Abstract
Human APOBEC3H (A3H) has one cytidine deaminase domain (CDD) and inhibits the replication of retrotransposons and human immunodeficiency virus type 1 (HIV-1) in a Vif-resistant manner. Human A3H has five single amino acid polymorphisms (N15Δ, R18L, G105R, K121D, and E178D), and four haplotypes (I to IV) have previously been identified in various human populations. Haplotype II was primarily found in African-derived populations, and it was the only one that could be stably expressed. Here, we identified three new haplotypes from six human population samples, which we have named V, VI, and VII. Haplotypes V and VII are stably expressed and inhibit HIV-1 replication. Notably, haplotype V was identified in samples from all African-, Asian-, and Caucasian-derived populations studied. Using haplotype VII, we investigated the A3H anti-HIV-1 mechanism. We found that A3H virion packaging is independent of its CDD but dependent on a 112YYXW115 motif. This motif binds HIV-1 nucleocapsid in an RNA-dependent manner, and a single Y112A mutation completely disrupts A3H virion incorporation. We further studied the mechanism of A3H resistance to Vif. Although the previously identified APOBEC3G Vif-responsive motif 128DPDY131 is not conserved in A3H, placement of this motif into A3H does not make it become less resistant to HIV-1 Vif. We conclude that stably expressed A3H haplotypes may be more broadly distributed in humans than previously realized, and A3H protein is resistant to Vif. These results have important implications for the role of A3H in retrotransposon and HIV-1 inhibition.
Human APOBEC3 (A3) cytidine deaminases encoded by A3A, A3C, and A3H have one copy of a cytidine deaminase domain (CDD) (HXEX23-28PCX2-4C), and those encoded by A3B, A3DE, A3F, and A3G have two copies (CDD1 and CDD2). All of them have antiretroviral activity, although the level of this activity can vary among different retroviral targets (26, 36, 38). In particular, A3B, A3DE, A3F, A3G, and A3H inhibit human immunodeficiency virus type 1 (HIV-1) replication, with A3G having the most potent anti-HIV activity (2, 6, 7, 34, 43, 47). The CDD1 of A3G is enzymatically inactive, but it interacts in an RNA-dependent manner with the HIV-1 nucleocapsid (NC), which packages A3G into virions (25). In addition, a 124YYFW127 domain is also required for A3G virion packaging (15). Virion incorporation allows A3G to inhibit viral replication by two mechanisms. The first is the cytidine deamination-dependent mechanism, where the enzymatically active CDD2 of A3G catalyzes dC-to-dU deamination of the de novo-synthesized minus-strand viral cDNAs during reverse transcription (RT), converting them into nonfunctional viral genomes (13, 19, 21, 45). The second is the cytidine deamination-independent mechanism, where A3G directly inhibits viral RT at multiple stages, including tRNALys3 priming, initiation and elongation of minus-strand DNA synthesis, and plus-strand DNA transfer (3, 11, 23). A3G also modifies viral cDNAs and blocks their integration (24). In spite of such potent antiretroviral activity, HIV-1 still infects its hosts because of the protective function of its Vif protein. Vif binds both A3G and a Cul5/EloB/EloC E3 ubiquitin ligase (44), causing A3G to be degraded by proteasomes (22, 35, 37). Vif may also inhibit A3G activity via other proteasome-independent mechanisms (30, 33, 37). Vif also neutralizes A3DE and A3F, but not A3B and A3H. Although A3F and A3G share 50% amino acid sequence identity, they use different domains to interact with HIV-1 Vif. Vif binds the A3G N-terminal 124YYFWDPDYQEALR136 domain via three critical residues, 128DPD130, whereas it binds the A3F C-terminal 283CAGEVAEFLARHSNVNLT300 domain for neutralization (15, 32).
Both human A3B and A3H are poorly expressed in vivo. The A3B mRNAs are barely detectable in white blood cells (8), and this gene is also frequently deleted in certain human populations (16). A3H mRNAs are more easily detectable (28), but no expressed protein has been observed (20). We found that the human A3H gene contains a premature termination codon (PTC) in its fifth (last) exon, compared to the A3H gene of Old World monkeys, and has significantly reduced levels of mRNA. After repairing this PTC, we detected a higher level of A3H protein in cell culture and a strong Vif-resistant anti-HIV activity (6). Later, others reported five single amino acid polymorphisms (N15Δ, R18L, G105R, K121D, and E178D) in the human A3H gene, and based on the five single nucleotide polymorphisms (SNPs) that underlie these protein variants, four haplotypes designated with Roman numerals (I, II, III, and IV) were reported to be present in different human populations (27). Both the N15Δ and G105R mutations can independently cause a dramatic decrease in A3H expression, and only the haplotype II (hap II) that does not have these two mutations is stably expressed (27). hap II was most common in the African-American population compared to Asian and European populations and potently inhibits HIV-1 replication (27). It is still unclear if there are any other haplotypes in humans, particularly those that can be stably expressed. So far, several groups have observed the anti-HIV-1 activity of human A3H hap I and II (6, 12, 18, 27, 39), and two groups found that A3H also inhibits the replication of non-long terminal repeat (LTR) retrotransposons, including long interspersed nuclear element 1 (LINE-1) and the short interspersed nuclear element (SINE) Alu (17, 39). However, it is still unclear how A3H is packaged into virions and whether human A3H can be neutralized by HIV-1 Vif. The A3H gene used in our previous studies was hap I, which contains the NRGKE polymorphisms (6). Although it is less stable than hap II, we were still able to express this gene by using the VR vector. We and others (6, 12, 18) reported that HIV-1 Vif could not neutralize human A3H, although we found that Vif from simian immunodeficiency virus (SIV) that infects rhesus macaques (SIVmac) or African green monkeys (AGM) could (6). Recently, Li et al. (20) confirmed that A3H hap I is completely resistant to HIV-1 Vif, but they reported that the A3H hap II (NRRDD) protein was partially sensitive to HIV-1 Vif. It was concluded that the increased Vif sensitivity was due to a single K121D mutation in the A3H protein (20, 46). However, in the investigation carried out by Harari et al. (12), A3H hap II was completely resistant to HIV-1 Vif.
Here, we performed genotype and haplotype analysis of A3H in samples from six major human populations and identified additional haplotypes that can be stably expressed. In addition, we identified the critical domain for A3H virion packaging and found that the K121D mutation does not reduce A3H resistance to HIV-1 Vif.
MATERIALS AND METHODS
Plasmids.
The HIV-1 proviral or subviral constructs pNL4-3, pNL4-3Δvif, pNL-Luc, pNL-LucΔvif, pNL-LucΔenv, pNL-LucΔenvΔvif, pNL-A1 HIV-1 Vif-HA, and pNL-A1 SIVmac Vif-HA were previously used (6). The mam- malian A3 expression vectors pcDNA3.1-humanA3G-V5-6×His, pcDNA3.1-humanA3H-V5-6×His, pcDNA3.1-humanA3H-L-V5-6×His, pcDNA3.1-agmA3H-V5-6×His, and VR-humanA3H were used as before (6, 7). A3H-L is the longer form of A3H produced by converting the PTC, compared to that in Old World monkeys, which we described previously. The pcDNA3.1-humanA3H-FLAG-HA expression vector was created from the pcDNA3.1-humanA3H-V5-6×His vector by inserting a FLAG-tagged hemagglutinin (FLAG-HA) linker after NotI/XbaI digestion. The residues at positions 15, 18, 105, 121, 140, and 178 in our human A3H starting template were Asn (N), Arg (R), Gly (G), Lys (K), Lys (K), or Glu (E), respectively; and those in human A3H-L were N, R, G, K, E, or E. Using these human A3H or agmA3H genes as templates, human A3H hap I to VII, hap II and VII mutants, the agmA3H mutant, huA3H-L mutants, and other human A3H mutants were created by the QuikChange XL site-directed mutagenesis kit (Stratagene). pcDNA3.1 vectors expressing Gag precursor, matrix (MA), capsid (CA), NCp6, or NC fused with a glutathione S-transferase (GST)-V5 tag were used as before (41).
A3H genotype and haplotype analysis.
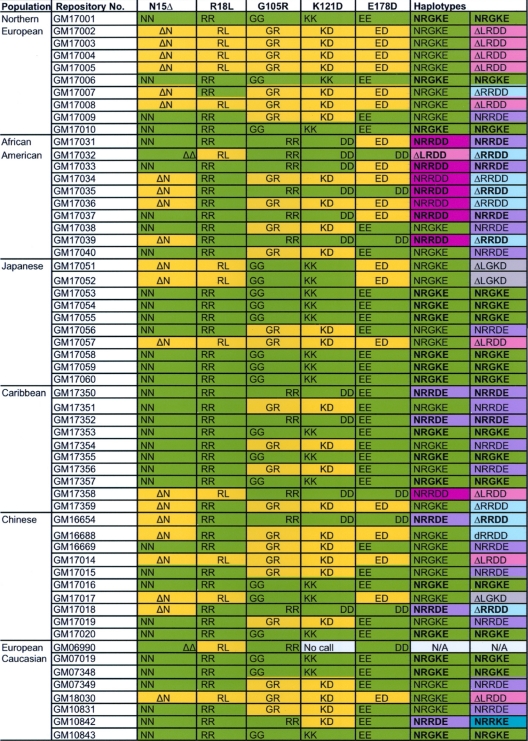
DNA samples from human variation panels (8 to 10 individuals each) were obtained from the Coriell Institute for Medical Research. The panel designations, originating populations, and sample identification (ID) numbers are listed in Table 1. The Northern European and Caucasian European panels are derived from slightly different geographical regions of Europe (see http://www.coriell.org/ for specific details). Previously identified SNPs that cause the amino acid polymorphisms R18L, G105R, K121D, and E178D, as well as the E140K SNP, were genotyped by means of restriction enzyme digestion assays. Assays were based upon naturally occurring restriction sites that could distinguish between alleles for some of the polymorphisms or, for those SNPs with no naturally occurring sites, production of restriction sites by primer mutagenesis. Control digestion sites common to both alleles were included for each assay to verify that complete digestion had occurred. The amplification conditions for the reactions were 5 min at 95°C, followed by 45 cycles of 1 min at 95°C, 2 min at 60°C, and 3 min at 72°C and 1 cycle of 10 min at 72°C. Betaine (1 M) was included for three primer sets because of the high GC content of the amplicon. Digestions were performed in the original amplification reaction with 5 U of restriction enzyme and 50 mM MgCl2 added to bring the final concentration to ∼10 mM. Based upon time course experiments, digestions were complete within 1 h, except for the R18L digestion with Fnu4HI, which requires an overnight digestion to ensure completion (see Fig. S2 in the supplemental material). Sequence analysis of exon 2 for 16 samples showed complete concordance with the results of the diagnostic assay for R18L. Primer sequences, restriction enzymes, and the diagnostic band sizes are given in Fig. S2, S3, S4, S5, and S6 in the supplemental material. For the N15Δ polymorphism, primers were designed to produce a small PCR product that could be genotyped on 15% polyacrylamide gels (see Fig. S1 in the supplemental material). Haplotypes were directly inferred for individuals that were homozygous for all SNPs or had only one heterozygous SNP. The haplotypes for individuals that were heterozygous for 2 or more SNPs were inferred by the method of Clark (4). The haplotypes for 28 of the individuals studied could be directly inferred because they were homozygous for each SNP or all but one SNP.
TABLE 1.
Human A3H genotype and haplotype analysis in six human populationsa
The first two columns show the populations examined and the Coriell Institute codes for the individual samples. Genotypes for the individual positions are either shaded green for homozygotes or yellow for heterozygotes. Haplotypes were either directly determined from the genotypes (with one or zero heterozygous position for the individual indicated by bold letters) or inferred by the method of Clark (4), using the entire 58 samples to determine the directly known haplotypes that are most common. The seven haplotypes are shaded in various colors as follows: I (NRGKE), green; II (NRRDD), dark pink; III (ΔRRDD), light blue; IV (ΔLRDD), pink; V (NRRDE), purple; VI (ΔLGKD), gray; and VII (NRRKE), blue-green.
Vif activity assay.
Vif activity was measured by its ability to rescue ΔVif HIV-1 virus infectivity in the presence of A3G or A3H. Viruses were produced from 293T cells by a standard calcium phosphate transfection. Typically, 21 μg of plasmid DNAs containing 5 μg pNL-LucΔenvΔvif, 5 μg Vif expression vector, 1 μg vesicular stomatitis virus glycoprotein (VSV-G) expression vector, and 10 μg A3 expression vector was transfected into 293T cells in a 100-mm culture dish with 20% confluence. The production of HIV-1 was quantified by p24Gag capture enzyme-linked immunosorbent assay (ELISA). Equal amount of viruses were used to infect GHOST-R3/X4/R5 cells. Thirty-six hours later, cells were lysed in a buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 3 mM EDTA, and 1% Triton X-100. After the nuclei were removed, the cytosolic fraction was used to determine luciferase activity with a luciferase assay kit (Promega).
Virion incorporation assay.
293T cells cultured in 6-well plates were transfected with 3 μg HIV-1 proviral construct pNL4-3Δvif and 3 μg A3 expression vector by a standard calcium phosphate transfection. Supernatants were harvested 48 h after transfection and clarified by low-speed centrifugation at 5,000 × g for 10 min at 4°C. Virions were further purified by spinning the clarified supernatants through a 20% sucrose cushion at 296,000 × g for 30 min at 4°C with the S100AT6 rotor (Sorvall). Pellets were dissolved in phosphate-buffered saline (PBS) and analyzed by Western blotting.
GST pulldown.
The interaction between Gag and A3H was determined by GST pulldown. Briefly, human A3H and various Gag-GST fusion proteins were expressed in 293T cells. Cells were then lysed with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM EDTA, 5 μg/ml aprotinin, 5 μg/ml leupeptin). The cytosolic fraction was precleared with Sepharose 4B beads. Samples were either treated or untreated with RNase A at 100 μg/ml for 1 h at 4°C and then rocked with prewashed glutathione (GSH)-Sepharose beads for 4 h at 4°C. After being extensively washed with PBS, bead-associated proteins were analyzed by Western blotting.
Western blotting.
Horseradish peroxidase (HRP)-conjugated anti-HA antibody (Roche) or HRP-conjugated anti-V5 antibody (Invitrogen) was used to directly detect the expression of A3G, A3H, and Vif proteins. The anti-green fluorescent protein (GFP) monoclonal antibody and anti-actin polyclonal antibody (C-11) were from Santa Cruz Biotechnology. The anti-human A3H monoclonal antibody was obtained from M. Emerman (20). The anti-HIV-1 Gag monoclonal antibody (no. 3537) was from the NIH AIDS Research and Reference Reagent Program. HRP-conjugated anti-human, anti-rabbit, or anti-mouse immunoglobulin G secondary antibodies were from Pierce. HRP-conjugated antibody was detected with an enhanced chemiluminescence detection kit (Amersham Bioscience).
RESULTS
Genotype and haplotype analysis of the human A3H gene.
To further understand how SNPs affect A3H expression, we genotyped each of the five N15Δ, R18L, G105R, K121D, and E178D polymorphic sites in six major human populations by PCR-based assays. In addition, there is another site, E140K, reported to be polymorphic in the Database of Single Nucleotide Polymorphisms (dbSNP; http://www.ncbi.nim.nih.gov/projects/SNP/) (42), which prompted us to also genotype this reported polymorphism. Human genomic DNA samples from Northern European, African American, Japanese, Caribbean, Chinese, and European Caucasian variation panels were purchased from the Coriell Institute for Medical Research. The genomic region containing each polymorphic site was amplified by PCR with specific primers. After that, the N15Δ deletion was detected by size difference on a 15% polyacrylamide gel, and the alleles for R18L, G105R, K121D, E178D, and E140K were detected by restriction enzyme digestion with Fnu4HI, HhaI, XmnI, SmlI, and BstNI, respectively (see Fig. S1 to S6 in the supplemental material). The frequencies of the genotypes for all SNPs studied were not significantly different from Hardy-Weinberg equilibrium expectations within and across all populations.
The assay for K121D(E) is ambiguous with the respect to the possible D or E alleles. However, a search of GenBank for independently derived and naturally occurring genomic and cDNA alleles suggests that the D allele is much more common than the potential E allele, if it exists at all (data not shown), which is in agreement with the results of a previous study (27). Genotypes for all six sites were called for 57 of the 58 individuals studied, but for one individual (Table 1; GM06990) the K121D(E) assay did not produce an interpretable pattern, and so the SNP for this individual was not included in the allele frequency calculation for this SNP, and the individual was not used in the determination of haplotype frequencies reported in the following section. Individual genotypes and inferred haplotypes are shown in Table 1, and allele frequencies for each SNP are shown in Table 2. Although the potential E140K variable site is reported in the dbSNP to have a K allele frequency of 0.92, we were surprised to find none of the 50 samples from five populations (Northern European, African-American, Japanese, Caribbean, and Chinese) that we genotyped had this allele. Because the K140 allele in dbSNP was reportedly found in the European Caucasian samples from the Coriell Institute, we tested 8 samples from this population but we still did not find a K140 allele.
TABLE 2.
Allele frequencies and average heterozygosities from this study and dbSNP
| Reference SNP | Allele | Frequency |
Allele relationship | Avg heterozygosity |
||
|---|---|---|---|---|---|---|
| This work | dbSNPa | This work | dbSNPa | |||
| rs79323350 | 15N | 0.78 | NAb | Ancestral | 0.36 | NA |
| 15Δ | 0.22 | NA | Derived | |||
| rs139293 | 18R | 0.88 | 0.99 | Ancestral | 0.24 | 0.03 |
| 18L | 0.12 | 0.01 | Derived | |||
| rs139297 | 105R | 0.44 | 0.51 | Ancestral | 0.44 | 0.47 |
| 105G | 0.56 | 0.49 | Derived | |||
| rs139298/99 | 121K | 0.57 | 0.49 | Derivedc | 0.46 | 0.32 |
| 121D | 0.43 | 0.51 | Derivedc | |||
| rs139300 | 140E | 1.00 | 0.08 | Ancestral | 0.00 | NA |
| 140K | 0.00 | 0.92 | Derived | |||
| rs139302 | 178E | 0.72 | 0.49 | Ancestral | 0.33 | 0.37 |
| 178D | 0.28 | 0.51 | Derived | |||
Shown are the allele frequencies reported in dbSNP (Database of Single Nucleotide Polymorphisms; http://www.ncbi.nlm.nih.gov/projects/SNP/) (42), based upon the HapMap allele frequencies for 418 individuals from 4 human populations, except for rs139300, which was examined in 148 individuals. The Coriell Institute European Caucasian sample we studied is drawn from the 148 individuals used for rs139300. Average heterozygosities by dbSNP were obtained from the dbSNP website.
NA, not available in dbSNP.
The ancestral allele (inferred from the chimp and orangutan reference sequences) for 121K and 121D is 121E. To date, there is no evidence that the 121E allele still exists in extant humans, although genomic evidence suggests it did exist in Neanderthal hominoids up to their extinction about 30,000 years ago (see the Discussion).
Of the 32 potential haplotypes (25) for five biallelic SNPs, we identified seven haplotypes among the 57 individuals from six populations included in this part of the study (Table 3). The first four haplotypes were previously described in detail in other studies (27), and the last three are new ones studied here (Table 3). The new haplotypes hap VI and hap VII appear to be rarer than the previously reported four haplotypes, whereas the new haplotype hap V is widely present among the 57 samples. Like hap II, both hap V and VII were stably expressed (see below), but hap V was found in all six population samples at an overall frequency of 0.202, which is greater than those of hap II and VII, which had frequencies of only 0.061 and 0.009, respectively (Table 3). We detected hap II mainly in the African-American population (Table 3), which has previously been shown to retain significantly stable expression and anti-HIV-1 activity (27). Interestingly, hap V is more frequently detected in not only the African-American population, but also the Caribbean and Chinese populations. In contrast, hap VII was found only in the European Caucasian sample. Based upon the stability data given in the following sections, among the African-derived populations, African-American and Caribbean, 9 of 10 and 6 of 10 individuals, respectively, had at least one active allele; 5 of 10 Chinese had active alleles; and only 1 individual each among the 10 Northern European and 10 Japanese samples had active alleles.
TABLE 3.
Summary of human A3H haplotype counts and their frequencies among six populations
| Haplotype no. | Haplotype | Haplotype count |
Frequency | |||||
|---|---|---|---|---|---|---|---|---|
| Northern European | African-American | Japanese | Caribbean | Chinese | European Caucasian | |||
| I | NRGKE | 13 | 4 | 16 | 10 | 10 | 9 | 0.526 |
| II | NRRDD | 0 | 5 | 0 | 0 | 0 | 0 | 0.061 |
| III | ΔDRRDD | 1 | 5 | 0 | 1 | 3 | 0 | 0.070 |
| IV | ΔLRDD | 5 | 1 | 1 | 1 | 1 | 1 | 0.088 |
| V | NRRDE | 1 | 5 | 1 | 8 | 5 | 3 | 0.202 |
| VI | ΔLGKD | 0 | 0 | 2 | 0 | 1 | 0 | 0.026 |
| VII | NRRKE | 0 | 0 | 0 | 0 | 0 | 1 | 0.009 |
A3H protein expression among different haplotypes.
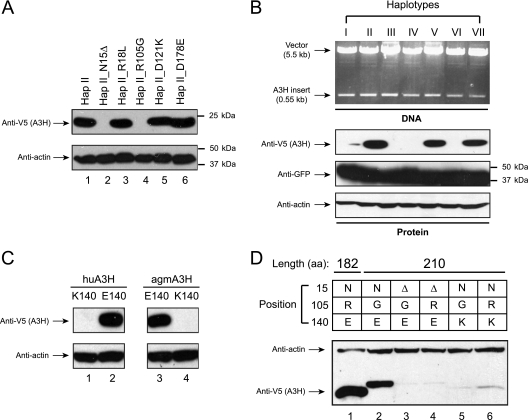
To understand how these polymorphisms affect A3H protein expression, we first introduced five mutations, including N15Δ, R18L, R105G, D121K, and D178E, into the stably expressed hap II gene and determined how its expression was altered. It was found that the wild-type hap II as well as its R18L, D121K, and D178E mutants were all stably expressed, but the expression of both N15Δ and R105G mutants was undetectable (Fig. 1A). This result confirms that either N15Δ or R105G mutation can completely disrupt A3H expression (12, 27, 46). We next compared levels of A3H protein expression among different haplotypes. Each of the seven different haplotype expression vectors was transfected with a GFP expression vector into 293T cells, and A3H and GFP expression was detected by Western blotting. GFP expression was detected from all transfected cells at the same levels, indicating that cells were transfected at a similar efficiency (Fig. 1B). However, A3H expression was only detected from cells transfected with hap II, V, and VII expression vectors, but not the others. Thus, only hap II, V, and VII could be stably expressed. This result is not surprising, because all of these stably expressed haplotypes do not have either of the N15Δ and R105G mutations (Table 3). We also determined whether the E140K mutation affects A3H expression. Although our genotyping data indicate this residue is not polymorphic despite the polymorphic frequency of the K allele being reported in dbSNP, it is possible that the K allele exists as a rare variant. When this mutation was introduced into the hap VII or AGM A3H gene, the expression of both proteins became undetectable by Western blotting (Fig. 1C). Thus, we found that like N15Δ and R105G mutations, the E140K mutation also disrupts A3H protein expression.
FIG. 1.
Expression of human A3H genes in 293T cells. A3H genes containing various mutations at the six polymorphic sites N15Δ, R18L, G105R, K121D, E178D, and E140K were inserted into the pcDNA3.1 vector with a V5 tag and were transiently expressed in 293T cells. Protein expression was detected by Western blotting with the indicated antibodies. Expression of A3H hap II and its mutants is shown in panel A, expression of the seven A3H haplotypes is shown in panel B, expression of A3H hap VII and AGM A3H bearing an E140K mutation is shown in panel C, and expression of A3H-L and its mutants is shown in panel D. In addition, the insert of the seven A3H gene fragments was sliced out from the pcDNA3.1 vector by HindIII/XhoI digestion and analyzed by agrose gel electrophoresis (B).
To understand whether these three mutations have different levels of activity in disruption of A3H expression, we introduced an N15Δ, R105G, or E140K mutation into a long version of the human A3H gene. Although simian A3H genes encode 210 amino acids, the human A3H gene encodes only 183 amino acids due to the presence of a PTC on the fifth exon. Previously, we repaired this PTC in the hap I gene to produce a full-length 210-amino-acid protein, A3H-L, and found that the hap I gene was then stably expressed (6). We confirmed that the A3H-L protein was expressed at similar levels to the A3H hap II protein (Fig. 1D, lanes 1 and 2). Since A3H-L already contains an R105G mutation, this result indicates that this mutation does not affect A3H-L protein expression. We further introduced N15Δ and E140K mutations and found that a single N15Δ or E140K mutation could completely disrupt A3H-L expression (Fig. 1D, lanes 3 to 6). Taken together, we found that among the three new haplotypes, two of these, hap V and hap VII, are stably expressed. In addition, although N15Δ, R105G, and E140K mutations all disrupt A3H expression, the N15Δ and E140K mutations may have a more dramatic effect than the R105G mutation to disrupt A3H expression.
Anti-HIV-1 activity of A3H haplotypes.
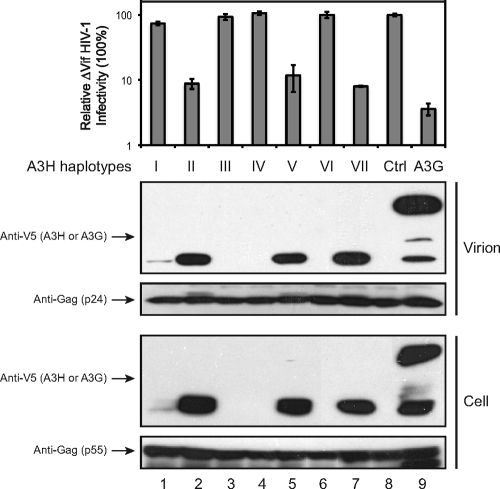
Having compared levels of protein expression among seven A3H haplotypes, we determined their anti-HIV-1 activities. Vif-defective (ΔVif) HIV-1 virions were produced from 293T cells in the presence of different A3H haplotypes or A3G expression and collected for protein and infectivity analysis. Initially, we determined levels of A3H virion packaging, since virion incorporation is an essential step for viral inhibition. It was found that the stably expressed A3H hap II, V, and VII were packaged into virions as efficiently as the A3G protein, whereas the levels of poorly expressed A3H hap I, III, IV, and VI were barely detectable in virions (Fig. 2). Next, we measured the infectivity of virions containing these different A3 proteins in GHOST cells. It was found that compared to the control, A3G reduced ΔVif HIV-1 infectivity approximately 30-fold, and A3H hap II, V, and VII reduced viral infectivity approximately 10-fold (Fig. 2, top panel). Taken together, we conclude that levels of A3H virion packaging are dependent on its levels of expression in viral producer cells, and all stably expressed A3H haplotypes can be efficiently packaged into virions and inhibit HIV-1 replication.
FIG. 2.
Anti-HIV-1 activity of the seven A3H haplotypes. 293T cells were transfected with ΔVif HIV-1 expression vector pNL4-3Δvif and A3H- or A3G-expressing pcDNA3.1 vector, or a control (Ctrl). Virions were collected from cell culture, and their infectivity was analyzed in the HIV-1 luciferase reporter cell line TZM-bl. In addition, virions were further purified by ultracentrifugation, and proteins in virions and viral producer cells were analyzed by Western blotting with the indicated antibodies. The standard errors of the means (SEMs) were calculated from three independent experiments.
Contribution of A3H CDD to its anti-HIV-1 activity.
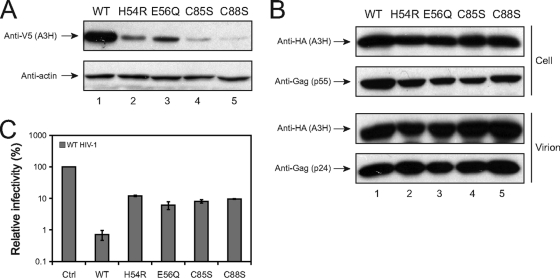
A3G has two CDDs with separate activities: CDD1 mediates A3G virion packaging, and CDD2 mediates cytidine deamination. We wanted to know whether the single CDD in A3H has both activities. Accordingly, we mutated the 54HXEX27PCXXC88 motif in the A3H hap VII gene, creating four single-point mutants: H54R, E56Q, C85S, and C88S. When these mutants were expressed from pcDNA3.1 mammalian expression vector, we found that all of these mutations impaired A3H protein expression (Fig. 3A), which made it impossible for further study. We then introduced these mutations into the VR-A3H-L expression vector and found that all of these mutants were stably expressed (Fig. 3B, upper panels). After that, we determined whether these mutations could affect A3H virion packaging. It was found that all these A3H mutants could be packaged into HIV-1 virions as efficiently as the wild-type protein (Fig. 3B, lower panels). However, we found that these mutants showed a reduced anti-HIV-1 activity compared to the wild-type protein (Fig. 3C). Thus, we conclude that A3H CDD does not determine A3H virion packaging, but it is required for A3H antiviral activity. These observations agree with a previous study (12).
FIG. 3.
Anti-HIV-1 activity of A3H CDD mutants. (A) Expression of A3H hap VII CDD mutants from the pcDNA3.1 vector. Indicated CDD mutants with a V5 tag were created from the A3H hap VII-expressing pcDNA3.1 vector and were transfected into 293T cells. Protein expression was determined by Western blotting with the indicated antibodies. WT, wild type. (B) Expression of A3H CDD mutants from the VR vector and their incorporation into virions. Indicated CDD mutants with an HA tag were created from the A3H hap I-expressing VR vector (VR-A3H-L). 293T cells were transfected with the indicated A3H expression constructs plus the HIV-1 proviral vector pNL4-3. Virions were purified by ultracentrifugation, and the presence of A3H protein in cells and in virions was determined by Western blotting with the indicated antibodies. (C) The anti-HIV-1 activity of A3H CDD mutants. HIV-1 reporter viruses were produced from 293T cells by transfection of pNL-Luc with VR vector expressing the indicated A3H hap I CDD mutants. Viral infectivity was analyzed in GHOST cells. The SEMs were calculated for three independent experiments.
A3H incorporation into virions.
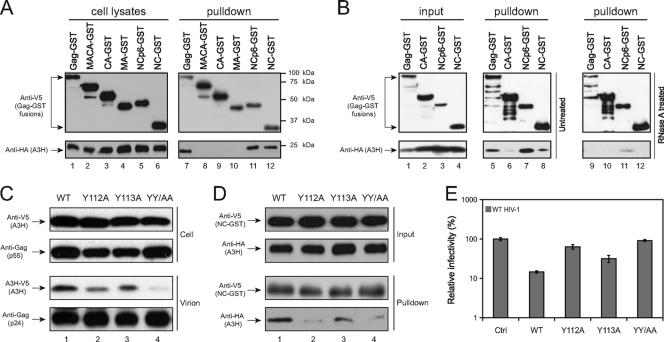
As mentioned in the introduction, A3G has two motifs that determine its virion packaging: CDD1 and 124YYFW127. In the 124YYFW127 motif, both Y124 and W127 residues were reported to be critical for A3G virion packaging (15), and they also mediate A3G oligomerization (14). In addition, like the A3G protein, A3B and A3F also interact with HIV-1 NC for virion incorporation (8, 40). To understand how A3H is packaged into virions, we first determined whether A3H interacts with HIV-1 Gag and where it binds. Previously, we created a panel of HIV-1 Gag-GST fusion expression vectors containing a C-terminal V5 tag, including Gag-GST, MA-CA-GST, CA-GST, NCp6-GST, and NC-GST (41). These Gag-GST fusion proteins were expressed with A3H hap VII protein in 293T cells, and their interactions were determined by GST pulldown. It was found that Gag-GST, NCp6-GST, and NC-GST could pull A3H down, whereas the others could not (Fig. 4A). These interactions required RNAs since the binding was lost in the presence of RNase A treatment (Fig. 4B). Thus, we conclude that A3H binds HIV-1 NC in an RNA-dependent manner, which is consistent with a recent study (29).
FIG. 4.
Mechanism of A3H virion packaging. Mapping of the interaction between A3H and HIV-1 Gag. 293T cells were transfected with the pcDNA3.1-A3H Hap VII-FLAG-HA vector and another pcDNA3.1 vector expressing the indicated HIV-1 Gag proteins with a C-terminal GST-V5 tag into 293T cells. The Gag and A3H interaction was then determined by GST pulldown in the absence (A) or presence (B) of RNase A. (C) Virion incorporation of the A3H 112YYHW115 mutants. 293T cells were transfected with pNL4-3 and the indicated pcDNA3.1-A3H Hap VII-V5-expressing vectors. Virions were purified by ultracentrifugation. The levels of A3H in cells and virions were determined by Western blotting with the indicated antibodies. (D) Interaction of the A3H 112YYHW115 mutants with HIV-1 NC. 293T cells were transfected with the pcDNA3.1-NC-GST-V5 vector and indicated pcDNA3.1-A3H Hap VII-FLAG-HA vectors. The interaction between A3H mutants and HIV-1 NC was determined by GST pulldown as in panel A. (E) Anti-HIV-1 activity of the A3H 112YYHW115 mutants. HIV-1 reporter viruses (pNL-Luc) were produced in the presence of each A3H hap VII CDD mutant, and viral infectivity was determined as in Fig. 3C. The SEMs were calculated from three independent experiments.
Next, we determined whether A3H contains a similar YYFW motif that serves as the packaging signal. Indeed, a quite similar 112YYHW115 motif is present in A3H (Fig. 5A). To understand its function, we mutated residues Y112, Y113, and W115 in the A3H hap VII gene, creating four single-residue mutations: Y112A, Y113A, W115A, and W115L. In addition, we created another double-residue mutation, YY/AA, with both Y112 and Y113 changed to alanines. Among these, the Y112A, Y113A, and YY/AA mutants were well expressed, but both W115A and W115L mutants were not expressed (Fig. 4C, upper panels) (data not shown). We then compared the levels of Y112A, Y113A, and YY/AA virion incorporation with that of the wild-type A3H protein. It was found that all of these mutants showed much reduced levels of virion packaging, and this reduction was particularly apparent for the Y112A and YY/AA mutants (Fig. 4C, lower panel). Moreover, we compared their interactions with HIV-1 NC by GST pulldown. It was found that the Y112A and YY/AA mutants bound NC poorly, and levels of Y113A mutant binding to NC were also reduced (Fig. 4D). Furthermore, we compared the anti-HIV-1 activities of these mutants. It was found that the Y112A and YY/AA mutants almost completely lost anti-HIV-1 activity, and the Y113A mutant partially lost this activity (Fig. 4E). Taken together, these results indicate that the 112YYHW115 motif determines A3H virion packaging via an interaction with HIV-1 NC, and among these residues, Y112 is particularly critical for NC interaction and virion packaging.
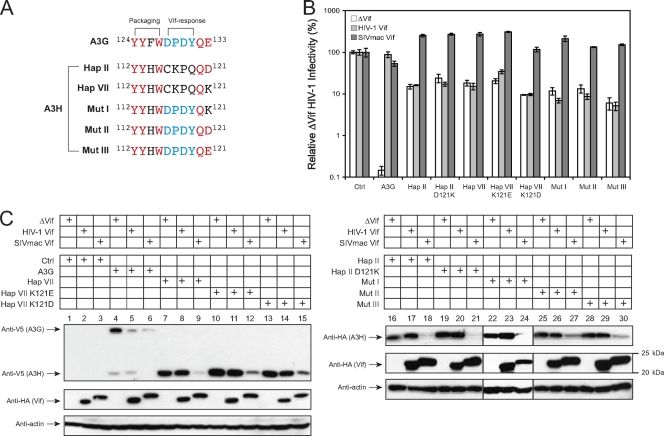
FIG. 5.
Analysis of human A3H sensitivity to HIV-1 and SIVmac Vif. (A) Amino acid sequence alignment of the A3G virion packaging and Vif-responsive domains with the corresponding A3H region. The conserved residues are shown in red, and residues in the Vif-responsive domain are shown in blue. Mut I, II, and III were created from A3H hap VII. (B) The sensitivity of A3H to Vif was measured by a single-round HIV-1 replication assay. 293T cells were transfected with the HIV-1 reporter construct pNL-LucΔenvΔvif, pNL-A1 expressing HIV-1 Vif-HA or SIVmac Vif-HA, a VSV-G expression vector, and a pcDNA3.1 vector expressing the indicated V5-tagged A3G or A3H proteins. The infectivity of the virus produced from the transfected cells was determined by infection of GHOST cells. The SEMs were calculated from three independent experiments. (C) The sensitivity of A3H to Vif was measured by Western blotting in viral producer cells. 293T cells were transfected with the indicated A3 expression vector plus pNL-A1ΔVif, pNL-A1 HIV-1 Vif-HA, or pNL-A1 SIVmac Vif-HA. Levels of A3G, A3H, Vif, and actin expression were determined by Western blotting with the indicated antibodies.
The sensitivity of human A3H to HIV-1 Vif.
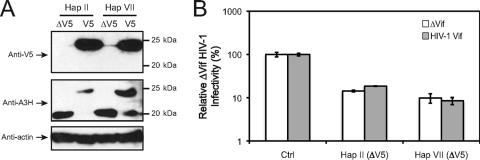
We reported that the A3H antiviral activity is resistant to HIV-1 Vif neutralization, which was confirmed by Harari et al. (12). When we compared A3G and A3H amino acid sequences, we found that the previously identified A3G Vif-responsive motif 128DPDY131 is not conserved in A3H (15). We then created an A3H hap VII mutant that contains this motif, which was called Mut I (Fig. 5A). Recently, two groups reported that A3H could become partially sensitive to HIV-1 Vif if the K121 residue was changed to an aspartate (K121D) (20, 46). We therefore introduced a K121D or K121E mutation into Mut I, creating another two mutants, Mut II and III. Since A3H hap II and VII have a respective D121 or K121 residue, we also introduced a D121K mutation into hap II and a K121E or K121D mutation into hap VII, creating the Hap II D121K, Hap VII K121E, and Hap VII K121D mutants. We then determined the Vif sensitivity of these A3H mutants by a single-round viral replication assay. It was found that both HIV-1 and SIVmac Vif proteins could completely neutralize the activity of A3G (Fig. 5B), confirming that A3G is very sensitive to both Vif proteins. The SIVmac Vif could also completely neutralize the activity of all of these A3H proteins (Fig. 5B). However, the HIV-1 Vif showed barely detectable activity to neutralize any of these A3H proteins (Fig. 5B). We further verified these results by determining A3H protein expressions in viral producer cells by Western blotting. It was found that unlike A3G, whose expression was decreased by both HIV-1 Vif and SIVmac Vif, the expression of all of these A3H proteins was selectively decreased by SIVmac Vif, but not HIV-1 Vif (Fig. 5C). This result is consistent with that from the viral infectivity assay. Since all of these different A3H mutants contained a V5 epitope tag at the C terminus, we wondered whether the V5 tag could contribute to A3H resistance to HIV-1 Vif protein. We then created expression vectors to express untagged A3H hap II [Hap II (ΔV5)] and hap VII [Hap VII (ΔV5)] proteins by site-directed mutagenesis. When these proteins were expressed in HIV-1 producer cells (Fig. 6A), they were still resistant to HIV-1 Vif (Fig. 6B). Thus, the V5 tag does not contribute to A3H Vif resistance. Taken together, we were unable to make human A3H become sensitive to HIV-1 Vif by introducing a K121D mutation and/or implanting an A3G Vif-responsive motif into A3H protein.
FIG. 6.
(A) Expression of untagged (ΔV5) or V5-tagged A3H Hap II and Hap VII from pcDNA3.1 vector in 293T cells as determined by Western blotting using the indicated antibodies. (B) Sensitivity of A3H Hap II (ΔV5) and Hap VII (ΔV5) to HIV-1 Vif as measured by a single-round HIV-1 replication assay as in Fig. 5B. The SEMs were calculated from three independent experiments.
DISCUSSION
We have examined human A3H genotypes and haplotypes among six major human populations. Our estimated allele frequencies for these six nonsynonymous SNPs are similar to those reported by dbSNP, with two notable exceptions. First, our frequency for the L18 allele is considerably greater than that contained in dbSNP (allele frequency of 0.13 versus 0.01), but is less than that reported by OhAinle et al., which was 0.20 (27). Second, we found no K140 allele among the 58 individuals we genotyped, although dbSNP reports this allele to be present at a frequency of 0.92 (Table 2). One possible explanation for the first difference is that the N15Δ allele is very close to the L18 allele, and it is possible that this deletion interfered with the correct genotyping calls for L18 used for the high-throughput method in dbSNP, as opposed to the sequencing method of OhAinle et al. (27) and the restriction enzyme method used here. The second difference may be due to the pooled genotyping method used to detect the K140 allele that may have overestimated the frequency of this allele in the European Caucasian population that was examined (1). It seems unlikely that the restriction enzyme-based assays that we used here could result in our missing the K140 allele, because this allele is not digested by the restriction enzyme used and all samples in our study were completely digested (i.e., were all homozygous for the E140 allele). Furthermore, we examined the SNP position among 13 informative expressed sequence tags contained in GenBank, as well as the six currently available genomes contained in the 1000 Genome Project, in addition to the Watson and Venter genomes. All showed only the E140 allele. The only other sequence that shows the presence of the K140 allele, besides the data in dbSNP, is the single reference genome itself. Because quality scores are not easily obtainable for this sequence, the reference sequence allele could either be a sequencing error or might indicate that this allele does exist, but at an apparently low frequency. As shown in the results and discussed later, the K140 allele is damaging to the protein.
It is also interesting to note that at amino acid position 121, there appear to be two alleles (K121 and D121), but each of these was independently derived from the ancestral E121 allele. The E121 allele was not found in the available GenBank sequences, and it appears that no other group has detected this allele among the samples they have studied (12, 27). This allele is present in the single Neanderthal sequence that has been studied, which is observable in the UCSC Genome Browser. Neanderthals are estimated to have diverged from humans about 500,000 years ago and became extinct about 30,000 years ago (9, 10). Thus, it appears that the E121 allele was replaced by K121 and D121 in the evolutionarily short period of time since the divergence of our species from the Neanderthals.
Out of 32 theoretically possible haplotypes for the 5 biallelic nonsynonymous sites studied here, we found seven haplotypes, including the previously reported four haplotypes (I to IV) that have been studied in some detail (27). Six of these haplotypes were observed in individuals that were either homozygous for all five SNPs or were heterozygous at only a single SNP, and this provides very high confidence for the existence of these haplotypes among humans (Table 3). One of them, hap V (NRRDE), is relatively frequent, particularly in the Caribbean population, and this haplotype is stably expressed and exhibits anti-HIV activity. The two rarer haplotypes, hap VI and hap VII, were found only within one or two of the populations examined (Table 3). The relatively high number of haplotypes within the short chromosomal span between amino acids 15 and 178 (1,714 bp) compared to the more commonly observed 2 to 4 haplotypes observed among humans at other sites in the genome is in keeping with the active evolutionary dynamics of this region thought to be due to strong host-pathogen interactions (5, 27, 31). This relatively large number of haplotypes also indicates that it will be necessary to examine the activity of the haplotypes found in different populations, as some of these haplotypes provide good A3H activity, whereas others do not. Additional unnumbered haplotypes have been inferred by using the PHASE 2.1.1 computer program and reported in the supplemental materials of the study carried out by OhAinle et al. (27). However, at least some of these haplotypes seem unlikely to exist. For example, haplotypes with N15-R18, Δ15-R18, N15-L18, and Δ15-L18 are all present among the unnumbered haplotypes. However, based upon the suggested phylogeny of OhAinle et al. (27), the presence of the Δ15-R18 combination would require either a recombination or gene conversion event within the 9 bp separating the two polymorphic sites or a recurrent mutation event, all of which are very unlikely.
We have compared the expression and anti-HIV-1 activity levels of these seven A3H haplotypes. We confirmed that two N15Δ and R105G polymorphisms seriously disrupt A3H expression (12, 27). In addition, we found that an E140K mutation could also disrupt A3H expression. Among these three N15Δ, R105G, and E140K mutations, the destructive effect of the R105G mutation was suppressed in the context of the full-length 210-amino-acid protein. This result indicates that the N15Δ and E140K mutations are more deleterious for A3H expression. Interestingly, we also observed that four CDD mutants H54R, E56Q, C85S, and C88S as well as the two W115A and W115L mutants were poorly expressed in cell culture, indicating that these mutations also disrupt A3H expression. It is still unclear why the human A3H gene is so easily damaged by point mutations. Previously, we found that although a PTC disrupts A3H expression, this disruption is not caused by protein degradation (6). When cells were treated with proteasomal and lysosomal inhibitors, the expression of A3H was increased only marginally. Others also reported a marginal increase of A3H expression by proteasomal inhibitors (27). Notably, when we measured A3H mRNA, we found that its levels were significantly decreased when a PTC was present (6). PTCs in the last exon of a gene generally do not cause nonsense-mediated mRNA decay, and so we speculated that human A3H mRNA could have a very unique secondary structure, which can be easily disrupted by mutations, resulting in decrease in A3H mRNA stability and loss of protein expression.
Unlike the A3B, A3DE, A3F, and A3G proteins, A3H has only one CDD, but it still inhibits HIV-1 replication. We have studied the function of A3H CDD and found that it is required for A3H anti-HIV-1 activity, but it is not required for virion packaging. Since the deaminase activity of A3H has been demonstrated in Escherichia coli (28, 29), we believe it is most likely that the deaminase activity is required for A3H anti-HIV-1 activity. Although its virion packaging is independent of its CDD, it is dependent on an YYXW motif, which was identified as a packaging signal for A3G. Indeed, this motif is conserved in all anti-HIV-1 A3 proteins, indicating that these anti-HIV A3 proteins may use a universal mechanism for virion packaging. Our results showed that this motif specifically interacts with HIV-1 NC, which further supports its important role in virion incorporation. Since the YYXW motif is enough for A3H virion packaging, it is not clear why A3G virion packaging requires its CDD1. Recently, Ooms et al. reported that although A3H hap I protein was poorly expressed in cells, it could be still packaged efficiently into HIV-1 virions. They also presented evidence that A3H hap I specifically interacted with MA-CA other than NC for virion packaging (29). We found that A3H hap I was poorly packaged into virions because it was not expressed in the cell, and, therefore, we cannot confirm this observation.
We have studied the mechanism of A3H resistance to Vif. Initially, we thought that the resistance of human A3H to HIV-1 Vif could be due to the lack of a similar A3G Vif-responsive domain, DPDY. However, even when this domain was placed in the corresponding region, A3H was still completely resistant to HIV-1 Vif (Fig. 5). Next, we tried to replicate work showing the K121D mutation could reduce A3H resistance to HIV-1 Vif, as reported by others (20, 46). However, we found that both hap II and VII, which have a D121 and K121 residue, respectively, were equally resistant to HIV-1 Vif (Fig. 5). Even when these two residues were exchanged between hap II and VII and when a K121D or K121E mutation was introduced into Mut I, we still could not change the resistance of human A3H to HIV-1 Vif (Fig. 5). From these results, we conclude that human A3H is completely resistant to HIV-1 Vif. Our results further highlight the potential role of human A3H in inhibition of HIV-1 infection.
Supplementary Material
Acknowledgments
We thank K. Strebel, M. Emerman, and the NIH AIDS Research and Reference Reagent Program for reagents.
Y.-H.Z. was supported by grants AI063944 and AI080225 from the National Institutes of Health.
Footnotes
Published ahead of print on 26 January 2011.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Anonymous. 2005. A haplotype map of the human genome. Nature 437:1299-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop, K. N., et al. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 3.Bishop, K. N., M. Verma, E. Y. Kim, S. M. Wolinsky, and M. H. Malim. 2008. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 4:e1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark, A. G. 1990. Inference of haplotypes from PCR-amplified samples of diploid populations. Mol. Biol. Evol. 7:111-122. [DOI] [PubMed] [Google Scholar]
- 5.Daly, M. J., J. D. Rioux, S. F. Schaffner, T. J. Hudson, and E. S. Lander. 2001. High-resolution haplotype structure in the human genome. Nat. Genet. 29:229-232. [DOI] [PubMed] [Google Scholar]
- 6.Dang, Y., et al. 2008. Human cytidine deaminase APOBEC3H restricts HIV-1 replication. J. Biol. Chem. 283:11606-11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang, Y., X. Wang, W. J. Esselman, and Y. H. Zheng. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 80:10522-10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doehle, B. P., A. Schafer, and B. R. Cullen. 2005. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 339:281-288. [DOI] [PubMed] [Google Scholar]
- 9.Green, R. E., et al. 2010. A draft sequence of the Neandertal genome. Science 328:710-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green, R. E., et al. 2008. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell 134:416-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo, F., S. Cen, M. Niu, J. Saadatmand, and L. Kleiman. 2006. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 80:11710-11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harari, A., M. Ooms, L. C. Mulder, and V. Simon. 2009. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J. Virol. 83:295-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris, R. S., et al. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 14.Huthoff, H., F. Autore, S. Gallois-Montbrun, F. Fraternali, and M. H. Malim. 2009. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 5:e1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huthoff, H., and M. H. Malim. 2007. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and virion encapsidation. J. Virol. 81:3807-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kidd, J. M., T. L. Newman, E. Tuzun, R. Kaul, and E. E. Eichler. 2007. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 3:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinomoto, M., et al. 2007. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 35:2955-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larue, R. S., J. Lengyel, S. R. Jonsson, V. Andresdottir, and R. S. Harris. 2010. Lentiviral Vif degrades the APOBEC3Z3/APOBEC3H protein of its mammalian host and is capable of cross-species activity. J. Virol. 84:8193-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 20.Li, M. M., L. I. Wu, and M. Emerman. 2010. The range of human APOBEC3H sensitivity to lentiviral Vif proteins. J. Virol. 84:88-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mangeat, B., et al. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 22.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 23.Mbisa, J. L., et al. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 81:7099-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mbisa, J. L., W. Bu, and V. K. Pathak. 2010. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J. Virol. 84:5250-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Navarro, F., et al. 2005. Complementary function of the two catalytic domains of APOBEC3G. Virology 333:374-386. [DOI] [PubMed] [Google Scholar]
- 26.Niewiadomska, A. M., and X. F. Yu. 2009. Host restriction of HIV-1 by APOBEC3 and viral evasion through Vif. Curr. Top. Microbiol. Immunol. 339:1-25. [DOI] [PubMed] [Google Scholar]
- 27.OhAinle, M., J. A. Kerns, M. M. Li, H. S. Malik, and M. Emerman. 2008. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4:249-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.OhAinle, M., J. A. Kerns, H. S. Malik, and M. Emerman. 2006. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J. Virol. 80:3853-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ooms, M., S. Majdak, C. W. Seibert, A. Harari, and V. Simon. 2010. The localization of APOBEC3H variants in HIV-1 virions determines their antiviral activity. J. Virol. 84:7961-7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Opi, S., et al. 2007. Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant. J. Virol. 81:8236-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patil, N., et al. 2001. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294:1719-1723. [DOI] [PubMed] [Google Scholar]
- 32.Russell, R. A., J. Smith, R. Barr, D. Bhattacharyya, and V. K. Pathak. 2009. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J. Virol. 83:1992-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santa-Marta, M., F. A. da Silva, A. M. Fonseca, and J. Goncalves. 2005. HIV-1 Vif can directly inhibit apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G-mediated cytidine deamination by using a single amino acid interaction and without protein degradation. J. Biol. Chem. 280:8765-8775. [DOI] [PubMed] [Google Scholar]
- 34.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 35.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 36.Smith, J. L., W. Bu, R. C. Burdick, and V. K. Pathak. 2009. Multiple ways of targeting APOBEC3-virion infectivity factor interactions for anti-HIV-1 drug development. Trends Pharmacol. Sci. 30:638-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 38.Strebel, K., J. Luban, and K. T. Jeang. 2009. Human cellular restriction factors that target HIV-1 replication. BMC Med. 7:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan, L., P. T. Sarkis, T. Wang, C. Tian, and X. F. Yu. 2009. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB J. 23:279-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, T., C. Tian, W. Zhang, P. T. Sarkis, and X. F. Yu. 2008. Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging. J. Mol. Biol. 375:1098-1112. [DOI] [PubMed] [Google Scholar]
- 41.Wang, X., et al. 2010. Moloney leukemia virus 10 (MOV10) protein inhibits retrovirus replication. J. Biol. Chem. 285:14346-14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wheeler, D. L., et al. 2007. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 35:D5-D12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, X., et al. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, H., et al. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhen, A., T. Wang, K. Zhao, Y. Xiong, and X. F. Yu. 2010. A single amino acid difference in human APOBEC3H variants determines HIV-1 Vif sensitivity. J. Virol. 84:1902-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng, Y. H., et al. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.