Abstract
The spike protein of murine leukemia virus, MLV, is made as a trimer of the Env precursor. This is primed for receptor-induced activation of its membrane fusion function first by cellular furin cleavage in the ectodomain and then by viral protease cleavage in the endodomain. The first cleavage separates the peripheral surface (SU) subunit from the transmembrane (TM) subunit, and the latter releases a 16-residue-long peptide (R) from the TM endodomain. Here, we have studied the distribution of R peptide cleavages in the spike TM subunits of Moloney MLV preparations with partially R-peptide-processed spikes. The spikes were solubilized as trimers and separated with an R peptide antibody. This showed that the spikes were either uncleaved or cleaved in all of its TM subunits. Further studies showed that R peptide cleavage-inhibited Env mutants, L649V and L649I, were rescued by wild-type (wt) Env in heterotrimeric spikes. These findings suggested that the R peptide cleavages in the spike are facilitated through positive allosteric cooperativity; i.e., the cleavage of the TM subunit in one Env promoted the cleavages of the TMs in the other Envs. The mechanism ensures that protease cleavage in newly released virus will generate R-peptide-cleaved homotrimers rather than heterotrimeric intermediates. However, using a cleavage site Env mutant, L649R, which was not rescued by wt Env, it was possible to produce virus with heterotrimers. These were shown to be less fusion active than the R-peptide-cleaved homotrimers. Therefore, the cooperative cleavage will speed up the maturation of released virus for fusion competence.
The spike protein of murine leukemia virus (MLV) is assembled in the endoplasmic reticulum (ER) of the producer cell from three copies of the Env precursor protein gp80 (10, 18). The trimeric spike undergoes two proteolytic cleavage events to prepare it for receptor-induced activation of the membrane fusion process. The first one is mediated by the furin of the host cell and takes place when the spike passes the trans-Golgi network on its way from the ER to the cell surface (3, 6). This cleavage separates the polypeptide of the peripheral surface (SU) subunit from that of the transmembrane (TM) subunit (7). It also releases the Env fusion peptide at the membrane-distal amino-terminal region of the TM (26). The second cleavage is mediated by the viral protease and takes place inside newly formed particles. Here, the protease cleaves not only the Gag and the Gag-Pol precursors into the mature internal proteins (matrix, p12, capsid, and nucleocapsid proteins) and the viral enzymes (protease, polymerase, and integrase) but also the endodomain of the TM subunit of Env. A 16-residue-long C-terminal peptide, the R peptide, is cleaved off, generating p15E from the TM precursor, Pr15E (5, 7, 17). By studying cell-associated Env with an R peptide truncation, it was shown that the R peptide inhibited the Env from becoming activated for membrane fusion by the receptor (14, 15).
The R peptide cleavage site QAL649/VLT of Moloney MLV (MoMLV) represents a type 2 retroviral protease cleavage site (13). This is characterized in particular by the preference of Leu over Phe and Tyr at the P1 position and by the exclusion of charged amino acids and amino acids with side chains branched at the beta-carbon (Ile and Val) at this position. Consequently, we along with others have created L649R (LR), L649I (LI), and L649V (LV) Env mutants and shown that these were inhibited in R peptide cleavage when assembled into virus particles, which were also shown to be noninfectious (8, 15, 20). Earlier, we used such mutant virus to characterize the mechanism of fusion facilitation by the R peptide cleavage (9). In MoMLV Env the fusion activation process is controlled by the disulfide that links the SU subunit with the TM (21). The disulfide Cys of SU is associated with a CXXC motif, where the other Cys residue is free. When the SU binds to the virus receptor on the cell surface, the free thiol is activated to attack the intersubunit disulfide and rearrange it into a disulfide isomer within the motif (21). This liberates the metastable TM to activate membrane fusion through refolding into a stable conformation. However, if the R peptide has not been released by cleavage, as in the mutants, this isomerization reaction proceeds inefficiently, and the ability of the particles to support cell-cell fusion from without is impaired. This suggests that the cleavage of the R peptide in the endodomain of the spike induces a structural change in the ectodomain of the spike that facilitates receptor-induced activation of the isomerization reaction and the subsequent fusion reaction. Such changes have been confirmed by mapping of the Env surface using antibody binding and biotin labeling (1).
A protease cleavage-mediated structural change in a homooligomeric protein like the trimeric Env would probably be more efficient if the cleavage of one subunit facilitated the cleavages of the other two. If mixed oligomers with only partially changed subunits persisted, it would probably result in less stable or less active protein complexes. Therefore, a relevant question appears to be whether protease cleavage of the R peptide in one subunit of the Env trimer promotes cleavage of the R peptides of the other two subunits. In the present study we approached this question using MoMLV with partially cleaved R peptide and an R-peptide-specific antibody (αR) that immunoprecipitated intact Env trimers. The rationale was that cooperativity in cleavage should ensure the generation of trimers with either completely cleaved or completely uncleaved R peptides. Our results supported cooperative cleavage.
MATERIALS AND METHODS
Cells and reagents.
293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 4.5 g/liter glucose (Gibco BRL) supplemented with sodium pyruvate, nonessential amino acids (MEM NEAA; Gibco BRL), l-glutamine, and 10% fetal calf serum (FCS). MoMLV-producing MOV-3 cells (G. Schmidt, GSF-National Research Center for Environment and Health, Neuherberg, Germany) were maintained similarly but without the NEAA and with 20 mM HEPES added to the medium. XC cells were maintained as 293T cells, but the medium included 1 g/liter glucose and 20 mM HEPES. Plasmids containing the provirus DNA for wild-type (wt) and mutant viruses have been described previously (9). The anti-R peptide antibody, αR, is an affinity-purified polyclonal antibody from rabbits immunized with a 17-amino-acid-long peptide corresponding to the MoMLV R peptide, NH2-CVLTQQYHQLKPIEYEP-CONH2. An extra Cys was added to the N terminus as a conjugation site (Innovagen, Lund, Sweden). The MoMLV-specific rabbit antiserum, αMLV (HE863), and the MoMLV SU-specific rabbit antiserum, αSU (HE699), originated from Viromed Biosafety Laboratories, Camden, NJ. Antibody against the influenza hemagglutinin (αHA) tag was from Roche (Basel, Switzerland). The viral protease inhibitor amprenavir was obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
Construction of HA-tagged Env.
The DNA sequence of the HA tag (YPYDVPDYA) was inserted into the polyproline encoding region (after Pro306) of the L649I and L649R mutant Env genes of the MoMLV provirus DNA (9, 12). This was done by fusion PCR using the pNCA plasmid as a template (2).
Production of labeled virus.
293T cells were transfected with 20 μg of proviral DNA in a 75-cm2 culture flask using calcium phosphate precipitation. The culture was labeled with 100 μCi of [35S]Cys/ml in cysteine-free DMEM (National Veterinary Institute, Uppsala, Sweden) 24 to 48 h after transfection. In some experiments the viral protease inhibitor amprenavir was present during the labeling. The culture supernatant was collected, and cell debris was separated by low-speed centrifugation. Virus-producing MOV-3 cells in a culture dish (diameter, 6 cm) were incubated in cysteine-free DMEM for 30 min before being washed with Ca2+- and Mg2+-supplemented phosphate-buffered saline (PBS+) and then labeled with 100 μCi of [35S]Cys/ml for 4 h. The dish was washed three times with PBS+, and virus was collected in cell culture medium for 20 min.
Virus isolation.
Virus-containing supernatant from transfected 293T cells was layered on top of a step gradient composed of 1 ml of 50% and 4.5 ml of 20% sucrose (wt/wt) in HNC buffer (50 mM HEPES, 100 mM NaCl, 1.8 mM CaCl2, pH 7.4,) and centrifuged at 4°C for 2 h at 93,000 × g (22,000 rpm) in a Beckman SW28.1 rotor. Virus was collected from the 20/50% sucrose interphase.
Analyses of viral proteins.
Virus isolated in the 20/50% sucrose step gradient was lysed in HNC buffer containing 0.15% Triton X-100 on ice for 10 min. Viral proteins were then analyzed directly or after complexing with αR antibodies on ice for 2 h by blue native polyacrylamide gel electrophoresis (BN-PAGE) as described previously (19). Alternatively, viral proteins in lysate were reacted with antisera or antibodies over night at +4°C and precipitated with protein A-Sepharose (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) for reducing SDS-PAGE (11). Immunoprecipitates were washed once with a buffer containing 10 mM Tris, 150 mM NaCl, 2 mM EDTA, and 0.2% Triton X-100, pH 7.5, once with a buffer containing 10 mM Tris, 0.5 M NaCl, 2 mM EDTA, and 0.2% Triton X-100, pH 7.5, and once with 10 mM Tris, pH 7.5, before being mixed with SDS-containing sample buffer and incubated at 70°C for 3 min. In some cases the virus was lysed in HN buffer (50 mM HEPES, 100 mM NaCl, pH 7.4) containing 0.15% Triton X-100, 10 mM EDTA, and 20 mM N-ethylmaleimide (NEM) for 30 min at 37°C. The EDTA chelates Ca2+ from the spike, which activates it for membrane fusion. However, the activation is interrupted at a stage before intersubunit disulfide isomerization by the NEM. This mediates modification (alkylation) of the isomerization active thiol in the SU subunit. The isomerization-arrested stage (IAS) intermediate represents a significantly more stable trimer than the native form (19). IAS intermediates were immunoprecipitated as describe above.
Fusion from without.
Virus in HNC buffer containing 20 μg/ml Polybrene was spinoculated at 850 × g for 1 h at 4°C onto confluent cultures of XC cells in 24-well plates in a Beckman JS5.9 rotor. The buffer was exchanged to prewarmed buffer (37°C), and the cultures were incubated at this temperature for 15 min to allow virus-mediated cell-cell fusion. After this, the remaining fusion-active spikes were inactivated by treatment with a buffer containing 40 mM sodium citrate, 10 mM KCl, and 135 mM NaCl, pH 3.0, for 1 min at room temperature. XC cell medium was added, and the cultures were incubated for 2 h at 37°C to let fused cells form polykaryons. These were visualized by staining with Giemsa (Sigma). To estimate fusion efficiency, we calculated the relative number of nuclei that were localized in polykaryons as a percentage of the total number of nuclei. For each experiment five microscope fields (about 1,000 nuclei) of each sample were analyzed with the help of the ImageJ plug-in Cell Counter.
Other methods.
The fraction of mixed Env trimers formed by two types of Env in an ideal situation with equal synthesis and random mixing in the rough endoplasmic reticulum (RER) is expected to be 75% as there are eight possible SU-TM combinations in a trimer, and six of them contain both wt and mutant subunit pairs. Each homotrimer fraction will be 12.5%. The fractions of heterotrimers comprised of SU-Pr15E and SU-p15E (SU-Pr15E/SU-p15E) and SU-Pr15E and SU-p15E homotrimers in our cotransfection experiments were estimated by the αR immunoprecipitation. The αR will react with all trimers containing at least one Env with the R peptide. The amount of coprecipitating SU-p15E, which lacks the R peptide, corresponds to the amount of this complex in heterotrimers. In the case of about equal synthesis of the two Env proteins, the amounts of different trimers can be estimated by measuring the amounts of Pr15E and p15E captured by αR. The heterotrimer fraction will correspond to the amount of p15E times two, and each homotrimer fraction will correspond to the amount of Pr15E with the amount of p15E subtracted.
RESULTS
The anti-R peptide antibody (αR) makes stable complexes with Env trimers.
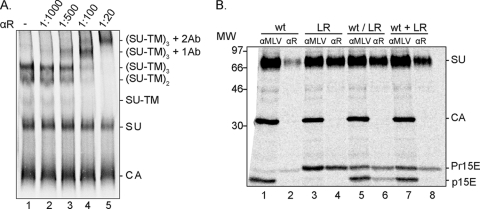
For our analyses we needed an antibody against the R peptide that maintained the spike trimer upon complex formation. Therefore, we raised a polyclonal antibody in rabbits against the MoMLV R peptide (αR) and tested its reactivity against the spike using BN-PAGE. To this end [35S]Cys-labeled L649R mutant virus (LR) was isolated in a 20/50% sucrose step gradient, solubilized in HNC buffer containing 0.3% Triton X-100 on ice for 10 min, and then reacted with αR on ice for 2 h. The LR mutant was chosen for this experiment because it was almost totally inhibited in R peptide cleavage (see below). In the absence of antibody, the BN-PAGE revealed the solubilized Env mostly as native SU-TM (in this case, SU-Pr15E) trimers (∼240 kDa), a significant amount of SU-TM dimers (∼160 kDa), free SU (∼70 kDa), and a small amount of SU-TM monomers (∼80 kDa) as described previously (Fig. 1 A, lane 1) (23). In addition, the capsid (CA) protein was resolved on the gel. Reaction of the Env with increasing amounts of αR before BN-PAGE resulted in a decreasing amount of the SU-TM complexes and an increasing amount of two complexes much larger than the SU-TM trimers (Fig. 1A, lanes 2 to 5). This is consistent with the idea that these represent Env trimers in complex with one and two antibodies, respectively. We concluded that the reaction of αR with the trimeric Env harboring SU-Pr15E resulted in stable complexes of the Env trimers with αR and no significant trimer dissociation.
FIG. 1.
The αR antibody reacts with the R peptide in SU-Pr15E homotrimeric and SU-Pr15E/SU-p15E heterotrimeric spikes. (A) BN-PAGE of αR-complexed LR Env. [35S]Cys-labeled LR mutant virus was solubilized with Triton X-100 on ice, and native spikes were reacted with αR antibody at increasing concentrations (dilutions of 1:1,000 to 1:20) for 2 h on ice and analyzed by BN-PAGE. A control sample was incubated without antibody. Viral proteins and protein oligomers are indicated. 1Ab and 2Ab indicate the numbers of antibodies associated with SU-TM trimers. (B) Immunoprecipitation of Env trimers with mixed R-peptide-containing and -lacking SU-TM complexes. 293T cells were cotransfected or transfected separately with wt and LR Env provirus DNA and labeled with [35S]Cys. The resulting virus (wt/LR, wt, and LR) was isolated and solubilized with Triton X-100 on ice for Env analysis by αR and αMLV immunoprecipitation and subsequent reducing SDS-PAGE. As a control, solubilized wt and LR viruses were mixed before immunoprecipitation (wt+LR). Note the immunoprecipitation of mixed trimers with αR from the solubilized wt/LR virus preparation but not from the other ones. Viral proteins and the migration of molecular weight (MW; in thousands) standards are indicated. The figure represents phosphorimages of the gels.
αR immunoprecipitates SU-Pr15E/SU-p15E heterotrimers.
The αR-mediated immunoprecipitation of Env heterotrimers was then tested. For this we used a virus preparation produced in 293T cells cotransfected with wt and LR Env provirus DNA (yielding wt/LR virus) and isolated in a 20/50% sucrose step gradient. We expected that the wt and the LR Envs should form mixed trimers during biosynthesis in the ER. These should be furin cleaved and assembled into virus, where the wt but not the LR TM subunits (Pr15E) of the trimers should be processed by the viral protease into p15E. Indeed, earlier studies had demonstrated that R-peptide-truncated Env formed mixed trimers with wt Env in cells cotransfected with corresponding DNAs (16, 25). The wt/LR virus was solubilized and reacted with αR as described and then precipitated with protein A for Env analysis by reducing SDS-PAGE. This showed that αR mediated precipitation of not only the R-peptide-containing SU-Pr15E complexes but also a significant amount of SU-p15E complexes lacking the R peptide, i.e., SU-Pr15E/SU-p15E heterotrimers (Fig. 1B, lane 6). The amount of coprecipitated p15E of total immunoprecipitated TM was 25% (±0.6%; n = 4). Immunoprecipitation with an anti-MLV antiserum, αMLV, showed that the wt/LR virus contained roughly equal amounts of SU-Pr15E and SU-p15E complexes (Fig. 1B, lane 5). In this analysis the CA protein is also revealed. We also immunoprecipitated a mixed lysate containing wt and LR mutant virus, each produced separately in 293T cells, as a control for αR specificity and a test for possible subunit mixing between trimers in solution. Also the mixed lysate contained about equal amounts of the SU-Pr15E and SU-p15E complexes, as shown by the αMLV immunoprecipitation, but only SU-Pr15E complexes were captured by αR, suggesting both high αR antibody specificity and trimer stability (Fig. 1B, lanes 7 and 8). The specificity of the αR antibody was also apparent in the analyses of wt and LR mutant virus separately (Fig. 1B, lanes 1 to 4).
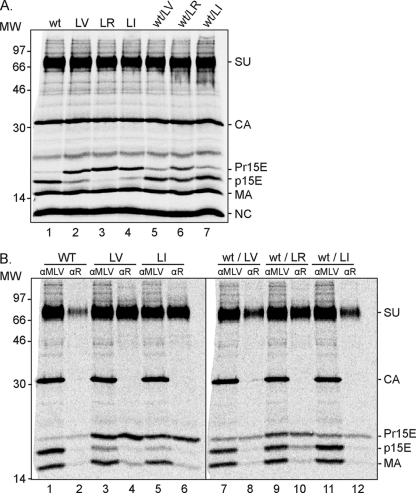
The restricted R peptide cleavage in the LV and LI mutants and the apparent rescued cleavage in the wt/LV and wt/LI virus are cooperative.
To analyze the possible cooperative cleavage of the Env R peptide in virus particles, we needed to study virus where the cleavage process was still incomplete. We first determined the cleavage efficiencies of our wt and mutant virus L649V (LV), L649I (LI), and LR preparations. When analyzed by reducing SDS-PAGE, they showed a range of R peptide cleavage efficiencies, as calculated from the amounts of Pr15E and p15E: wt, 95% ± 2% (n = 3); LV, 18% ± 5% (n = 3); LR, 3% ± 1% (n = 3); and LI, 11% ± 4% (n = 3) (Fig. 2 A, lanes 1 to 4). Thus, the question was whether the residual uncleaved SU-Pr15E complexes in the wt were present in trimers together with SU-p15E and whether the minor fractions of cleaved SU-p15E complexes in the LV and the LI mutants were together with SU-Pr15E. Two additional virus preparations were useful for our study. These were viruses produced by cotransfection of 293T cells with wt and LV or LI Env provirus DNA. To our surprise, and in contrast to virus produced by the cotransfection of wt and LR Env provirus DNA, these appeared to contain mixed trimers where the cleavage of the R peptide in mutant Env was largely rescued by the presence of the wt Env of the complex. This was shown by analyzing the virus preparations by reducing SDS-PAGE (Fig. 2A, lanes 5 to 7). The possibility of cleavage rescue was studied in detail later.
FIG. 2.
wt, LV, LR, LI, wt/LV, and wt/LI virus, but not wt/LR, contain their SU-p15E and R-peptide-containing SU-Pr15E complexes in separate homotrimers. (A) 293T cells were cotransfected or transfected separately with wt and LV, LR, or LI Env provirus DNA and labeled with [35S]Cys. The resulting virus (wt, LV, LR, LI, wt/LV, wt/LR, and wt/LI) was isolated and analyzed by reducing SDS-PAGE. (B) The virus preparations were solubilized with Triton X-100 on ice, and native spikes were subjected to immunoprecipitation with αR and αMLV and subsequent analyses by reducing SDS-PAGE. Note that in all preparations but wt/LR, αR precipitates Env only with SU-Pr15E complexes, i.e., complexes with the R peptide. MW, molecular weight (in thousands).
All virus preparations were solubilized as native spikes and subjected to immunoprecipitation by the αR and the αMLV antibodies. Reducing SDS-PAGE showed that, with one exception, the αR antibody captured virtually only trimers with SU-Pr15E complexes, i.e., complexes with R peptides left (Fig. 2B, lanes 2, 4, 6, 8, and 12). The exception was the wt/LR preparation, where both SU-Pr15E and SU-p15E complexes were captured as shown before (Fig. 2B, lane 10). Corresponding analyses with the αMLV antibody showed the total amount of Env subunits present in the various samples (Fig. 2B, lanes 1, 3, 5, 7, 9, and 11). It also revealed the CA and matrix (MA) proteins. We concluded that all virus preparations except the wt/LR contained Env trimers where the R peptides were either completely cleaved or completely uncleaved, suggesting cooperativity in the cleavage reaction.
Cooperative R peptide cleavage in newly assembled virus.
To follow the possible cooperative cleavage of the R peptide in Env trimers of newly assembled wt MoMLV, we collected virus for 20 min after a 4-h labeling of MOV-3 cells with [35S]Cys. MOV-3 is a 3T3 cell line chronically infected with MoMLV. Samples of the culture supernatant were incubated for 0 to 24 h at 37°C to allow progressive cleavage of the R peptide in viral spikes. The samples were divided into two parts, one of which was used to immunoprecipitate all spikes by αSU antibody for reducing SDS-PAGE and the other of which was used to capture only those with R peptide by the αR antibody. For the former analyses, the spikes were solubilized at 37°C in the presence of EDTA and NEM. Under these conditions the spike is activated but stalled by CXXC thiol alkylation in the SU at an intersubunit disulfide isomerization arrested stage (IAS) with increased trimer stability (19, 21). This facilitated the use of the αSU antibody, an SU-specific polyclonal antibody, to capture all the spikes. The analyses showed that there was about 40% R-peptide-cleaved TM (p15E) in the pulse-labeled virus preparation and that this fraction increased to about 65% during the 24-h-long incubation at 37°C (Fig. 3, lanes 1, 3, 5, and 7). Immunoprecipitation of Env trimers with the αR was done using solubilized native spikes. These analyses showed that virtually all of the SU-Pr15E complexes were captured, but none of the SU-p15E complexes, in all of the samples (Fig. 3, lanes 2, 4, 6, and 8). This indicated that the captured trimers retained the R peptide in all of their TM subunits and that the noncaptured ones had the R peptide cleaved in all their TM subunits. Therefore, we concluded that there is cooperative cleavage of the R peptide in the Env trimer of newly assembled virus.
FIG. 3.
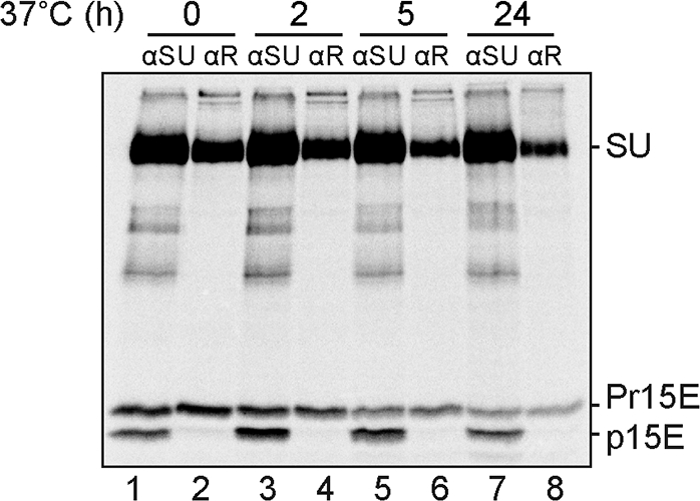
R peptide cleavage cooperativity in newly made virus. MOV-3 cells were pulse labeled for 4 h with [35S]Cys, and then virus was collected in the culture medium for 20 min. Medium samples were incubated for 0 to 24 h at 37°C before solubilization of the virus under conditions that generated either IAS Env trimers or native trimers. The former samples were immunoprecipitated with αSU antibodies to capture total Env trimers, and the latter were incubated with αR antibodies to capture spikes with R peptide(s) for reducing SDS-PAGE. Env subunits are indicated. Note the increased Pr15E cleavage into p15E with increasing time of incubation in the analyses of total Env and the virtually exclusive capture of R-peptide-containing SU-Pr15E trimers with the αR antibody.
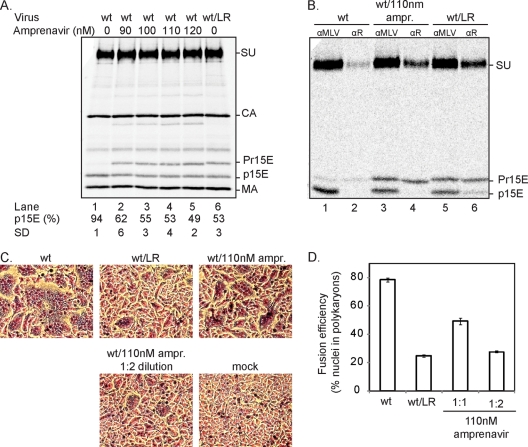
SU-p15E homotrimers are more fusion active than SU-p15E/SU-Pr15E heterotrimers.
In order to compare the fusion activity of the SU-p15E homotrimers with that of the SU-p15E/SU-Pr15E heterotrimers, we prepared virus with the respective spikes and analyzed their fusion-from-without activity with rat XC cells. The virus with heterotrimers was made by cotransfecting 293T cells with wt and LR proviral DNAs. The wt/LR virus contained about equal amounts of SU-Pr15E and SU-p15E complexes, as shown by αSU analyses of IAS Env (Fig. 4 A, lane 6). Analysis with the αR antibody, using solubilized native spikes, showed the presence of a significant fraction of SU-p15E that coprecipitated with SU-Pr15E, indicative of heterotrimers, as described previously (Fig. 4B, lane 6). Quantification and subsequent calculation (see Materials and Methods) showed that the preparation contained about 56% heterotrimeric spikes and 22% SU-p15E homotrimeric and 22% SU-Pr15E homotrimeric spikes. A virus preparation containing the corresponding amount of R-peptide-cleaved Envs was made by transfecting the 293T cells with wt provirus DNA in the presence of amprenavir. This is a drug developed to inhibit the HIV-1 protease, but it also works for the MoMLV protease (4). A concentration of about 110 nM of the drug caused a 50% inhibition of the R peptide cleavage without any significant effect on Gag cleavage (Fig. 4A, lane 4). Analysis of the spike trimer composition using the αR antibody showed that only SU-Pr15E complexes were precipitated (Fig. 4B, lane 4). Consequently, the virus preparation contained only homotrimers, with or without R peptide, in about equal amounts; i.e., the amprenavir-restricted cleavage still occurred cooperatively. Corresponding amounts of these two and a wt virus preparation, estimated by CA protein concentration, were then tested for fusion activity. To this end, the virus was spinoculated on XC cells at 4°C and then incubated at 37°C for 15 min to allow fusion from without. After low-pH inactivation of residual unused viral spikes, virus-mediated cell-cell fusion was visualized by polykaryon formation in a subsequent 2 h of incubation at 37°C. Fusion efficiency was measured as the relative number of nuclei in polykaryons. This showed that the heterotrimer-containing virus preparation (wt/LR) was about half as efficient (25% fusion) as the homotrimer-containing one (wt with 110 nM amprenavir) (49% fusion) (Fig. 4C and D). Considering that the former preparation contained 22% fully fusion-competent SU-p15E homotrimers and 56% heterotrimers, most of the fusion activity of this preparation must be derived from the former fraction of spikes. Indeed, the fusion efficiency of a 1:2 diluted homotrimer preparation, which should contain about as many homotrimers as the undiluted heterotrimer preparation, had a fusion efficiency of 27% (Fig. 4C and D). We concluded that the fusion activity of the heterotrimeric spike is significantly reduced compared to the activity of the R-peptide-cleaved homotrimeric one.
FIG. 4.
R-peptide-cleaved homotrimeric spikes are more efficient in fusion than heterotrimeric ones. (A) Preparation of wt virus with the same R peptide cleavage efficiency as wt/LR virus. 293T cells were transfected with wt or a mixture of wt and LR provirus DNA and labeled for 24 h. The wt DNA-transfected cells were labeled in the presence of 90 to 120 nM amprenavir. The released virus was purified in a sucrose step gradient and analyzed by reducing SDS-PAGE. The efficiency of R peptide cleavage in each virus preparations is given as the relative amount of p15E (percentage of the total TM ± standard deviation [SD]; n = 3) below each lane. Note that amprenavir at the given concentrations inhibits R peptide cleavage without affecting Gag cleavage in any significant way. Note also that producing wt virus in the presence of 110 nM amprenavir (lane 4) results in a preparation with the same R peptide cleavage efficiency as the wt/LR virus (lane 6). (B) Analysis of spike composition. The wt virus, the wt virus prepared in the presence of 110 nM amprenavir, and the wt/LR virus shown in panel A, lanes 1, 4, and 6, respectively, were lysed under conditions generating IAS Env trimers or native trimers. The former samples were used for immunoprecipitation of total Env with αSU, and the latter were used for capture of R-peptide-containing Env trimers with αR antibody. Immunoprecipitates were analyzed by reducing SDS-PAGE. Note the coprecipitation of p15E in the αR reaction of the wt/LR spike preparation, which is indicative of heterotrimeric spikes. (C and D) The fusion efficiencies of the wt virus, the virus produced in the presence of 110 nM amprenavir, and the L/R virus are shown in panel A. The virus preparations were spinoculated on XC cells at 4°C and then incubated at 37°C for 15 min to allow virus-mediated cell-cell fusion. After this, the cultures were treated with pH 3.0 buffer to inactivate still-functional spikes and incubated for 2 h to allow fused cells to form polykaryons (C). The average fraction of cell nuclei in polykaryons was calculated (n = 3) and used as an indicator of fusion efficiency (D). A 1:2 dilution of the wt virus produced in the presence of amprenavir was also analyzed.
wt Env rescue of LV and LI Env R peptide cleavage in heterotrimers.
The apparent rescue of mutant Env R peptide cleavage in the wt/LV and the wt/LI virus preparations was most intriguing. It cannot be explained by overexpression of the wt provirus since all proviruses were expressed about equally in separate transfections (Fig. 2A, lanes 1 to 4), and the cotransfection of wt and LR Env mutant viruses resulted in expression of both constructs at about the same levels (Fig. 2A, lane 6). In the results shown in Fig. 5, we have quantified the R peptide cleavage in the wt/LV, wt/LR, and the wt/LI virus preparations as a mean of four to six experiments. The measured cleavage efficiencies (Fig. 5, filled diamonds) were compared with those expected, assuming random wt and mutant Env mixing in the RER at equal concentrations (12.5% wt homotrimers, 75% heterotrimers, and 12.5% mutant homotrimers) and either wt-like cleavage of all Envs in the heterotrimers (i.e., rescue of the mutant subunits) (Fig. 5, open triangles) or wt-like cleavage of the wt Envs and mutant-like cleavage of the mutant Envs in the heterotrimers (i.e., no rescue of the mutant subunits) (Fig. 5, open squares). We found that the cleavage efficiencies of the wt/LV and the wt/LI viruses were similar to the theoretical values, assuming cleavage rescue. In contrast, the cleavage of wt/LR virus was similar to the expected value assuming no cleavage rescue.
FIG. 5.
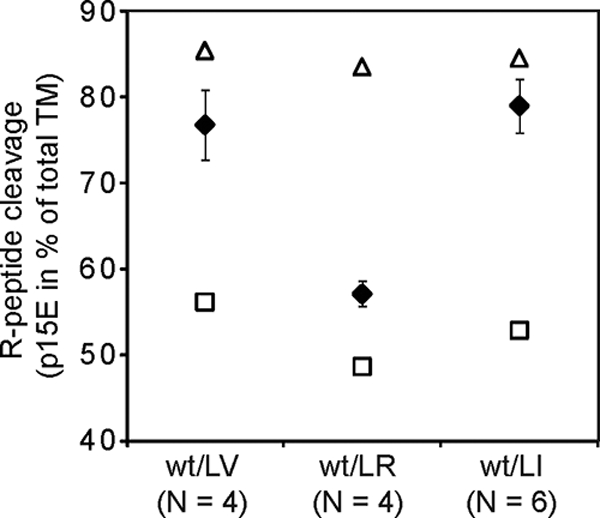
R peptide cleavage efficiencies in wt/LV and wt/LI virus suggest wt-mediated cleavage rescue. The mean R peptide cleavage efficiencies in four to six preparations of wt/LV, wt/LI, and wt/LR virus were calculated (filled diamonds) and compared with the expected efficiencies assuming random wt and mutant Env mixing at equal concentrations in the ER and either wt-like R-peptide cleavage of both wt and mutant Envs in the mixed trimers (▵, cleavage rescue) or wt-like cleavage of only the wt Envs of the heterotrimers (□, no cleavage rescue). The cleavage efficiencies are given as percent p15E where total TM (p15E plus Pr15E) in the preparation is 100%. Note the apparent rescue of the R peptide cleavage in LV and LI Env by wt Env in the wt/LV and wt/LI virus.
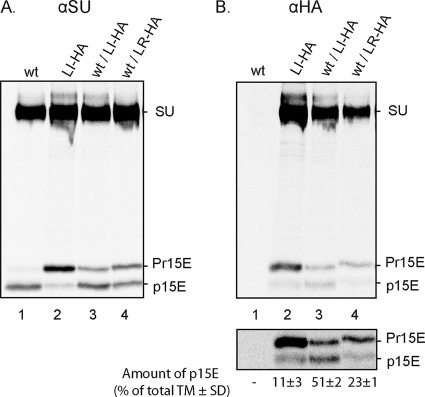
In order to get direct evidence for the R peptide cleavage rescue, we tagged the LI Env with an influenza virus hemagglutinin epitope (LI-HA) and followed its processing in the spikes of the wt/LI-HA virus using a corresponding antibody (αHA). We showed before that wt/LI virus contained essentially only homotrimeric spikes, i.e., trimers with the R peptide cleaved or retained in all its TM subunits (Fig. 2B, lanes 11 and 12). In the case of rescue, initially formed mixed wt and mutant trimers with R peptide should be cooperatively cleaved into mixed wt and mutant trimers without R peptide. Thus, the immunoprecipitation of any Env with R peptide cleaved using the αHA from a wt/LI-HA virus should indicate R peptide cleavage rescue. As a negative control, we used the wt/LR virus with no apparent cleavage rescue of mutant Env, which contains a fraction of heterotrimeric spikes, i.e., spikes with the R peptide cleaved in one or two of its TM subunits (Fig. 2B, lanes 9 and 10). If the LR Env was HA tagged, then the αHA should capture much less Env without R peptide from wt/LR-HA virus than from wt/LI-HA virus. The HA tag sequence was engineered into the polyproline-rich region separating the N-terminal receptor binding domain and the C-terminal disulfide isomerase-containing domain of SU. wt/LI-HA and wt/LR-HA virus were produced as described and solubilized at conditions generating IAS spikes. The reducing SDS-PAGE of the αHA immunoprecipitates revealed the p15E, Pr15E, and SU proteins of both samples (Fig. 6 B, lanes 3 and 4). Quantifications showed that 51% ± 2% of TM was present as p15 in anti-HA-reactive spikes from the wt/LI-HA virus compared to 23% ± 1% in spikes captured from the wt/LR-HA virus. We concluded that the R peptide cleavage of LI-HA Env present in wt/LI-HA heterotrimers has been rescued. Additional controls showed the lack of reactivity of the αHA antibody toward the wt Env (Fig. 6B, lane 1) and the αHA immunoprecipitation of mostly R-peptide-containing Env from LI-HA virus (Fig. 6B, lane 2). The αSU analyses of total Env content in all virus preparations are shown in Fig. 6A. The results concur with those described above for the corresponding virus preparations with nontagged Env.
FIG. 6.
wt Env rescues the R peptide cleavage of HA-tagged LI Env in wt/LI-HA virus. An HA tag was inserted into the polyproline-rich region of SU in LI and LR Env, and the wt, LI, wt/LI-HA, and wt/LR-HA viruses were prepared. The virus preparations were solubilized with Triton X-100 at 37°C in the presence of EDTA and NEM to transform the spikes into the stabile IAS Env trimers. These were immunoprecipitated with αSU to capture total Env for reducing SDS-PAGE (A) and with αHA (B) to follow trimers containing tagged Env only. A longer exposure of the lower part of the αHA gel analysis with the p15E and Pr15E protein bands is included. The relative amount of p15E (percentage of total TM ± SD; n = 3) is also shown. Note the presence of significantly more R-peptide-cleaved Env (SU-p15E) in the wt/LI-HA virus than in wt/LR-HA virus, suggesting wt Env-mediated cleavage rescue in the former but not in the latter virus. Note also that the reaction of αHA with the tagged Env is weaker than that with αSU.
DISCUSSION
According to our model the R peptides maintain the trimeric endodomain of Env in a configuration that is rather resistant to the viral protease because of partial hiding of the cleavage site. However, when the protease gets the chance to cleave the R peptide in one TM subunit, there is an allosteric change in the other two that reveals their cleavage sites. Therefore, the cleavage of the R peptide from one TM subunit of the Env trimer will be followed very rapidly by the cleavages of the other two TM subunits. Thus, the R peptide cleavage process in the Env trimer appears to be controlled by positive allosteric cooperativity (22).
The model is supported by the surprising rescue of the R peptide cleavage in the LV and LI Env mutants by the wt Env in heterotrimers. The cleavage site of the Pr15E is apparently much less sensitive in the endodomain of the mutant homotrimers than it is in the wt homotrimer. Accordingly, the cleavage of the mutant homotrimers will be inhibited. However, in a wt/mutant heterotrimer, the wt cleavage site will be sensitive to proteolysis, and this will through allostery mediate the sensitivity also to the mutant cleavage site(s).
The structure of the endodomain of the Env trimer with the R peptide is not known, but it has been modeled as a coiled coil (20). This has been suggested to stabilize the prefusion structure of the spike. The model has been supported by the phenotypes of mutants designed to perturb the proposed coiled-coil interaction (20, 24). In such a structure the cleavage of the R peptide of one TM subunit is expected to disturb the coiled-coil interaction, possibly resulting in the release of the uncleaved R peptide tails of the other two subunits in a protease-sensitive form.
The consequence of the cooperative R peptide cleavage mechanism is that newly released virus will generate an increasing fraction of R-peptide-cleaved SU-p15E homotrimeric spikes rather than SU-Pr15E/SU-p15E heterotrimers. As the latter spikes were shown to be much less fusion active than the R-peptide-cleaved homotrimers, it will mean that the cooperative cleavage will facilitate the maturation of the virus to fusion competence. The heterotrimers, in particular the ones containing two copies of R-peptide-cleaved SU-p15E complexes, are probably not completely inhibited in mediating virus-cell fusion. This follows the fact that heterotrimerization-mediated complementation of two fusion-defective Envs has been observed in cells (16, 25). In these complementation studies, one of the defective Envs could not interact with the receptor but was devoid of the fusion-inhibiting R peptide, whereas the other one was receptor binding positive but carried the R-peptide.
Previous studies have shown that there is an exclusion of charged amino acids and amino acids with side chains branched at the beta-carbon (Ile and Val) at the P1 position of the viral protease cleavage site (13). The fact that that the L649V and L649I Env mutants were so efficiently cleaved when presented together with the wt in heterotrimers clearly violates this rule about cleavage site specificity. Apparently, the specificity can be modulated by the endodomain configuration. However, the L649R Env mutant was not cleaved when presented in the same context. This supports previous results and suggests that charged residues in the P1 position of the cleavage site cannot be tolerated by the MoMLV protease, independent of the context in which it is presented. Our experiments with the protease inhibitor amprenavir showed that inhibition of the R peptide cleavage in Env required a much lower drug concentration than cleavage of the Gag precursor. It is possible that the R peptide cleavage site is less accessible to the viral protease, requiring higher concentrations of active protease for cleavage (and lower concentrations of amprenavir for cleavage inhibition) than the sites in the Gag precursor.
The cleavage of the R peptide induces a structural change in the Env trimer that facilitates the receptor-induced activation of the spike for membrane fusion (1, 9). The cooperativity that we found for the R peptide cleavage reaction in the Env trimer helps explain how symmetry and functionality can be maintained during this structural transition. We anticipate that similar cooperativity will characterize the other maturation cleavage event of the Env trimer, i.e., the furin-mediated cleavage of the Env precursor gp80 in the virus-infected cell. The mechanism of protease cleavage cooperativity might also apply to the cleavage of the Gag precursors to generate the submembranous MA layer, the capsid, and the nucleocapsid structures of the mature virus.
Acknowledgments
Swedish Science Foundation grant 2778 and Swedish Cancer Foundation grant 0525 to H.G. supported this study.
Footnotes
Published ahead of print on 12 January 2011.
REFERENCES
- 1.Aguilar, H. C., W. F. Anderson, and P. M. Cannon. 2003. Cytoplasmic tail of Moloney murine leukemia virus envelope protein influences the conformation of the extracellular domain: implications for mechanism of action of the R Peptide. J. Virol. 77:1281-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colicelli, J., and S. P. Goff. 1988. Sequence and spacing requirements of a retrovirus integration site. J. Mol. Biol. 199:47-59. [DOI] [PubMed] [Google Scholar]
- 3.Dong, J. Y., J. W. Dubay, L. G. Perez, and E. Hunter. 1992. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein define a requirement for dibasic residues for intracellular cleavage. J. Virol. 66:865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feher, A., et al. 2006. Characterization of the murine leukemia virus protease and its comparison with the human immunodeficiency virus type 1 protease. J. Gen. Virol. 87:1321-1330. [DOI] [PubMed] [Google Scholar]
- 5.Green, N., et al. 1981. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc. Natl. Acad. Sci. U. S. A. 78:6023-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallenberger, S., et al. 1992. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360:358-361. [DOI] [PubMed] [Google Scholar]
- 7.Henderson, L. E., R. Sowder, T. D. Copeland, G. Smythers, and S. Oroszlan. 1984. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described gag and env cleavage products. J. Virol. 52:492-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubo, Y., and H. Amanuma. 2003. Mutational analysis of the R peptide cleavage site of Moloney murine leukaemia virus envelope protein. J. Gen. Virol. 84:2253-2257. [DOI] [PubMed] [Google Scholar]
- 9.Löving, R., K. Li, M. Wallin, M. Sjöberg, and H. Garoff. 2008. R-peptide cleavage potentiates the fusion controlling isomerization of the intersubunit disulfide in Moloney murine leukemia virus Env. J. Virol. 82:2594-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng, V. L., T. G. Wood, and R. B. Arlinghaus. 1982. Processing of the env gene products of Moloney murine leukaemia virus. J. Gen. Virol. 59:329-343. [DOI] [PubMed] [Google Scholar]
- 11.Opstelten, D. J., M. Wallin, and H. Garoff. 1998. Moloney murine leukemia virus envelope protein subunits, gp70 and Pr15E, form a stable disulfide-linked complex. J. Virol. 72:6537-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ou, W., N. Lu, S. S. Yu, and J. Silver. 2006. Effect of epitope position on neutralization by anti-human immunodeficiency virus monoclonal antibody 2F5. J. Virol. 80:2539-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pettit, S. C., et al. 1991. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J. Biol. Chem. 266:14539-14547. [PubMed] [Google Scholar]
- 14.Ragheb, J. A., and W. F. Anderson. 1994. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J. Virol. 68:3220-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rein, A., J. Mirro, J. G. Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 68:1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rein, A., C. Yang, J. A. Haynes, J. Mirro, and R. W. Compans. 1998. Evidence for cooperation between murine leukemia virus Env molecules in mixed oligomers. J. Virol. 72:3432-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schultz, A., and A. Rein. 1985. Maturation of murine leukemia virus env proteins in the absence of other viral proteins. Virology 145:335-339. [DOI] [PubMed] [Google Scholar]
- 18.Shapiro, S. Z., M. Strand, and J. T. August. 1976. High molecular weight precursor polypeptides to structural proteins of Rauscher murine leukemia virus. J. Mol. Biol. 107:459-477. [DOI] [PubMed] [Google Scholar]
- 19.Sjöberg, M., B. Lindqvist, and H. Garoff. 2008. Stabilization of TM trimer interactions during activation of Moloney murine leukemia virus Env. J. Virol. 82:2358-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor, G. M., and D. A. Sanders. 2003. Structural criteria for regulation of membrane fusion and virion incorporation by the murine leukemia virus TM cytoplasmic domain. Virology 312:295-305. [DOI] [PubMed] [Google Scholar]
- 21.Wallin, M., M. Ekstrom, and H. Garoff. 2004. Isomerization of the intersubunit disulphide-bond in Env controls retrovirus fusion. EMBO J. 23:54-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitty, A. 2008. Cooperativity and biological complexity. Nat. Chem. Biol. 4:435-439. [DOI] [PubMed] [Google Scholar]
- 23.Wu, S. R., et al. 2008. Turning of the receptor-binding domains opens up the murine leukaemia virus Env for membrane fusion. EMBO J. 27:2799-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang, C., and R. W. Compans. 1997. Analysis of the murine leukemia virus R peptide: delineation of the molecular determinants which are important for its fusion inhibition activity. J. Virol. 71:8490-8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao, Y., S. Lee, and W. F. Anderson. 1997. Functional interactions between monomers of the retroviral envelope protein complex. J. Virol. 71:6967-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu, N. L., P. M. Cannon, D. Chen, and W. F. Anderson. 1998. Mutational analysis of the fusion peptide of Moloney murine leukemia virus transmembrane protein p15E. J. Virol. 72:1632-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]