indicated above each lane on the top panel. The migration position of each detected polypeptide species is shown to the right. (B) Kinase activities present in MT immunoprecipitates. Each of the MT species shown in panel A was immunoprecipitated and incubated with [γ-32P]ATP, separated by SDS-PAGE, and autoradiographed. The mutant used is indicated above each lane, and the migration positions of MT and its associated proteins are indicated to the right.
The observation that ER-located MT does not transform suggests that, once the protein is inserted into the ER membrane, there must be signals in wild-type MT to target it to additional membrane locations. In other tail-anchored proteins, the hydrophobic sequence alone is thought to determine the membrane site where the polypeptide is finally located (3). To determine whether the MT hydrophobic sequence directed it just to the ER or specified movement to additional membranes, we replaced the MT TMD with a region that promotes ER location only. The sequence chosen was the hydrophobic area from the principle component of the complex involved in cotranslational insertion of the membrane protein murine Sec61β (18) (Fig. 3A, MTSec61β), which is generally found in ER membranes only. This mutant, MTSec61β, transformed less well than wt MT, forming a smaller number of foci (Fig. 3B), and the protein was located mainly in the ER, with some protein found in outer membranes (Fig. 4). An HA+ sequence added to the C terminus of MTSec61β showed that this MT species does translocate its C-terminal amino acids similarly to wt MT but at lower levels (data not shown). These results partially fit with the hypothesis that the MT TMD has a role in targeting membranes other than the ER although they suggest that another property of MT outside the hydrophobic region also contributes to membrane targeting as the MTSec61β inhibition is not complete.
Association with PP2A is required for MT to migrate out of the ER.
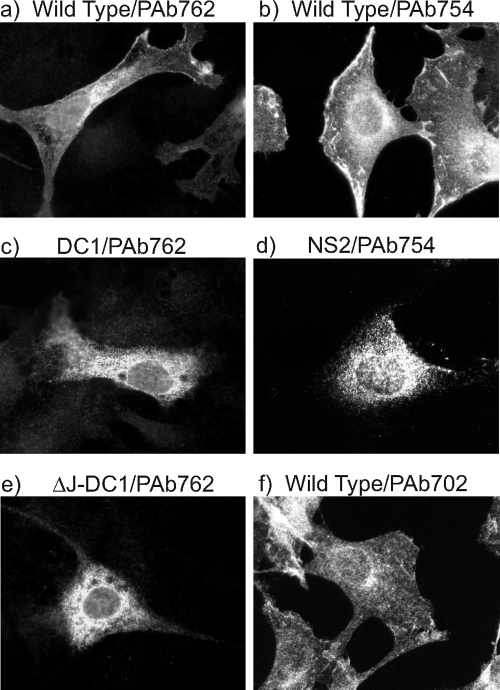
We next examined whether another MT property could play a role in the relocation of MT from the ER to additional membrane sites. We have previously reported that an MT polypeptide lacking the ability to bind PP2A was retained in cytoplasmic membranes, probably the ER (4). To determine whether other MT mutants also failed to reach the plasma membrane, the location of the MT polypeptide in cell lines expressing PP2A-binding-defective mutants NS2, DC1, and DC2 (11) were examined by immunofluorescence using a number of anti-MT monoclonal antibodies (Fig. 6 and data not shown). Mutant NS2 has an insertion of an Ile residue between amino acids 178 and 179 in MT (7), mutant DC1 involves a deletion of the sequence CSCILC at amino acids 120 to 125 (26), and mutant DC2 has a deletion of CFCLEC at positions 148 to 153 in MT (26). In all cases, MT mutants defective for binding PP2A were observed to be present only on cytoplasmic membranes and not on the plasma membrane, whether antibody PAb762 (which recognizes the N-terminal region of MT) or PAb754 (14) (which recognizes the C-terminal region of MT) was used (Fig. 6c and d, and data not shown). With both antibodies, significant amounts of wild-type MT were detected at the plasma membrane (Fig. 6a and b). It has been suggested that the binding of Hsc70 to the J domain in PP2A-binding-defective mutants of MT might account for this cytoplasmic retention (17). To determine whether this was correct, we next created a mutant that contained both a deletion of the HPDKGG sequence at position 42 to 47 in MT that is required for the J domain interaction with Hsc70 (46) and the DC1 mutation that disrupts PP2A binding (26) (mutant ΔJ-DC1). This mutant was then transfected into Rat2 cells, and the MT was detected by immunofluorescence with antibody PAb762 after a 48-h incubation. The results show clearly that ΔJ-DC1 MT does not locate to the plasma membrane, and the location resembles that of DC1 (Fig. 6e). It seems likely, therefore, that Hsc70 binding to the J domain is not responsible for maintaining PP2A-binding-defective MT mutants within cytoplasmic membranes. Finally, to examine the role of PP2A binding in MT subcellular distribution in the absence of a mutation, we performed immunofluorescence analysis on wild-type MT-expressing cells using monoclonal antibody PAb702. This antibody reacts with the N-terminal region of MT and fails to bind to the MT-PP2A complex (14), so the MT detected by PAb702 represents the small proportion of wild-type MT that is not associated with PP2A. This PP2A-free MT shows very little plasma membrane location (Fig. 6f), though this is not as discrete as in the PP2A-binding-defective mutants. However, the small amount of plasma membrane staining with PAb702 probably represents MT that was associated with PP2A but subsequently lost binding once it located at the outer membrane. Taken together, our data suggest strongly that association with PP2A is required for MT to relocate from the ER to the plasma membrane.
FIG. 6.
Association with PP2A is required for MT migration to the plasma membrane. Immunofluorescence analysis was performed on cells expressing various mutants of MT using anti-MT monoclonal antibodies that react with different regions of the molecule. Cells were plated onto coverslips, fixed, and then reacted with individual antibodies. Images were then taken at high magnification. Above each panel, the MT mutant expressed is noted, followed by the monoclonal antibody used to detect the MT. Mutants DC1 and NS2 fail to bind PP2A and, consequently, most of the other signaling proteins; mutant ΔJ-DC1 fails to bind Hsc70 and PP2A. Antibody PAb762 reacts with the N terminus of MT, PAb754 reacts with the C terminus, and PAb702 reacts with an N-terminal site that is required for binding PP2A.
DISCUSSION
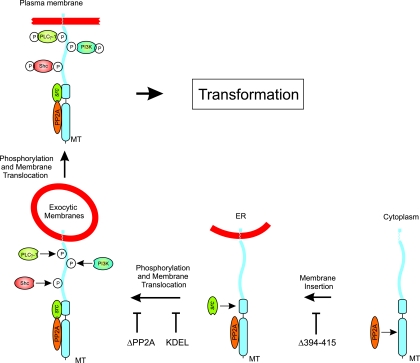
We set out here to investigate how polyomavirus MT inserts into membranes and the role of subcellular distribution in cell transformation. To achieve this, we used additions to the C terminus of MT to show that the hydrophobic domain of MT is a TMD and that the six amino acids after this region are exposed on the outside of the cell. A similar approach using the addition of a KDEL motif demonstrated that MT is probably inserted in the ER membranes but cannot generate a mitotic signal here as it does not associate with the complete series of signaling proteins. To exit the ER, MT uses a combination of the properties of the TMD and PP2A binding. Characterization of a number of different mutants demonstrates that the plasma membrane-located MT complex assembles in a defined series of steps, with signaling proteins being bound at discrete membrane sites in the MT maturation process (represented schematically in Fig. 7).
FIG. 7.
A schematic representation of the sequence of events thought to occur during the maturation of a transforming MT complex. Starting in the bottom right-hand corner, where MT is synthesized in the cytoplasm on free polysomes, the stages in MT maturation through the ER to exocytic membranes and eventually to the plasma membrane are shown. At each stage, the cellular proteins added to the MT complex are illustrated. A representative mutant that blocks each individual step is also indicated.
MT is an integral membrane protein.
The first step in assembly of the MT plasma membrane complex occurs in the cytoplasm, where MT is translated on free polysomes and binds to PP2A before association with membranes. This is indicated by the observation that MT mutants lacking the hydrophobic domain still bind to PP2A (27) but not pp60c-src (6, 16, 24) and that the hydrophobic domain is so close to the C terminus that it must be released from the polysome before it can interact with the membrane translocation machinery. MT-PP2A is then inserted into membranes. In this report, incorporation of an epitope tag on the C terminus of MT was used to show that at least some of the MT is inserted as a transmembrane protein, with the N-terminal bulk of the polypeptide within the cytoplasm and the six-amino-acid segment at the C terminus translocated across the membrane. The C-terminal tail is then exposed to the membrane lumen and eventually presented on the cell exterior (Fig. 2). Therefore, MT belongs to the group of tail-anchored (TA) membrane proteins that are inserted into the membrane posttranslationally (3). MT does not spontaneously integrate into membranes (20) and so probably uses the recently discovered ATP-requiring system involving TRC40/Asna-1/GET (36, 39). It is not yet clear whether all of the MT is transmembranal or just a portion or whether this polypeptide structure plays a role in the transforming properties of MT. The three amino acids immediately C-terminal to the TMD are required for correct functioning of MT when an HA tag is added to the C terminus of MT (Fig. 2) although they may not be required when there are no other amino acids added (9). This mutant MT (Fig. 1, Δ416-421MTHA+) is located mainly in cytoplasmic membranes (data not shown), so this could reflect a role either for the positively charged amino acids immediately proximal to the membrane when other residues are present that need to be translocated across the membrane during insertion or for trafficking to other membranes.
MT inserts at the ER but must migrate to the plasma membrane to generate a mitogenic signal.
Most TA proteins insert into the ER membrane, so to determine whether this was the case for MT and to examine what role ER-located MT has in mitogenic signaling, we inserted a KDEL ER retention signal at the MT C terminus. This prevented MT from exiting the ER and cis-Golgi compartment of the cell (Fig. 4), thus indicating that MT does, indeed, insert primarily into the ER as the KDEL would be ineffective if it inserted in any other organelle. The MTKDEL mutant has intriguing properties. It interacts with the core dimer of PP2A and with pp60c-src, activating the Src kinase which then phosphorylates a number of tyrosines in MT. However, it fails to bind the SH2 or PTB domains in the signaling molecules ShcA, PI3K, and PLC-γ1 (Fig. 5), so these proteins probably do not have access to the cytoplasmic ER and associate with MT only after it has migrated to another compartment, possibly the plasma membrane. The very small amount of PI3K observed in the in vitro kinase assays could represent either a slight leakiness of the KDEL receptor, which allows a small amount of MTKDEL out of the ER compartment, or some protein binding occurring in the lysate after lysis. As a consequence of this lack of binding, the MTKDEL mutant fails to transform cells in culture. Therefore, not only must MT bind to membranes to transform (6, 31), it must also exit the ER and migrate to another membrane compartment, possibly the cell periphery. Zhu et al. (48) have reported that newly synthesized MT is found in a perinuclear membrane population that appears to be separate from the bulk ER. They showed that ShcA and c-Src may also be concentrated at this location following MT expression, so this could reflect another post-ER membrane population where additional signaling proteins interact with the MT.
These results correlate well with reports in two other systems involving growth factor receptor-mediated transformation. Expression of a KDEL-tagged single-chain antibody that reacts with an oncogenic ErbB-2 receptor retains a significant proportion of the receptor in the ER and inhibits transformation though in this case tyrosine kinase activity was also reduced (2). Additionally, a KDEL motif added to the E5 protein of bovine papillomavirus, which transforms by activating the PDGF receptor, resulted in retention in the ER and inhibition of transforming activity (38). In both cases, the signaling molecules bound to each ER-located receptor were not examined, but our results suggest that at least part of the transforming defect could be caused by a failure to interact with such polypeptides.
MT exit from the ER requires PP2A binding and the TMD.
The subcellular distribution of TA proteins is thought to be principally determined by the nature of the TMD sequence. To investigate whether the MT TMD specifies the movement of the MT-PP2A-pp60c-src complex out of the ER to the plasma membrane, we replaced the MT transmembrane sequence with that from Sec61β, a component of the ER protein translocon Sec61 complex that is mainly ER located (18). This mutation inhibits but does not completely abolish MT transformation. Similarly, the MT mutant protein is mainly but not exclusively ER located. This suggests that either migration out of the ER is partially dependent upon the MT transmembrane sequence with perhaps the contribution of another component of the complex or that the Sec61β sequence is not as efficient as the native TMD in generating a transmembranal MT.
We have reported previously that there may be a role for PP2A binding in the eventual location of MT at the plasma membrane (4), and in confirmation of this, we found that all of the MT mutants examined that fail to bind PP2A (and consequently, c-Src, ShcA, PI3K, and PLC-γ1) were found located exclusively on cytoplasmic membranes, probably the ER. In the absence of binding to PP2A, MT associates with Hsc70 (44), so it is feasible that this acts to tether MT within the ER, thus preventing migration to the plasma membrane (17). However, a mutant MT defective for binding both PP2A and Hsc70 via the MT J domain still located only to cytoplasmic membranes (Fig. 6), suggesting that Hsc70 tethering to the J domain does not explain the location of PP2A-binding-defective MTs. This conclusion was also supported by results using the monoclonal antibody PAb702, which showed that wild-type MT that is not associated with PP2A is largely excluded from the plasma membrane (Fig. 6).
Our results add support to the evidence that MT and certain growth factor receptors can transform fibroblasts only if correctly trafficked to the plasma membrane (2, 28, 38). Further investigation of this process promises to uncover novel targets for cancer drug development.
Acknowledgments
We are grateful to Aylin Hanyaloglu, Malcolm Parker, and Kate Hardy for their comments and help with the manuscript.
The work was supported by grants from the Association for International Cancer Research, the BBSRC UK, and Belgian Science Policy Organization under the Interuniversity Attraction Poles grant P6/26 to S.M.D., by grants from the Genesis Research Trust to S.M.D. and N.J.D., and by grants LC545, 1M0506, and MSM0021620858 from the MSMT of the Czech Republic to J.F. and V.Z.
A.Y.Z., N.I., A.A, and V.Z. performed the experiments; J.F., N.J.D., and S.M.D. directed the research and wrote the manuscript.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print on 12 January 2011.
REFERENCES
- 1.Atkin, S. J., B. E. Griffin, and S. M. Dilworth. 2009. Polyoma virus and simian virus 40 as cancer models: history and perspectives. Semin. Cancer Biol. 19:211-217. [DOI] [PubMed] [Google Scholar]
- 2.Beerli, R. R., W. Wels, and N. E. Hynes. 1994. Intracellular expression of single chain antibodies reverts ErbB-2 transformation. J. Biol. Chem. 269:23931-23936. [PubMed] [Google Scholar]
- 3.Borgese, N., S. Brambillasca, and S. Colombo. 2007. How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 19:368-375. [DOI] [PubMed] [Google Scholar]
- 4.Brewster, C. E., H. R. Glover, and S. M. Dilworth. 1997. pp60c-src binding to polyomavirus middle T-antigen (MT) requires residues 185 to 210 of the MT sequence. J. Virol. 71:5512-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, K. S., et al. 1994. Polyoma middle tumor antigen interacts with SHC protein via the NPTY (Asn-Pro-Thr-Tyr) motif in middle tumor antigen. Proc. Natl. Acad. Sci. U. S. A. 91:6344-6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmichael, G. G., B. S. Schaffhausen, D. I. Dorsky, D. B. Oliver, and T. L. Benjamin. 1982. Carboxy terminus of polyoma middle-sized tumor antigen is required for attachment to membranes, associated protein kinase activities, and cell transformation. Proc. Natl. Acad. Sci. U. S. A. 79:3579-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, S. H., W. Markland, A. F. Markham, and A. E. Smith. 1986. Mutations around the NG59 lesion indicate an active association of polyoma virus middle-T antigen with pp60c-src is required for cell transformation. EMBO J. 5:325-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courtneidge, S. A., and A. E. Smith. 1983. Polyoma virus transforming protein associates with the product of the c-src cellular gene. Nature 303:435-439. [DOI] [PubMed] [Google Scholar]
- 9.Dahl, J., U. Thathamangalam, R. Freund, and T. L. Benjamin. 1992. Functional asymmetry of the regions juxtaposed to the membrane-binding sequence of polyomavirus middle T antigen. Mol. Cell. Biol. 12:5050-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dilworth, S. M. 2002. Polyoma virus middle T antigen and its role in identifying cancer-related molecules. Nat. Rev. Cancer 2:951-956. [DOI] [PubMed] [Google Scholar]
- 11.Dilworth, S. M., et al. 1994. Transformation by polyoma virus middle T-antigen involves the binding and tyrosine phosphorylation of Shc. Nature 367:87-90. [DOI] [PubMed] [Google Scholar]
- 12.Dilworth, S. M., and B. E. Griffin. 1982. Monoclonal antibodies against polyoma virus tumor antigens. Proc. Natl. Acad. Sci. U. S. A. 79:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dilworth, S. M., et al. 1986. Subcellular localisation of the middle and large T-antigens of polyoma virus. EMBO J. 5:491-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dilworth, S. M., and V. P. Horner. 1993. Novel monoclonal antibodies that differentiate between the binding of pp60c-src or protein phosphatase 2A by polyomavirus middle T antigen. J. Virol. 67:2235-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckhart, W., M. A. Hutchinson, and T. Hunter. 1979. An activity phosphorylating tyrosine in polyoma T antigen immunoprecipitates. Cell 18:925-933. [DOI] [PubMed] [Google Scholar]
- 16.Elliott, J., M. D. Jones, B. E. Griffin, and N. Krauzewicz. 1998. Regulation of cytoskeletal association by a basic amino acid motif in polyoma virus middle T antigen. Oncogene 17:1797-1806. [DOI] [PubMed] [Google Scholar]
- 17.Fluck, M. M., and B. S. Schaffhausen. 2009. Lessons in signaling and tumorigenesis from polyomavirus middle T antigen. Microbiol. Mol. Biol. Rev. 73:542-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartmann, E., et al. 1994. Evolutionary conservation of components of the protein translocation complex. Nature 367:654-657. [DOI] [PubMed] [Google Scholar]
- 19.Ito, Y., J. R. Brocklehurst, and R. Dulbecco. 1977. Virus-specific proteins in the plasma membrane of cells lytically infected or transformed by polyoma virus. Proc. Natl. Acad. Sci. U. S. A. 74:4666-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, P. K., F. Janiak-Spens, W. S. Trimble, B. Leber, and D. W. Andrews. 1997. Evidence for multiple mechanisms for membrane binding and integration via carboxyl-terminal insertion sequences. Biochemistry 36:8873-8882. [DOI] [PubMed] [Google Scholar]
- 21.Kornbluth, S., M. Sudol, and H. Hanafusa. 1987. Association of the polyomavirus middle-T antigen with c-Yes protein. Nature 325:171-173. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Li, N., et al. 1993. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature 363:85-88. [DOI] [PubMed] [Google Scholar]
- 24.Markland, W., S. H. Cheng, B. A. Oostra, and A. E. Smith. 1986. In vitro mutagenesis of the putative membrane-binding domain of polyomavirus middle-T antigen. J. Virol. 59:82-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markland, W., et al. 1986. Site-directed mutagenesis of polyomavirus middle-T antigen sequences encoding tyrosine 315 and tyrosine 250. J. Virol. 59:384-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markland, W., and A. E. Smith. 1987. Mapping of the amino-terminal half of polyomavirus middle-T antigen indicates that this region is the binding domain for pp60c-src. J. Virol. 61:285-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Messerschmitt, A., C. Disela, S. Dilworth, A. G. Marti, and K. Ballmer-Hofer. 1996. Polyomavirus middle-T antigen lacking a membrane anchor sequence accumulates in the nucleus. J. Gen. Virol. 77:17-26. [DOI] [PubMed] [Google Scholar]
- 28.Morley, G. M., M. Uden, W. J. Gullick, and N. J. Dibb. 1999. Cell specific transformation by c-fms activating loop mutations is attributable to constitutive receptor degradation. Oncogene 18:3076-3084. [DOI] [PubMed] [Google Scholar]
- 29.Moyne, G., F. Harper, S. Saragosti, and M. Yaniv. 1982. Absence of nucleosomes in a histone-containing nucleoprotein complex obtained by dissociation of purified SV40 virions. Cell 30:123-130. [DOI] [PubMed] [Google Scholar]
- 30.Nicholson, P. R., S. Empereur, H. R. Glover, and S. M. Dilworth. 2001. ShcA tyrosine phosphorylation sites can replace ShcA binding in signalling by middle T-antigen. EMBO J. 20:6337-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novak, U., and B. E. Griffin. 1981. Requirement for the C-terminal region of middle T-antigen in cellular transformation by polyoma virus. Nucleic Acids Res. 9:2055-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pallas, D. C., et al. 1990. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell 60:167-176. [DOI] [PubMed] [Google Scholar]
- 33.Rozakis-Adcock, M., R. Fernley, J. Wade, T. Pawson, and D. Bowtell. 1993. The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature 363:83-85. [DOI] [PubMed] [Google Scholar]
- 34.Schaffhausen, B. S., H. Dorai, G. Arakere, and T. L. Benjamin. 1982. Polyoma virus middle T antigen: relationship to cell membranes and apparent lack of ATP-binding activity. Mol. Cell. Biol. 2:1187-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaffhausen, B. S., and T. M. Roberts. 2009. Lessons from polyoma middle T antigen on signaling and transformation: a DNA tumor virus contribution to the war on cancer. Virology 384:304-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuldiner, M., et al. 2008. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134:634-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soeda, E., J. R. Arrand, N. Smolar, and B. E. Griffin. 1979. Sequence from early region of polyoma virus DNA containing viral replication origin and encoding small, middle and (part of) large T antigens. Cell 17:357-370. [DOI] [PubMed] [Google Scholar]
- 38.Sparkowski, J., J. Anders, and R. Schlegel. 1995. E5 oncoprotein retained in the endoplasmic reticulum/cis Golgi still induces PDGF receptor autophosphorylation but does not transform cells. EMBO J. 14:3055-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stefanovic, S., and R. S. Hegde. 2007. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128:1147-1159. [DOI] [PubMed] [Google Scholar]
- 40.Su, W., W. Liu, B. S. Schaffhausen, and T. M. Roberts. 1995. Association of polyomavirus middle tumor antigen with phospholipase C-gamma 1. J. Biol. Chem. 270:12331-12334. [DOI] [PubMed] [Google Scholar]
- 41.Talmage, D. A., et al. 1989. Phosphorylation of middle T by pp60c-src: a switch for binding of phosphatidylinositol 3-kinase and optimal tumorigenesis. Cell 59:55-65. [DOI] [PubMed] [Google Scholar]
- 42.Templeton, D., A. Voronova, and W. Eckhart. 1984. Construction and expression of a recombinant DNA gene encoding a polyomavirus middle-size tumor antigen with the carboxyl terminus of the vesicular stomatitis virus glycoprotein G. Mol. Cell. Biol. 4:282-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Treisman, R., U. Novak, J. Favaloro, and R. Kamen. 1981. Transformation of rat cells by an altered polyoma virus genome expressing only the middle-T protein. Nature 292:595-600. [DOI] [PubMed] [Google Scholar]
- 44.Walter, G., A. Carbone, and W. J. Welch. 1987. Medium tumor antigen of polyomavirus transformation-defective mutant NG59 is associated with 73-kilodalton heat shock protein. J. Virol. 61:405-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walter, G., R. Ruediger, C. Slaughter, and M. Mumby. 1990. Association of protein phosphatase 2A with polyoma virus medium tumor antigen. Proc. Natl. Acad. Sci. U. S. A. 87:2521-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whalen, K. A., R. de Jesus, J. A. Kean, and B. S. Schaffhausen. 2005. Genetic analysis of the polyomavirus DnaJ domain. J. Virol. 79:9982-9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitman, M., C. P. Downes, M. Keeler, T. Keller, and L. Cantley. 1988. Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature 332:644-646. [DOI] [PubMed] [Google Scholar]
- 48.Zhu, W., A. Eicher, B. Leber, and D. W. Andrews. 1998. At the onset of transformation polyomavirus middle-T recruits shc and src to a perinuclear compartment coincident with condensation of endosomes. Oncogene 17:565-576. [DOI] [PubMed] [Google Scholar]