Abstract
Hepatitis C virus infections proceed to chronicity in the majority of cases. In patients, hepatitis C viruses exist as a dynamic and complex quasispecies. The dominant species at any one time arises in response to host immune pressure and other, incompletely understood factors. It is critical to understand all the mechanisms by which dominance is achieved, but this is difficult to study in vivo. Therefore, it would be useful to develop a cell culture system in which naturally occurring quasispecies could be studied. Hepatitis C virus glycoprotein genes E1 and E2 were PCR amplified as a cassette from the plasma of a chronically infected patient and shotgun cloned into a modified 1a/JFH1 infectious cDNA clone. Following transformation of bacteria, plasmids were batch harvested, transcribed, and transfected into Huh7.5 cells to produce a quasispecies of hypervariable region 1 (HVR1) that mimicked that circulating in vivo. Serial passage of the quasispecies in vitro resulted in replacement of the initially dominant species with a new HVR1 species coexisting with selected growth-enhancing mutations located outside HVR1. Antibody raised against one HVR1 sequence neutralized virus with the homologous HVR1 and cross-neutralized virus with a different sequence. Reciprocal swapping of the HVR1 regions between the two dominating species demonstrated that the HVR1 sequence affects the efficiency of replication and of neutralization by anti-HVR1 but that both processes are strongly influenced by regions outside HVR1.
Hepatitis C virus (HCV) is a small, enveloped RNA virus with a positive-sense genome that encodes nonstructural proteins required for replication and/or assembly, a capsid or core protein, and two glycoproteins, E1 and E2, that form heterodimers which comprise the virus receptor. The virus is a member of the Flaviviridae family, genus Hepacivirus (8), and is a significant human pathogen which is transmitted mainly through blood or blood products. One hundred seventy million people worldwide are infected by HCV, and it is the major cause of chronic hepatitis: severe sequelae include liver cirrhosis, hepatocellular carcinoma, and death (11). Presently, antiviral therapy includes interferon and ribavirin treatment (14, 18), but the efficacy of these drugs varies depending on the HCV genotype, host factors, and treatment dose and schedule (6, 9). Unfortunately, the side effects of the drug treatment can be severe. For these reasons, vaccine alternatives to drug treatment have been pursued vigorously but with limited success (16).
Vaccine development has been stymied by the extreme quasispecies diversity of HCV (3). Antibodies to the glycoproteins usually arise in the chronic phase of infection, and virus persists even in the presence of neutralizing antibodies, presumably because neutralization escape mutants are selected from the vast quasispecies. Hypervariable region 1 (HVR1), located at the N terminus of E2, is the most variable domain in HCV and may play a role in immune evasion (4, 15). The HVR1 region is thought to be involved in cell attachment through interaction with scavenger receptor class B type 1 (13). Although antibodies to HVR1 could neutralize virus after attachment to cultured hepatoma cells, they did not inhibit binding to CD81, another HCV receptor (19). However, viruses lacking HVR1 were no longer neutralized by anti-HVR1 and exhibited impaired fusion (1). A deletion mutant lacking the HVR1 region was viable, although attenuated, in chimpanzees (5). Antibodies directed against HVR1 can neutralize the virus, and it has been proposed to serve as a decoy to divert the humoral immune response away from more critical regions. The results of recent studies of JFH1-based recombinant viruses lacking HVR1 suggested that HVR1 shields important conserved neutralization epitopes (1, 10). Until recently, studies of HCV replication and neutralization had to be performed in chimpanzees, but the development of the HCV pseudoparticle system provided a more widely available system with which to study virus-cell interactions and neutralization (2). Even more useful was the subsequent development of the JFH1 cell culture system, since this genotype 2a virus strain could complete the entire replication cycle of HCV in vitro (20). Although the JFH1 virus remains the only strain to be successfully cultured, chimeric viruses which express the structural proteins of the other genotypes from the JFH1 backbone were subsequently developed, and all are available as infectious cDNA clones (7, 21). In the current proof-of-principle study, we have explored the possibility of utilizing the chimeric genotype 1a/JFH1 cDNA clone to reproduce and study a naturally occurring quasispecies of HVR1.
MATERIALS AND METHODS
Shotgun cloning.
Acute-phase plasma (40 μl) from patient H, containing 106.5 50% chimpanzee infectious doses of HCV per ml, was extracted with Trizol LS (Invitrogen, Carlsbad, CA), and HCV RNA was reverse transcribed with Superscript II reverse transcriptase (RTase; Invitrogen). The first step of nested PCR was performed as long PCR (17), with KlenTaq enzyme (Clontech, Mountain View, CA) and primers, of core and the 3′ untranscribed region as described previously (12). The second-step PCR used E1- and E2-specific primers. The sense primer (5′-TGCCCGCTTCAGCCTACCAAGTTCGAAATTCC-3′) contained an introduced unique Bsp119I restriction site (boldface) at the 5′ end of E1, and the antisense primer (5′-GATGCTGCATTGAGTATTACGAGGTTCTCCAGCGCT-3′) contained an introduced AfeI restriction site (boldface) at the 3′ end of E2; neither introduced restriction site altered the amino acid sequence.
The backbone plasmid, a gift from S. Lemon, consisted of the 1a/2a intergenotypic chimera H77-NS2/NS3-JFH1/Q125L, a chimera that had been adapted to grow in cell culture (21). A naturally occurring AfeI restriction site was abolished, and Bsp119I and a new AfeI restriction site were introduced by standard methods at the beginning of E1 and the end of E2. Following digestion of the plasmid and the plasma PCR products with Bsp119I and AfeI, the two components were purified on agarose gels, ligated together, and transformed into the JM109 strain (Promega, Fitchburg, WI) of Escherichia coli. Individual colonies were picked from one plate for sequencing to determine quasispecies distribution, while colonies on a duplicate plate, consisting of 115 well-separated colonies and about 50 overlapping colonies, were batch harvested as a mixture for the production of a quasispecies. Primer sequences for plasmid construction or PCR amplification are available on request.
Transfection of Huh7.5 cells.
The mixture of quasispecies plasmids was linearized with XbaI, and 50 μg of DNA was transcribed with T7 RNA polymerase (Promega) as described previously (12). The RNA reaction mixture was mixed with DMRIE-C transfection reagent (Invitrogen) and inoculated onto Huh7.5 cells. Transfected cells were grown at 37°C in the CO2 incubator, and medium was harvested at the indicated times for HCV sequence analysis and serial passage. The whole experiment was performed twice with very similar results; therefore, only data from the first experiment are shown.
Recovery of viral quasispecies and sequence analysis.
HCV was extracted with Trizol LS from 100-μl amounts of cell culture medium harvested at the times indicated below and was amplified with the E1 and E2 primers described above. The product was digested with Bsp119I and AfeI, cloned into the modified 1a/2a vector, and sequenced for either HVR1 or the entire E1E2 sequence. At least 18 colonies were analyzed for each time point.
Infectivity determinations.
Medium from cultured cells was plated on Huh7.5 cells growing in Lab-Tek 8-well chamber slides (Thermo Fisher Scientific, Waltham, MA) and incubated at 37°C. Three days postinfection, cells were fixed in 100% acetone and incubated for 20 min with mouse anticore or chimpanzee 1530 anti-HCV plasma. After 20 min, slides were rinsed and incubated with Alexa Fluor 568 antimouse or Alexa Fluor 488 antihuman serum (Invitrogen) to detect core or core and E1E2 proteins, respectively. Immunofluorescence microscopy was performed with an Axioskop 2-Plus Zeiss microscope and individual foci (focus-forming units [FFU]) were counted manually.
Neutralization by anti-HVR1 antibody.
LMF87 is a rabbit polyclonal antibody raised against a 21-amino-acid peptide containing the HVR1 sequence of species A (4). Triplicate aliquots of 100 μl each of virus in cell culture medium were mixed with control serum or anti-LMF87 serially diluted with complete Dulbecco's modified Eagle's medium (DMEM), incubated at 37°C for 1 h, and inoculated onto Huh7.5 cells growing in 8-well chamber slides. After 5 h, the inoculum was replaced with complete DMEM and incubation was continued for 2 days. Cells were stained for immunofluorescence microscopy as described above, and the foci were counted.
Mutagenesis.
Backbone mutations were reverted or HVR1 regions were swapped by standard techniques. Primers and detailed procedures will be provided on request.
RESULTS
Construction of a quasispecies library.
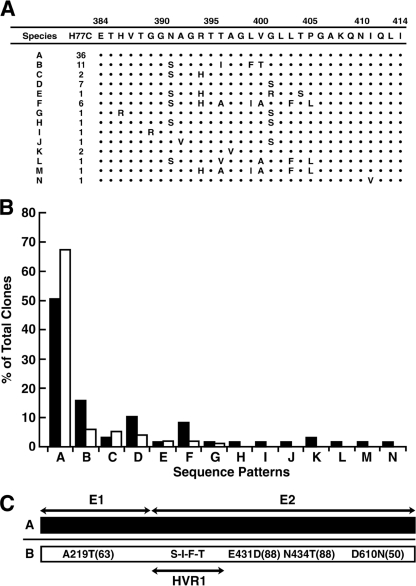
First, we determined whether the HCV glycoprotein quasispecies present in a human plasma could be faithfully reproduced as a cDNA population of full-length, replication-competent genomes that grew in cell culture. As a source of quasispecies, H77 plasma collected in 1977 from patient H during the early acute phase of posttransfusion hepatitis C was chosen. The E1E2 region of this genotype 1a HCV was amplified from the plasma by reverse transcription (RT)-PCR as a single fragment which was then shotgun cloned into an infectious cDNA chimeric clone that expressed genotype 1a structural proteins in cell culture; therefore, each E1E2 combination of glycoproteins represented a naturally occurring, coevolved gene pair. The cDNA mixture was transformed into E. coli cells, and plasmids from 72 colonies were sequenced to determine the HVR1 diversity. The HVR1 species distribution in the full-length cDNA clones was very similar to that obtained previously by cloning much shorter RT-PCR products from the same plasma (Fig. 1A and B). Therefore, the HVR1 quasispecies population of the cDNA chimera appeared to accurately mimic the quasispecies population in the human plasma. The sequence of the entire E1E2 region was determined for the two dominant species, A and B. Species A contained no consistent differences from H77C among 22 clones sequenced, whereas, in 7 of 8 clones, species B, in addition to the differences in HVR1, contained a set or subset of four new amino acids outside HVR1 (Fig. 1C).
FIG. 1.
Quasispecies analysis of shotgunned clones. (A) The HVR1 amino acid sequences in comparison to the sequence of H77C and the number of clones encoding each sequence are shown. (B) Comparison of HVR1 sequences in shotgunned clones (solid bars) with those found by Farci et al. (open bars) (4). Species H through N were not detected in the previous study. (C) The predominant amino acid sequences for the entire E1E2 region of species A and B are shown as differences from H77C. The original amino acid is followed by the amino acid position in H77C and the new amino acid; the percentage of clones with that amino acid is given in parentheses. S-I-F-T indicates mutations N391S, T396I, L399F, and V400T in HVR1 of species B.
Replacement of the dominant quasispecies during sequential passage in cell culture.
The quasispecies of HCV chimeras was prepared by pooling the library of E. coli colonies on a petri dish and isolating a single preparation of plasmid DNAs. The plasmid mixture was batch transcribed, and the library of viral genomes was transfected into Huh7.5 cells. Viruses released into the medium were serially passaged three times to naïve Huh7.5 cells. Viruses in the medium were sequenced at the time of passage (Fig. 2A). Even by the first time point, the complexity of the quasispecies mixture was reduced. The initial ratio of species A to B was maintained until the second serial passage, when species B gained dominance. After the third serial passage, species A was no longer detected and 100% of recovered sequences were species B.
FIG. 2.
Evolution of HVR1 and E1E2 quasispecies during long-term culture. (A) Huh7.5 cells were transfected with the quasispecies chimeras shown in Fig. 1A. The quasispecies viruses released into the medium at day 18 were sequenced and used to infect naïve Huh7.5 cells. This step was repeated at days 50 and 65, and the final sequences were obtained from the day-75 sample. Species A, B, D, and N were identified in the original inoculum, and the level of abundance of each is expressed as the percentage of viruses sequenced; species without a letter designation were not detected in the inoculum. (B) Consensus sequence of E1E2 of species A and B chimeras. See Fig. 1 legend for details.
A repeat experiment with a slightly modified schedule of virus collection produced similar results. Quasispecies diversity diminished with time, and the originally dominant species A disappeared and was replaced with species B (data not shown). Since the kinetics of the rise of species B to dominance suggested the presence of an adaptive mutation, the entire E1E2 region was sequenced.
In both experiments, at the time species B became dominant, the region outside HVR1 contained 5 (Fig. 2B) or 6 (not shown) amino acid changes in 100% of the clones sequenced. Four of the changes were present in both experiments, and 3 of the 4 (E431D, N434T, and D610N) were already present in all of the genomes from the earliest time sequenced (Fig. 2B). In both experiments, species A genomes also contained one or more amino acid differences after the first passage, but the level of species A still decreased thereafter.
Effects on growth of changes within or outside HVR1.
In order to estimate the relative importance to growth in cell culture of the selected mutations outside HVR1 compared to that of the differences within HVR1, the HVR1 sequence of species A was replaced with that of species B and vice versa. In both cases, the backbone of the species contained the mutations that were present when the species was at its highest level, according to the results shown in Fig. 2B. Four days posttransfection, the yield of infectious virus released into the medium was quantified and compared to that released by the wild-type 1a/2a chimera (Fig. 3A and B). Evolved species A and B (A-Evolved and B-Evolved), containing their respective homologous HVR1 regions, produced yields of infectious virus that were similar to each other and about 7-fold greater than that of the wild type. Swapping the species A and B HVR1 regions affected virus yield but not in the way expected. The species A HVR1, surprisingly, increased in virus yield when in the B backbone (B-Swapped), whereas the species B HVR1 decreased yield when in the A backbone (A-Swapped). These results suggested that the changes outside HVR1 were more important for growth than was the sequence of HVR1.
FIG. 3.
Effects of HVR1 sequence and non-HVR1 mutations on growth in Huh7.5 cells (A) Diagram of “evolved” species showing mutations that were selected during passage and were present in the backbone of the majority of genomes at the time of peak virus titer, and diagram of “swapped” species in which the HVR1 regions of species A and B were swapped by PCR mutagenesis. Species A and B sequences are shown in black and white, respectively. (B) Infectious viruses released into the medium at day 4 posttransfection were quantified (FFU) by a focus-forming assay. The wild-type 1a/2a chimera served as a standard and was assigned a value of 1. Species A and B regions are shown in black and white, respectively. Error bars show standard deviations.
The changes outside HVR1 in species B-Evolved were eliminated by site-directed mutagenesis, and the effects on virus yield were determined (Fig. 4). Back mutation of the amino acid at 219 had little effect, but back mutation of the amino acid pair at 431 and 434 or the amino acid at 348 or 610 decreased virus yield significantly (Fig. 4B). These same mutations were joined by A592T in the B-Evolved species from the second experiment. Back mutation of A592T did not decrease virus yield, whereas back mutation of the amino acids at 431 and 434 or at 610 again significantly decreased virus yield, demonstrating that they remained important in the context of a slightly different background (data not shown).
FIG. 4.
Effects of backbone mutations on growth of species B. (A) Diagram of backbone mutations present in the dominant B species at peak titer in the two experiments. (B) Ratio of released infectious test virus and the wild type. Wild-type virus (crosshatched) was set at 1. A delta indicates that the mutation was reverted back to the wild type. Error bars show standard deviations.
Neutralization by anti-HVR1 is affected by sequences outside HVR1.
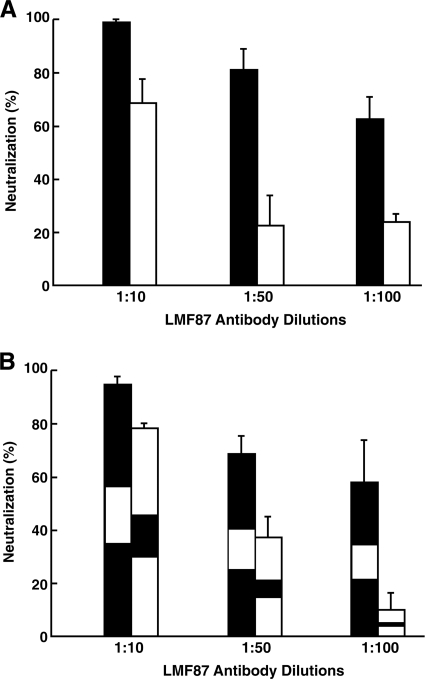
LMF87 antibody raised against a peptide of species A HVR1 was thought to be specific for species A, since treatment of the quasispecies (represented in Fig. 1) with this antibody resulted in the elimination of species A and the dominance of species B when the antibody-antigen mixture was tested for infectivity by inoculation into a chimpanzee (4). In order to determine whether LMF87 antibody was specific for species A or whether it cross-reacted with species B HVR1, species A-Evolved and species B-Evolved, as shown in Fig. 3, were individually mixed with LMF87 anti-HVR1 and infectivity was quantified by a focus-forming assay. Species A was neutralized in a dose-dependent manner, but species B neutralization, although less efficient, was significant (Fig. 5A). In order to rule out any effect of sequences outside the HVR1 region, the experiment was repeated with the swapped HVR1 species A and B viruses shown in Fig. 3. Surprisingly, the pattern was reversed; the chimera containing species A HVR1 in the species B backbone was neutralized less efficiently than the chimera containing the species B HVR1 in the species A backbone (Fig. 5B). Therefore, not only does species A anti-HVR1 cross-react with species B HVR1, but the extent of neutralization is greatly influenced by sequences outside HVR1.
FIG. 5.
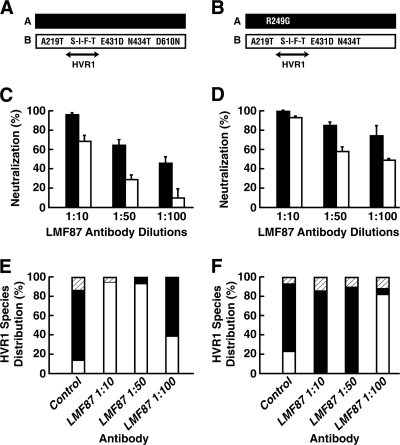
Neutralization of species A and B and “swapped” chimeras by rabbit antibody LMF87 raised against the HVR1 peptide of species A. (A) Huh7.5 cells were transfected with RNA transcripts from species A (black bars) or B (white bars), viruses in the medium at day 4 posttransfection were mixed with dilutions of LMF87 or control serum, and the number of infectious viruses was determined in a focus-forming assay. % Neutralization = 1 − (FFU in LMF87 sample/FFU in control sample) × 100. (B) HVR1 regions of species A (black) and species B (white) were swapped by PCR mutagenesis, and neutralization by LMF87 was determined as described for panel A. Error bars show standard deviations.
Outgrowth of viruses from a mixture incompletely neutralized by LMF87.
In order to better understand the implications of incomplete neutralization in the context of a quasispecies, we selected two versions of species A and two versions of species B (Fig. 6A and B) and tested them individually for neutralization by anti-HVR1.
FIG. 6.
Effects of backbone mutations on virus outgrowth following neutralization by LMF87 antibody raised against the HVR1 peptide of species A. (A and B) Diagrams of species A (black) and B (white) pairs tested. (C and D) Respective percentage of neutralization by LMF87. Error bars show standard deviations. (E and F) Mixtures (50:50) of species A and B as diagramed in panels A and B were incubated with control serum or with dilutions of LMF87 anti-HVR1 prior to inoculation onto Huh7.5 cells. After 42 days of culture, the HVR1 sequences of viruses in the medium were sequenced to determine the species distribution. Species A (black); species B (white); new mutation in HVR1 (hatched).
In each case, species B was more resistant to neutralization than species A, but inclusion of the D610N mutation in species B or removal of the R249G mutation from species A decreased the extent of neutralization for that species (Fig. 6C and D). The A and B species were then paired as indicated in the figure, and the mixtures were incubated with different concentrations of LMF87 antibody and inoculated onto Huh7.5 cells to permit amplification of nonneutralized virus. Forty-two days later, the HVR1 sequences of viruses in the medium were determined. Even though each of the two species differed from its counterpart only by a single amino acid, the results from the two sets of cultures were completely opposite (Fig. 6E and F). The antibody-negative controls for the two sets were virtually identical, and species A predominated. The combination of unmodified species A and species B-Evolved resulted in outgrowth of species B (Fig. 6E), as might be expected since species A was more efficiently neutralized and the mutations in species B enhanced growth, as shown in Fig. 4. However, the outgrowth of species A containing an R249G mutation (Fig. 6F) was more difficult to explain, because this mutant appeared to be neutralized even more efficiently than the other species A used (Fig. 6, compare C and D). However, note that species B in the right panels lacked the D610N growth-enhancing mutation (Fig. 4).
DISCUSSION
The complex E1E2 quasispecies circulating in a patient was reproduced as a chimeric quasispecies within a homogeneous HCV genomic backbone that grew in cell culture. This development provided an in vitro system with which to study factors resulting in the dominance of a particular glycoprotein species. Because tandem E1 and E2 genes were amplified as a single unit, the two glycoproteins encoded by each virus had coevolved and, thus, were expected to be functional. This assumption was confirmed by recovering viruses D, N, and K (Fig. 2A and repeat experiment, not shown) that were present in low copy number in the inocula but must have replicated successfully in order not to have been diluted out.
Although species A was 3-fold more abundant than species B in the transfection inoculum, species B eventually gained dominance, suggesting that the non-HVR1 mutations that distinguished species B from species A were candidates for adaptive mutations. Indeed, as the results of reversion mutagenesis indicated, the mutations at I348T, E431D and/or N434T, and D610N all individually contributed to species B's growth (Fig. 4). Since E341D/N434T and D610N were already colocalized within viral genomes in the patient, they most likely enhanced growth in vivo as well as in vitro. However, it was surprising that the A219T mutation did not seem to enhance growth in cell culture since it was detected in early passages in 100% of the species B clones in two independent experiments (Fig. 2 and repeat experiment, not shown), consistent with its conveying a growth advantage. The fact that 5 of the 8 species B clones isolated from the patient already contained this mutation suggested that it may have offered a selective advantage in vivo but was neutral in vitro and, therefore, was passively retained. The comparative release of virus into the medium by species A-Evolved and species B-Evolved viruses containing the mutations present at their peak prevalence in cell culture suggested that the I348S mutation in species A, like the I348T mutation in species B, was adaptive, since the growth of species A-Evolved virus was significantly greater than that of the wild-type 1a/2a control, which differed only in lacking this mutation (Fig. 3A and B). However, this mutation was not detected in the second experiment and was never present in 100% of the A species clones throughout the first experiment. This may explain why species B dominated in the long term even though species A-Evolved and B-Evolved viruses released similar amounts of progeny virus into the medium in the short term (Fig. 3). Note that since the data in Fig. 3 were collected 4 days posttransfection, they reflect the rate of replication, assembly, and egress but not infection. Therefore, the A-Evolved virus, as has been shown for other HCV mutants (12), might have been able to exit but not to enter cells efficiently and, thus, not able to spread as rapidly as the B-Evolved virus.
The fact that non-HVR1 mutations in E1 or E2 enhanced growth in cell culture was not surprising, since enhancing mutations have been demonstrated for many HCV variants. In contrast, it was surprising that these mutations had such a profound and nonpredictable effect when paired with the heterologous HVR1. Even though both species A-Evolved and B-Evolved viruses grew to similar extents, swapping the HVR1 regions from one background to the other led to decreased growth of the chimeric virus in one case (A-Swapped) and increased growth in the other (B-Swapped) (Fig. 3).
Even more remarkable was the effect that these non-HVR1 mutations had on virus neutralization by the LMF87 anti-HVR1 antibody. The fact that LMF87 antibody neutralized species A more efficiently than species B (Fig. 5A) was understandable, since the antibody was raised against a peptide with an amino acid sequence that perfectly matched the HVR1 of species A and differed from that of species B in 4 of 21 positions (4). The hierarchy of neutralization was reversed when the HVR1 regions were swapped between species A and B, such that the virus containing the homologous species A HVR1 region was neutralized less efficiently than that containing the cross-reactive species B sequence. These data suggest that although HVR1 occupies the terminus of the E2 protein, it does not function as an independent unit: the neutralization data comparing authentic E2 and HVR1-swapped E2 suggest that access to the relevant epitope(s) or avidity for the epitope(s) is greatly affected in unpredictable ways by mutations in other parts of the molecule. Recently, HVR1 was proposed to have different functional roles in different isolates (10). Reverse genetic studies with JFH1-based recombinant viruses of genotypes 1 to 6 demonstrated that the deletion of HVR1 from 3 of the 8 recombinants had little effect on virus spread or infectivity in vitro; in contrast, the deletion of HVR1 from 5 of the 8 recombinants produced viruses that were severely attenuated but, with one exception, could be rescued by the acquisition of adaptive mutations in E1 or in the remaining portion of E2 (10). In toto, these data suggest that ignoring distal variations in mutational studies and focusing on limited regions or motifs to the exclusion of genomic context may be unwise.
The confusing data from experiments examining the outgrowth of a virus quasispecies following incomplete neutralization (Fig. 6E and F) emphasize that dominance reflects a delicate balance between the initial concentration of viable virus (nonneutralized) and the fitness of the virus and that seemingly small differences can determine which species ultimately achieves dominance.
In summary, it is possible to create a quasispecies of HCV glycoproteins that represent a natural population and that can be analyzed in toto or in parts in an in vitro cell culture system. Although only genotype 1a was examined in the present study, this strategy should be applicable to almost any HCV strain for which a cDNA clone encoding a cell culture-adapted virus of that particular genotype exists. Gottwein and colleagues (7) have already developed cDNA clones of cell culture-adapted viruses for the remaining genotypes, 2 to 7, which, just like the genotype 1a clone used here, encode JFH1-based recombinant viruses expressing core/E1/E2/p7/NS2 of each genotype. Therefore, it should be straightforward to insert unique restriction sites abutting the E1E2 region to construct a cassette which can be used to generate an E1E2 quasispecies in the background of the homologous core and p7/NS2 proteins. The generation of a representative glycoprotein quasispecies is another step toward the development of a cell culture model that more closely reproduces the complexity of the in vivo situation. It provides a promising system to study virus replication and the dynamics of HCV replication and neutralization.
Acknowledgments
We thank Stanley Lemon (University of North Carolina, Chapel Hill, NC) for providing the 1a/2a chimera clones and Charles Rice (Rockefeller University, New York, NY) for providing the Huh7.5 cell lines.
This research was supported by the Intramural Research Program of NIH, Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 26 January 2011.
REFERENCES
- 1.Bankwitz, D., et al. 2010. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J. Virol. 84:5751-5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642.12615904 [Google Scholar]
- 3.Bukh, J., R. H. Miller, and R. H. Purcell. 1995. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin. Liver Dis. 15:41-63. [DOI] [PubMed] [Google Scholar]
- 4.Farci, P., et al. 1996. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. U. S. A. 93:15394-15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forns, X., et al. 2000. Hepatitis C virus lacking the hypervariable region 1 of the second envelope protein is infectious and causes acute resolving or persistent infection in chimpanzees. Proc. Natl. Acad. Sci. U. S. A. 97:13318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ge, D., et al. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461:399-401. [DOI] [PubMed] [Google Scholar]
- 7.Gottwein, J. M., et al. 2009. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology 49:364-377. [DOI] [PubMed] [Google Scholar]
- 8.Lindenbach, B. D., and C. M. Rice. 2007. Flaviviridae: the viruses and their replication, p. 1101-1152. In D. M. Knipe et al. (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 9.Mbow, M. L., and R. T. Sarisky. 2004. What is disrupting IFN-alpha's antiviral activity? Trends Biotechnol. 22:395-399. [DOI] [PubMed] [Google Scholar]
- 10.Prentoe, J., et al. 1 December 2010. Hypervariable region 1 differentially impacts viability of hepatitis C genotype 1-6 strains and impairs virus neutralization. J. Virol. doi: 10.1128/JVI.01594-10. [DOI] [PMC free article] [PubMed]
- 11.Ray Kim, W. 2002. Global epidemiology and burden of hepatitis C. Microbes Infect. 4:1219-1225. [DOI] [PubMed] [Google Scholar]
- 12.Russell, R. S., et al. 2008. Advantages of a single-cycle production assay to study cell culture-adaptive mutations of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 105:4370-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scarselli, E., et al. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimakami, T., R. E. Lanford, and S. M. Lemon. 2009. Hepatitis C: recent successes and continuing challenges in the development of improved treatment modalities. Curr. Opin. Pharmacol. 9:537-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimizu, Y. K., et al. 1996. A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell cultures. Virology 223:409-412. [DOI] [PubMed] [Google Scholar]
- 16.Stoll-Keller, F., H. Barth, S. Fafi-Kremer, M. B. Zeisel, and T. F. Baumert. 2009. Development of hepatitis C virus vaccines: challenges and progress. Exp. Rev. Vaccines 8:333-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tellier, R., J. Bukh, S. U. Emerson, R. H. Miller, and R. H. Purcell. 1996. Long PCR and its application to hepatitis viruses: amplification of hepatitis A, hepatitis B, and hepatitis C virus genomes. J. Clin. Microbiol. 34:3085-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson, A., K. Patel, H. Tillman, and J. G. McHutchison. 2009. Directly acting antivirals for the treatment of patients with hepatitis C infection: a clinical development update addressing key future challenges. J. Hepatol. 50:184-194. [DOI] [PubMed] [Google Scholar]
- 19.Vieyres, G., J. Dubuisson, and A. H. Patel. 17 November 2010. Characterization of antibody-mediated neutralization directed against the hypervariable region 1 of hepatitis C virus E2 glycoprotein. J. Gen. Virol. doi: 10.1099/vir.0.028092-0. [DOI] [PMC free article] [PubMed]
- 20.Wakita, T., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi, M., Y. Ma, J. Yates, and S. M. Lemon. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629-638. [DOI] [PMC free article] [PubMed] [Google Scholar]