Abstract
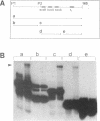
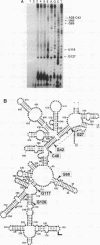
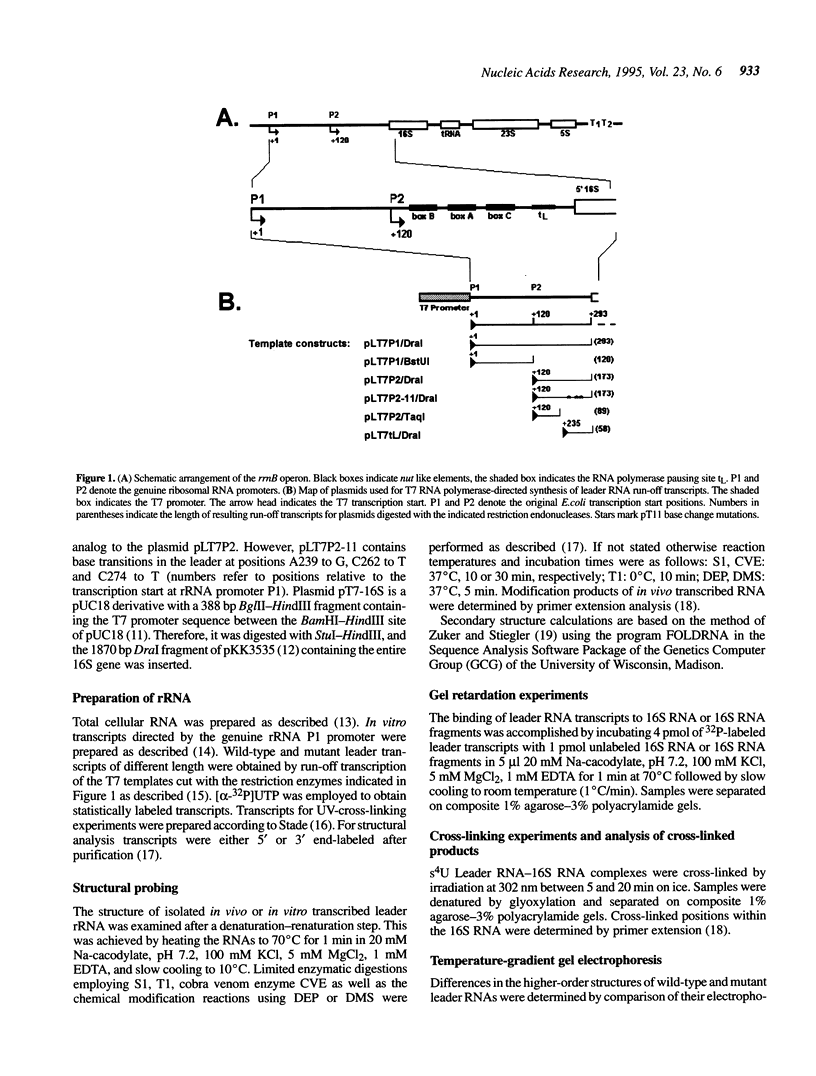
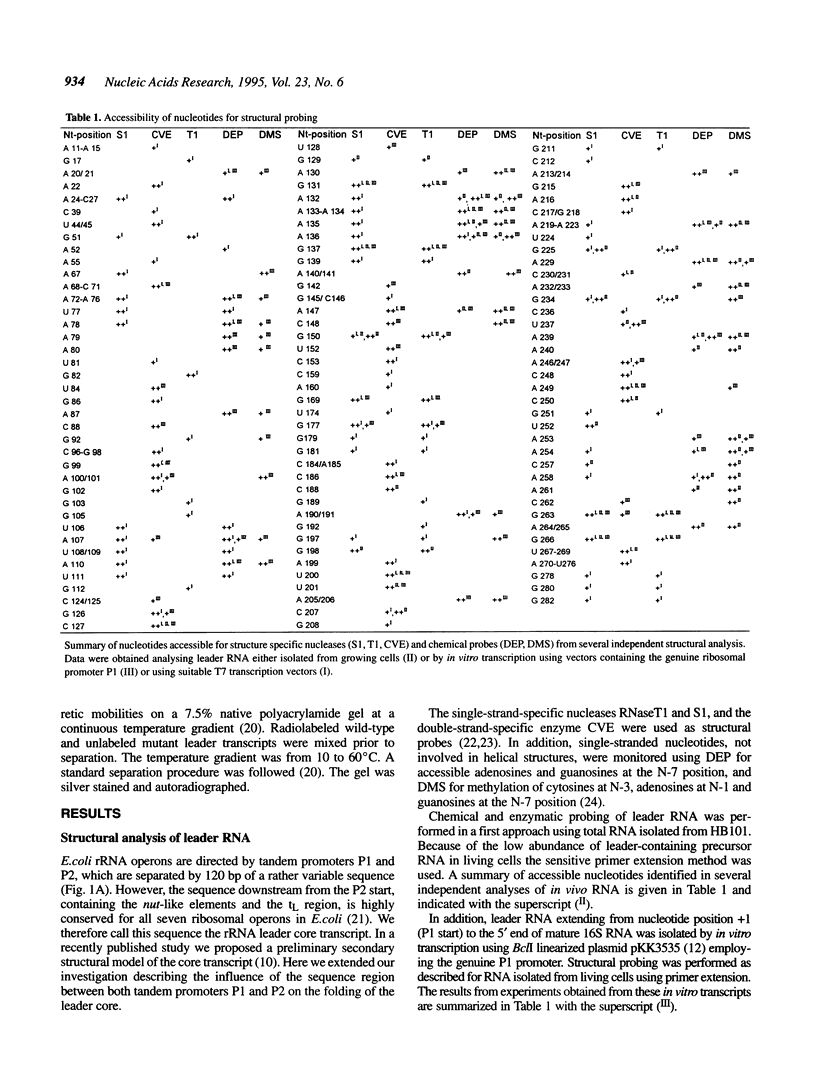
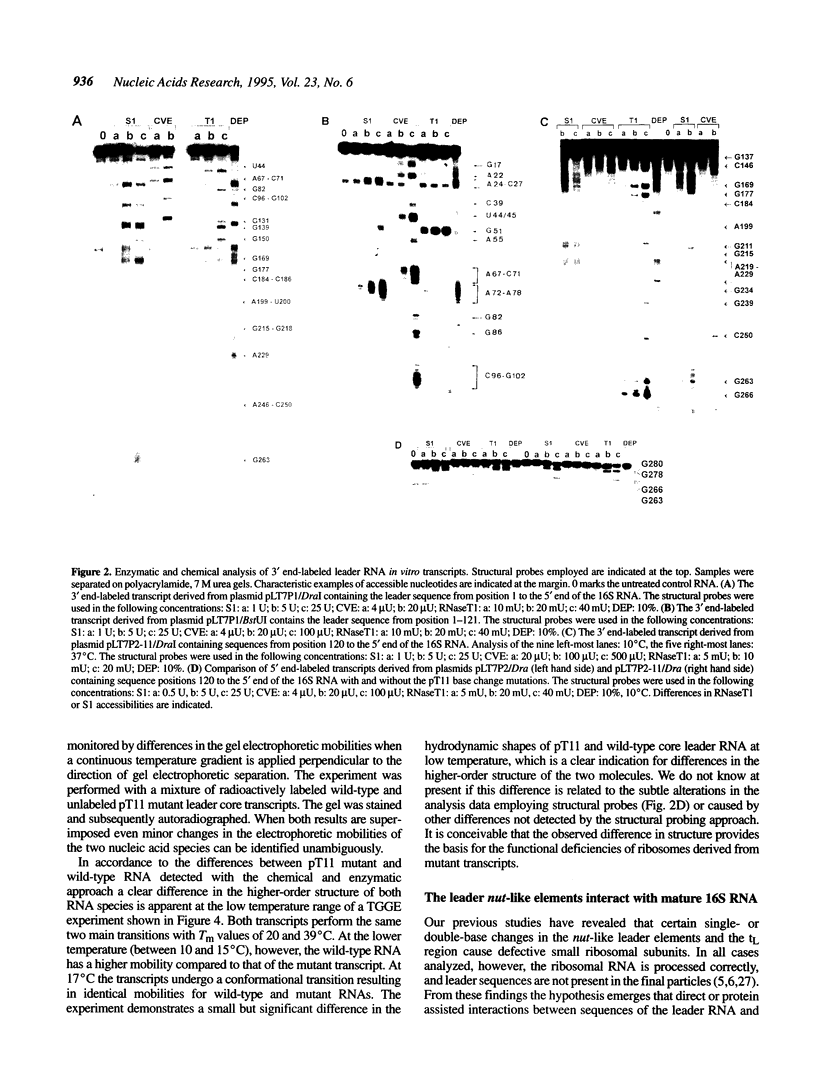
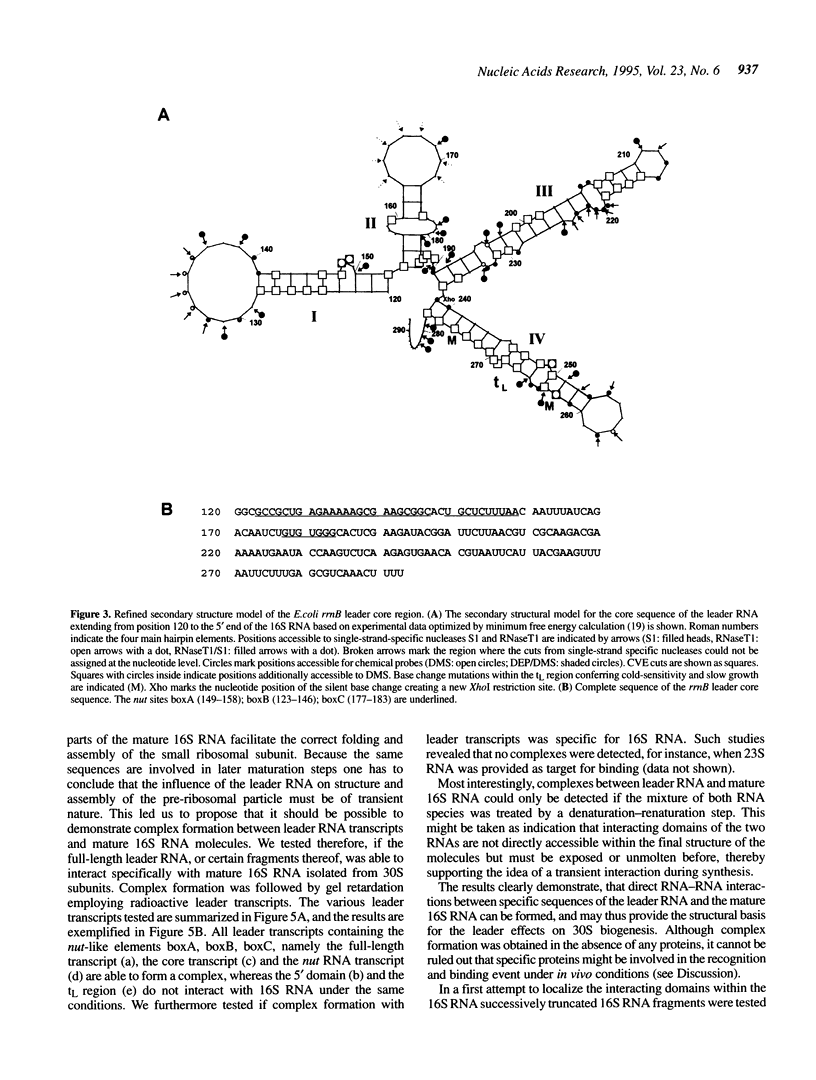
All ribosomal RNAs are preceded by leader sequences not present in the final ribosome particles. The highly conserved leader sequences of bacterial rRNAs are known to be important for the folding and assembly of functional ribosomes. Very likely transient binding of the leader to mature parts of the 16S RNA occurs during transcription. To better understand the mechanistic details of these functions we have performed a secondary structural analysis of E. coli ribosomal RNA leader transcripts by chemical modification and enzymatic hydrolysis studies. The data were combined with results from thermodynamic stability calculations to yield a generalized structural model. The same secondary structure of the leader core, comprising the nut-like sequences up to the mature 5' end of the 16S RNA, was deduced, irrespective if transcripts started at promoter P1 or 120 nucleotides downstream at P2. Employing gelshift and cross-linking studies we were able to demonstrate that a part of the leader core, namely the nut-like sequence elements bind directly to specific regions within the mature 16S RNA. The sites of RNA-RNA cross-linking could be localized by sequencing. They map in the 16S RNA 5' domain at nucleotide positions G27 to G42, C48, G68, G117 and G126. The results may explain the recently observed scaffolding function of the leader RNA during ribosome biogenesis.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atmadja J., Brimacombe R., Blöcker H., Frank R. Investigation of the tertiary folding of Escherichia coli 16S RNA by in situ intra-RNA cross-linking within 30S ribosomal subunits. Nucleic Acids Res. 1985 Oct 11;13(19):6919–6936. doi: 10.1093/nar/13.19.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Backman K. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- Brimacombe R. RNA-protein interactions in the Escherichia coli ribosome. Biochimie. 1991 Jul-Aug;73(7-8):927–936. doi: 10.1016/0300-9084(91)90134-m. [DOI] [PubMed] [Google Scholar]
- Brink M. F., Verbeet M. P., de Boer H. A. Formation of the central pseudoknot in 16S rRNA is essential for initiation of translation. EMBO J. 1993 Oct;12(10):3987–3996. doi: 10.1002/j.1460-2075.1993.tb06076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Dammel C. S., Noller H. F. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 1993 Apr;7(4):660–670. doi: 10.1101/gad.7.4.660. [DOI] [PubMed] [Google Scholar]
- Feunteun J., Monier R., Vola C., Rosset R. Ribosomal assembly defective mutants of Escherichia coli. Nucleic Acids Res. 1974 Jan;1(1):149–169. doi: 10.1093/nar/1.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinaro H., Domenjoud L., Jacob M. Structural study of the 5' end of a synthetic premessenger RNA from adenovirus. Evidence for a long-range exon-intron interaction. J Mol Biol. 1994 Jul 15;240(3):205–225. doi: 10.1006/jmbi.1994.1436. [DOI] [PubMed] [Google Scholar]
- Glotz C., Brimacombe R. An experimentally-derived model for the secondary structure of the 16S ribosomal RNA from Escherichia coli. Nucleic Acids Res. 1980 Jun 11;8(11):2377–2395. doi: 10.1093/nar/8.11.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greuer B., Osswald M., Brimacombe R., Stöffler G. RNA-protein cross-linking in Escherichia coli 30S ribosomal subunits; determination of sites on 16S RNA that are cross-linked to proteins S3, S4, S7, S9, S10, S11, S17, S18 and S21 by treatment with bis-(2-chloroethyl)-methylamine. Nucleic Acids Res. 1987 Apr 24;15(8):3241–3255. doi: 10.1093/nar/15.8.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göringer H. U., Szymkowiak C., Wagner R. Escherichia coli 5S RNA A and B conformers. Characterisation by enzymatic and chemical methods. Eur J Biochem. 1984 Oct 1;144(1):25–34. doi: 10.1111/j.1432-1033.1984.tb08426.x. [DOI] [PubMed] [Google Scholar]
- Kingston R. E., Chamberlin M. J. Pausing and attenuation of in vitro transcription in the rrnB operon of E. coli. Cell. 1981 Dec;27(3 Pt 2):523–531. doi: 10.1016/0092-8674(81)90394-9. [DOI] [PubMed] [Google Scholar]
- LeCuyer K. A., Crothers D. M. The Leptomonas collosoma spliced leader RNA can switch between two alternate structural forms. Biochemistry. 1993 May 25;32(20):5301–5311. doi: 10.1021/bi00071a004. [DOI] [PubMed] [Google Scholar]
- Lewicki B. T., Margus T., Remme J., Nierhaus K. H. Coupling of rRNA transcription and ribosomal assembly in vivo. Formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage T7 RNA polymerase. J Mol Biol. 1993 Jun 5;231(3):581–593. doi: 10.1006/jmbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- Lockard R. E., Kumar A. Mapping tRNA structure in solution using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res. 1981 Oct 10;9(19):5125–5140. doi: 10.1093/nar/9.19.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G., Carraway M., Frey A. Z., Brown L., Arraj J. A. Insertion mutations in the dam gene of Escherichia coli K-12. Mol Gen Genet. 1983;192(1-2):288–289. doi: 10.1007/BF00327681. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Mori H., Dammel C., Becker E., Triman K., Noller H. F. Single base alterations upstream of the E. coli 16S rRNA coding region result in temperature-sensitive 16S rRNA expression. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):323–327. doi: 10.1016/0167-4781(90)90189-9. [DOI] [PubMed] [Google Scholar]
- Nashimoto H., Held W., Kaltschmidt E., Nomura M. Structure and function of bacterial ribosomes. XII. Accumulation of 21 s particles by some cold-sensitive mutants of Escherichia coli. J Mol Biol. 1971 Nov 28;62(1):121–138. doi: 10.1016/0022-2836(71)90135-5. [DOI] [PubMed] [Google Scholar]
- Nodwell J. R., Greenblatt J. The nut site of bacteriophage lambda is made of RNA and is bound by transcription antitermination factors on the surface of RNA polymerase. Genes Dev. 1991 Nov;5(11):2141–2151. doi: 10.1101/gad.5.11.2141. [DOI] [PubMed] [Google Scholar]
- Pardon B., Thelen L., Wagner R. The Escherichia coli ribosomal RNA leader: a structural and functional investigation. Biol Chem Hoppe Seyler. 1994 Jan;375(1):11–20. doi: 10.1515/bchm3.1994.375.1.11. [DOI] [PubMed] [Google Scholar]
- Peattie D. A., Gilbert W. Chemical probes for higher-order structure in RNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij C. W., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985 Mar 11;13(5):1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T., Noller H. F. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991 Aug;10(8):2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesner D., Steger G., Zimmat R., Owens R. A., Wagenhöfer M., Hillen W., Vollbach S., Henco K. Temperature-gradient gel electrophoresis of nucleic acids: analysis of conformational transitions, sequence variations, and protein-nucleic acid interactions. Electrophoresis. 1989 May-Jun;10(5-6):377–389. doi: 10.1002/elps.1150100516. [DOI] [PubMed] [Google Scholar]
- Rosenbaum V., Riesner D. Temperature-gradient gel electrophoresis. Thermodynamic analysis of nucleic acids and proteins in purified form and in cellular extracts. Biophys Chem. 1987 May 9;26(2-3):235–246. doi: 10.1016/0301-4622(87)80026-1. [DOI] [PubMed] [Google Scholar]
- Squires C. L., Greenblatt J., Li J., Condon C., Squires C. L. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):970–974. doi: 10.1073/pnas.90.3.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A. K., Schlessinger D. Mechanism and regulation of bacterial ribosomal RNA processing. Annu Rev Microbiol. 1990;44:105–129. doi: 10.1146/annurev.mi.44.100190.000541. [DOI] [PubMed] [Google Scholar]
- Stade K., Rinke-Appel J., Brimacombe R. Site-directed cross-linking of mRNA analogues to the Escherichia coli ribosome; identification of 30S ribosomal components that can be cross-linked to the mRNA at various points 5' with respect to the decoding site. Nucleic Acids Res. 1989 Dec 11;17(23):9889–9908. doi: 10.1093/nar/17.23.9889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Changchien L. M., Craven G. R., Noller H. F. Interaction of proteins S16, S17 and S20 with 16 S ribosomal RNA. J Mol Biol. 1988 Mar 20;200(2):291–299. doi: 10.1016/0022-2836(88)90241-0. [DOI] [PubMed] [Google Scholar]
- Stern S., Moazed D., Noller H. F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Stern S., Powers T., Changchien L. M., Noller H. F. Interaction of ribosomal proteins S5, S6, S11, S12, S18 and S21 with 16 S rRNA. J Mol Biol. 1988 Jun 20;201(4):683–695. doi: 10.1016/0022-2836(88)90467-6. [DOI] [PubMed] [Google Scholar]
- Stiege W., Atmadja J., Zobawa M., Brimacombe R. Investigation of the tertiary folding of Escherichia coli ribosomal RNA by intra-RNA cross-linking in vivo. J Mol Biol. 1986 Sep 5;191(1):135–138. doi: 10.1016/0022-2836(86)90429-8. [DOI] [PubMed] [Google Scholar]
- Theissen G., Behrens S. E., Wagner R. Functional importance of the Escherichia coli ribosomal RNA leader box A sequence for post-transcriptional events. Mol Microbiol. 1990 Oct;4(10):1667–1678. doi: 10.1111/j.1365-2958.1990.tb00544.x. [DOI] [PubMed] [Google Scholar]
- Theissen G., Eberle J., Zacharias M., Tobias L., Wagner R. The tL structure within the leader region of Escherichia coli ribosomal RNA operons has post-transcriptional functions. Nucleic Acids Res. 1990 Jul 11;18(13):3893–3901. doi: 10.1093/nar/18.13.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Thelen L., Wagner R. Some base substitutions in the leader of an Escherichia coli ribosomal RNA operon affect the structure and function of ribosomes. Evidence for a transient scaffold function of the rRNA leader. J Mol Biol. 1993 Sep 20;233(2):203–218. doi: 10.1006/jmbi.1993.1500. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Gutell R. R. Evidence for several higher order structural elements in ribosomal RNA. Proc Natl Acad Sci U S A. 1989 May;86(9):3119–3122. doi: 10.1073/pnas.86.9.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurst R. M., Vournakis J. N., Maxam A. M. Structure mapping of 5'-32P-labeled RNA with S1 nuclease. Biochemistry. 1978 Oct 17;17(21):4493–4499. doi: 10.1021/bi00614a021. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zacharias M., Göringer H. U., Wagner R. Influence of the GCGC discriminator motif introduced into the ribosomal RNA P2- and tac promoter on growth-rate control and stringent sensitivity. EMBO J. 1989 Nov;8(11):3357–3363. doi: 10.1002/j.1460-2075.1989.tb08498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias M., Theissen G., Bradaczek C., Wagner R. Analysis of sequence elements important for the synthesis and control of ribosomal RNA in E coli. Biochimie. 1991 Jun;73(6):699–712. doi: 10.1016/0300-9084(91)90050-b. [DOI] [PubMed] [Google Scholar]
- Zacharias M., Wagner R. Deletions in the tL structure upstream to the rRNA genes in the E. coli rrnB operon cause transcription polarity. Nucleic Acids Res. 1987 Oct 26;15(20):8235–8248. doi: 10.1093/nar/15.20.8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]