Abstract
Integrase inhibitors are emerging anti-human immunodeficiency virus (HIV) drugs, and multiple retroviruses and transposable elements were evaluated here for susceptibilities to raltegravir (RAL) and elvitegravir (EVG). All viruses, including primate and nonprimate lentiviruses, a Betaretrovirus, a Gammaretrovirus, and the Alpharetrovirus Rous sarcoma virus (RSV), were susceptible to inhibition by RAL. EVG potently inhibited all lentiviruses and intermediately inhibited Betaretrovirus and Gammaretrovirus infections yet was basically ineffective against RSV. Substitutions based on HIV type 1 (HIV-1) resistance changes revealed that integrase residue Ser150 contributed significantly to the resistance of RSV. The drugs intermediately inhibited intracisternal A-particle retrotransposition but were inactive against Sleeping Beauty transposition and long interspersed nucleotide element 1 (LINE-1) retrotransposition.
Reverse transcription of retroviral RNA yields linear viral DNA (vDNA) containing a copy of the long terminal repeat (LTR) at each end. Integrase (IN) is an essential retroviral enzyme that catalyzes two reactions to insert the vDNA into cellular chromosomal DNA. IN prepares the LTR ends by hydrolyzing phosphodiester bonds adjacent to invariant CA dinucleotides, yielding reactive 3′ deoxyadenylate (dAOH) termini. In the nucleus, IN catalyzes DNA strand transfer by using the 3′ OHs to cut the chromosome in a staggered fashion, concomitantly joining the vDNA ends to 5′ phosphates. Host-mediated repair of the resulting DNA recombination intermediate completes the integration process. See reference 8 for an overview of retroviral reverse transcription and integration.
IN belongs to the polynucleotidyl transferase superfamily of nucleic acid-metabolizing enzymes (7). Conserved amino acid residues (typically Asp and Glu [32]) arranged commonly on an RNase H structural fold comprise active sites that coordinate divalent metal ions for in-line nucleophilic attack of phosphodiester bonds. Due to its critical role in replication, human immunodeficiency virus type 1 (HIV-1) IN has long been targeted for drug development, and the first-in-class inhibitor raltegravir (RAL) was licensed in 2007 (45). Because RAL and related compounds preferentially inhibit DNA strand transfer activity, the drugs are referred to as IN strand transfer inhibitors (INSTIs) (24). Elvitegravir (EVG) is another well-studied INSTI (41). Recently determined X-ray crystal structures revealed that the drugs work by ejecting the 3′ dA and its associated OH nucleophile from the IN active site (10, 11). Drug resistance occurs through mutations in the downstream region of the pol gene, in the region that encodes IN (reviewed in references 24 and 27).
Drugs discovered through their abilities to adversely affect HIV-1 replication show divergent activities against other retroviruses. Nonnucleoside reverse transcriptase (RT) inhibitors such as nevirapine are highly selective for HIV-1 (50), whereas the nucleoside RT inhibitor (NRTI) azidothymidine (AZT) inhibits infection by a variety of viruses, including the primate lentiviruses HIV-2 (35) and simian immunodeficiency virus (SIV) (47), the nonprimate lentiviruses bovine immunodeficiency virus (BIV) (46) and feline immunodeficiency virus (FIV) (31), gammaretroviruses (34, 39, 42, 43), and Spumavirus (29). Protease inhibitors harbor an intermediate phenotype which is highly active against HIV-2/SIV (18) but ineffective against the Gammaretrovirus xenotropic murine leukemia virus-related virus (42). RAL and EVG were previously shown to be effective against HIV-2/SIV (21, 38, 40), gammaretroviruses (1, 34, 40, 42, 43), and the Spumavirus prototype foamy virus (PFV) (48), suggesting that they, like NRTIs, might harbor pantropic antiretroviral activities. To comprehensively address this question, we determined RAL and EVG concentrations required to inhibit infection by vectors derived from five different lentiviruses, the Betaretrovirus Mason-Pfizer monkey virus (MPMV), the Alpharetrovirus Rous sarcoma virus (RSV), and the Gammaretrovirus Moloney murine leukemia virus (MLV). Moreover, we extended the analysis to nonviral elements that transpose intracellularly.
Experimental system.
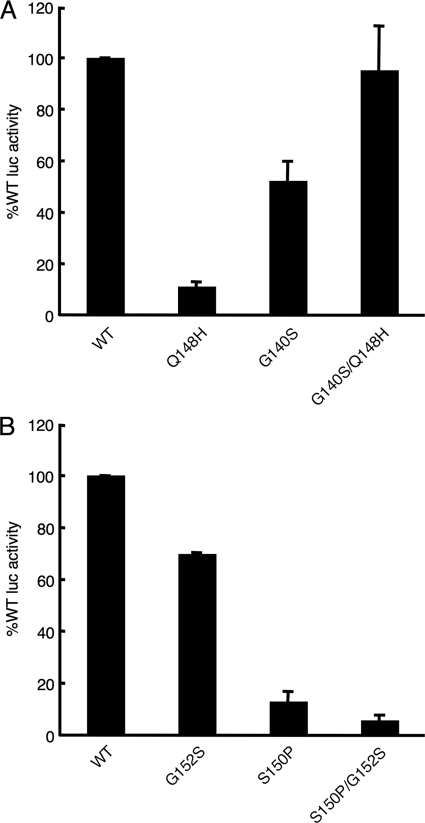
Resistance to RAL arises through one of three genetic pathways, Y143H/R/C, Q148H/R/K, or N155H (5), and substitutions at Gln148 confer significant cross-resistance to EVG (16). To start, an ex vivo infection assay (17) was calibrated to wide-ranging INSTI sensitivities by determining the 50% effective concentrations (EC50s) and EC95 doses of RAL and EVG using the wild-type (WT) IN and the Q148H, G140S, and Q148H/G140S mutants constructed in HIV-1NLX.Luc.R−, a single-round strain that expresses firefly luciferase from the HIV-1NL4-3 nef position (22). Viruses pseudotyped with vesicular stomatitis virus G (VSV-G) glycoprotein by cotransfecting 293T cells were measured by exogenous RT assay, and equal counts per minute (cpm) were applied in duplicate to HeLa-T4 cells (17). Two days after being infected, cells were lysed and resulting luciferase activities were corrected for total protein concentrations. The normalized level of infection by the Q148H IN mutant was reduced about 9-fold compared to that of the WT, whereas the G140S mutation conferred about a 2-fold defect in virus infectivity (Fig. 1 A). The double mutant, by contrast, infected cells at a level similar to that of the WT (Fig. 1A). The EC50 and EC95 values for RAL (AIDS Research and Reference Reagent Program [RRRP], Germantown, MD) and EVG (Selleck Chemicals, Houston, TX) against WT HIV-1NLX.Luc.R− were about 8 and 1.3 nM, respectively (Table 1). The G140S mutation elicited relatively small fold changes (FCs) in the EC50s of RAL and EVG, approximately 1.5 and 4.3, respectively. About 24- and 10-fold-higher concentrations of RAL and EVG, respectively, were required to thwart 50% of Q148H infectivity. The double mutation conferred significant resistance to both compounds, as the EC50s of RAL and EVG were about 1.9 μM and 2.3 μM, respectively (FC values, 241 and 1,762, respectively; Table 1). These results are fully consistent with those of previous studies that concluded the secondary G140S change increased the overall level of resistance to RAL conferred by the primary Q148H mutation and repaired an inherent IN catalytic defect (6, 26). Moreover, they established our ability to detect relatively large FCs in sensitivity to INSTIs (Table 1).
FIG. 1.
IN mutant virus infectivities. (A) Normalized levels of HIV-1 IN mutant infectivities compared to that of the WT, which was set at 100%. (B) Same as panel A, except that RSV was studied.
TABLE 1.
Activities of RAL and EVG against wild-type and IN mutant virusesa
| Virus | RALb |
EVGb |
||
|---|---|---|---|---|
| EC50 (μM) | EC95 (μM) | EC50 (μM) | EC95 (μM) | |
| HIV-1 | 0.008 ± 0.002 | 0.089 ± 0.004 | 0.0013 ± 0.0006 | 0.024 ± 0.010 |
| G140S mutant | 0.012 ± 0.002 (1.5) | 0.16 ± 0.11 (1.8) | 0.0056 ± 0.0022 (4.3) | 0.079 ± 0.003 (3.3) |
| Q148H mutant | 0.19 ± 0.10 (24) | 1.64 ± 0.89 (18) | 0.013 ± 0.005 (10) | 0.092 ± 0.007 (3.8) |
| G140S/Q148H mutant | 1.93 ± 0.24 (241) | 45.8 ± 6.8 (515) | 2.29 ± 0.65 (1,762) | >10 (>417) |
Means ± standard deviations obtained from three independent experiments, each conducted in duplicate.
The FC in drug resistance of the IN mutant relative to that of the wild-type is indicated in parentheses.
Inhibition of retroviral infection by INSTIs.
Cells were next challenged with VSV-G-pseudotyped single-round reporter constructs derived from HIV-2 strain ROD (54), SIV from macaques (SIVmac), RSV (51), FIV, BIV, MLV (20), or MPMV (30) in the absence or presence of AZT (AIDS RRRP), RAL, or EVG. FCs in EC50 and EC95 values for the drugs were determined based on the EC50s and EC95s against HIV-1NLX.Luc.R−, which was included in parallel infections. Accordingly, a virus for which the FC in EC50 was ≤5 was considered highly susceptible to the challenge compound, that for which the FC was >5 and <50, moderately drug sensitive, and that for which the FC was ≥50, relatively insensitive. As expected (28, 31, 35, 39, 46, 47), AZT effectively inhibited all retroviruses, yielding FC values that ranged from a low of 0.4 for RSV to a high of 2.4 for FIV (Table 2). In contrast, relatively large FC spectra were determined for the INSTIs. As previously reported (21, 38, 40), RAL and EVG each potently inhibited infection by HIV-2 and SIVmac. RAL potently inhibited MLV (1), while EVG acted moderately, with an FC of 39. RAL also potently inhibited MPMV, whereas EVG was particularly effective against all lentiviruses. RAL displayed intermediate strength against FIV, BIV, and RSV, while an intermediate dose of EVG was required to inhibit MPMV. Of all tested virus-drug combinations, one stood out as relatively ineffective: RSV naturally resisted EVG (Table 2).
TABLE 2.
Antiviral activities of INSTIs and AZTa
| Virus | Genus | RALb |
EVGb |
AZTb |
|||
|---|---|---|---|---|---|---|---|
| EC50 (μM) | EC95 (μM) | EC50 (μM) | EC95 (μM) | EC50 (μM) | EC95 (μM) | ||
| HIV-1 | Lentivirus | 0.0084 ± 0.0028 | 0.098 ± 0.0029 | 0.0019 ± 0.0013 | 0.035 ± 0.007 | 0.071 ± 0.027 | 0.81 ± 0.10 |
| HIV-2 | Lentivirus | 0.020 ± 0.008 (2.4) | 0.20 ± 0.04 (2.0) | 0.0031 ± 0.0011 (1.6) | 0.064 ± 0.018 (1.8) | 0.04 ± 0.009 (0.6) | 0.87 ± 0.13 (1.1) |
| SIVmac | Lentivirus | 0.009 ± 0.002 (1.1) | 0.11 ± 0.01 (1.1) | 0.003 ± 0.0003 (1.6) | 0.054 ± 0.017 (1.5) | 0.033 ± 0.007 (0.5) | 1.1 ± 0.4 (1.4) |
| FIV | Lentivirus | 0.17 ± 0.05 (20) | 3.8 ± 0.1 (39) | 0.0056 ± 0.0006 (2.9) | 0.095 ± 0.002 (2.7) | 0.17 ± 0.08 (2.4) | 7.9 ± 0.8 (9.8) |
| BIV | Lentivirus | 0.19 ± 0.006 (23) | 6.9 ± 1.4 (70) | 0.0057 ± 0.0011 (3.0) | 0.23 ± 0.08 (6.6) | 0.12 ± 0.03 (1.7) | 8.8 ± 0.2 (11) |
| RSV | Alpharetrovirus | 0.13 ± 0.005 (15) | 5.8 ± 1.8 (59) | 7.2 ± 0.8 (3,789) | >10 (>286) | 0.03 ± 0.009 (0.4) | 0.52 ± 0.10 (0.6) |
| MPMV | Betaretrovirus | 0.0086 ± 0.0007 (1.0) | 0.27 ± 0.03 (2.8) | 0.050 ± 0.0007 (26) | 0.99 ± 0.02 (28) | 0.066 ± 0.016 (0.9) | 3.0 ± 0.6 (3.7) |
| MLV | Gammaretrovirus | 0.0042 ± 0.0001 (0.5) | 0.21 ± 0.03 (2.1) | 0.075 ± 0.011 (39) | 1.14 ± 0.31 (33) | 0.043 ± 0.005 (0.6) | 0.66 ± 0.05 (0.8) |
| XMRVc | Gammaretrovirus | 0.0022 ± 0.0011 (0.3) | ND | 0.087 ± 0.029 (46) | ND | 0.06 ± 0.02 (0.9) | ND |
| PFVd | Spumavirus | 0.06 (7.1) | ND | 0.8 (421) | ND | ND | ND |
IN residue Ser150 contributes significantly to RSV resistance to EVG.
The majority of RAL and EVG resistance changes occur within the catalytic core domain (CCD) of HIV-1 IN and, moreover, cluster around Glu152 within its D,D-35-E active-site motif (27). To appreciate IN amino acid residues that might confer resistance to RAL or EVG, sequences from the heart of CCDs, corresponding to HIV-1 IN residues 91 to 169, were aligned (Fig. 2). To facilitate interpretation, residues that when altered can contribute to HIV-1 resistance were color coded green, and amino acid differences known to confer resistance, magenta (9, 16, 24, 40); gray represents changes whose contributions to drug resistance are unknown. BIV IN carries His at the position corresponding to Asn155 in HIV-1 IN, perhaps contributing to its 23-fold-reduced sensitivity to RAL (Fig. 2 and Table 2). We noted, however, that His at this position is not a universal resistance predictor, as MPMV was as sensitive to the drug as HIV-1. Based on acute gammaretroviral sensitivity to RAL, we concluded that Ser at the position corresponding to Gln148 in HIV-1 is insufficient to confer resistance in these contexts (Fig. 2). Though the FIV IN sequence does not harbor a change known to confer primary resistance, we speculate that Gly at the position corresponding to Tyr143 in HIV-1 IN is likely to contribute due to the lack of a phenolic side chain, which directly contacts the RAL oxadiazole group in PFV IN-vDNA co-crystal structures (10, 11).
FIG. 2.
IN sequence alignment and contribution of potential amino acid residues to resistance to INSTIs. Green indicates residues that when changed can confer resistance to RAL and/or EVG; magenta marks residues known to confer resistance when present at the analogous HIV-1 position; gray indicates residues with unknown effects on potential HIV-1 resistance; red indicates active-site residues; and blue highlights a conserved DNA binding residue (15). Numbers above the alignment indicate HIV-1 amino acid positions; those to the left and right mark positions in the respective IN or transposase protein sequences. Underlining marks the positions of secondary structural elements for HIV-1 (19), SIV (4), RSV (52), and PFV (10) INs and the positions of SB elements from a structure-based alignment with the related Mos1 transposase (37).
RSV IN harbors two amino acids, Ser150 and Gly152, that when present in HIV-1 as P145S or S147G confer significant resistance to EVG (16, 24, 40). To test if either or both of these residues contribute to the relative insensitivity to EVG, S150P and G152S changes were introduced into RSV IN either alone or in combination, and equivalent RT cpm of VSV-G-pseudotyped virions were used to infect cells. The G152S change elicited an approximate 30% drop in RSV infectivity, while the S150P mutant virus was only 13% as infectious as the WT. The S150P/G152S double mutant was more defective, retaining about 6% of WT function (Fig. 1B). Nearly equal doses of EVG were required to inhibit WT and G152S IN mutant infections (Table 3). Impressively, the S150P change increased RSV sensitivity to EVG approximately 74-fold, and the addition of the G152S change did not further impact this effect (Table 3). To ascertain if inherently weak S150P IN mutant infectivity contributed to increased sensitivity, experiments were repeated with a 10-fold-higher dose of the mutant than of the WT. Because this yielded an EC50 of 0.1 ± 0.02 μM, we concluded that Ser150 contributes significantly to the natural resistance of RSV to EVG.
TABLE 3.
Antiviral activities against wild-type and IN mutant RSVa
| Virus | RALb |
EVGb |
AZTb |
|||
|---|---|---|---|---|---|---|
| EC50 (μM) | EC95 (μM) | EC50 (μM) | EC95 (μM) | EC50 (μM) | EC95 (μM) | |
| RSV | 0.15 ± 0.03 | 7.2 ± 0.3 | 7.2 ± 0.3 | >10 | 0.036 ± 0.006 | 0.66 ± 0.0004 |
| G152S mutant | 0.85 ± 0.23 (5.7) | 9.2 ± 0.2 (1.3) | 7.7 ± 1.1 (1.1) | >10 (>1.0) | 0.031 ± 0.002 (0.9) | 0.55 ± 0.08 (0.8) |
| S150P mutant | 0.29 ± 0.04 (1.9) | 8.9 ± 0.8 (1.2) | 0.098 ± 0.014 (0.01) | 5.0 ± 0.4 (<0.5) | 0.037 ± 0.004 (1.0) | 0.67 ± 0.03 (1.0) |
| S150P/G152S mutant | 0.64 ± 0.30 (4.3) | 9.4 ± 1.0 (1.3) | 0.19 ± 0.03 (0.03) | >10 (>1.0) | 0.036 ± 0.002 (1.0) | 0.65 ± 0.01 (1.0) |
Means ± standard deviations of three independent experiments, each conducted in duplicate.
The FC in EC50 or EC95 relative to that of the wild-type is indicated in parentheses.
Nonviral sensitivities to INSTIs.
The abilities of RAL and EVG to inhibit intracellular transposition of noninfectious elements were tested next. Intracisternal A-particles (IAPs) are LTR retroelements that rely on RT and IN activities for retrotransposition (reviewed in reference 2) and are indigenous to mice. Sleeping Beauty (SB) is a member of the Tc1/mariner family of DNA transposable elements that moves from one genomic position to another via the activity of its transposase protein, which harbors a D,D-34-E active-site motif (Fig. 2), in the absence of an RNA intermediate (reviewed in reference 36). Though long interspersed nucleotide element 1 (LINE-1) retrotransposition occurs via an RNA intermediate, chromosomal DNA nicking in this case occurs via a functionally distinct endonuclease that is not a member of the polynucleotidyl transferase superfamily (49), and therefore LINE-1 served as a negative control in these assays.
Transposition was assessed using recombinant elements carrying the reporter gene for green fluorescence protein (GFP). Accordingly, IAP retrotransposition was scored in HeLa cells (30,000 plated the previous day in 48-plate wells) cotransfected with 0.1 μg of pDE1 bearing a GFP-intron cassette and 0.1 μg of pQ14CAG (13), which encodes IAP proteins, or 0.1 μg of pUC19 to define the assay background, using FuGENE 6 (Roche Molecular Biochemicals, Indianapolis, IN) in the absence or presence of RAL, EVG, or AZT. Seven days thereafter, GFP-positive cells were quantified using a FACSCanto flow cytometer equipped with FACSDiva software (BD, Franklin Lakes, NJ). For SB transposition, cells cotransfected with 0.15 μg of pT2βGFP (53) and 0.075 μg of pSB100X (14, 23) or 0.075 μg of pUC19 were developed similarly by flow cytometry at day 7. Cells transfected with 0.2 μg of LINE-1 bearing L1RP-EGFP(puro) or negative control pL1RP(JM111)-EGFP(puro) were also scored after 7 days, with the addition of puromycin (2 μg/ml) at 48 h posttransfection to enhance the selection of de novo events (33).
As expected, neither RAL nor EVG detectably inhibited LINE-1 retrotransposition (Table 4). Similarly, AZT was inactive against SB transposition. IAP retrotransposition was acutely inhibited by AZT, moderately inhibited by RAL, and somewhat less sensitive to EVG. Despite utilizing a DDE active site, SB transposase was not detectably inhibited by RAL or EVG (Table 4).
TABLE 4.
RAL and EVG activities against nonviral elementsa
| Element | RAL (μM) | EVG (μM) | AZT (μM) | Transposition rate of no drug controlb |
|---|---|---|---|---|
| LINE-1 | >10 (>1,190)c | >10 (>5,263) | >10d (>140) | 2.2 ± 0.2 |
| IAP | 0.37 ± 0.18 (44) | 0.33 ± 0.19 (174) | 0.020 ± 0.004 (0.3) | 2.7 ± 1.1 |
| SB | >10 (>1,190) | >10 (>5,263) | >10 (>140) | 22.0 ± 5.1 |
Means ± standard deviations from two to three independent experiments of concentrations required to inhibit 50% of the control transposition rate.
Percentage of GFP-positive cells after background correction.
The FC in concentration compared to that for HIV-1 (Table 2) is indicated in parentheses.
45% inhibition at 10 μM.
Conclusions.
The results of this study reveal that the prototype INSTIs RAL and EVG differentially inhibit the activities of LTR-containing retroviruses and retrotransposons yet are ineffective against the non-LTR LINE-1 retrotransposon and the DNA transposon SB. EVG was more lentiviral specific than RAL, and RSV rather impressively resisted EVG (Table 2), which was in large part attributable to IN residue Ser150 (Table 3). Based on these results, we speculate that element-specific amino acid sequences likely dictate sensitivities to INSTIs. We note that SB transposase, like RSV IN, harbors serine at the position analogous to Pro145 in HIV-1 IN (Fig. 2). Differences at transposase residues analogous to HIV-1 IN positions Tyr143, Gln148, and Asn155 might accordingly account for the RAL resistance of this element. Unlike LTR retroelements that harbor the strand transfer nucleophile as part of a recessed 3′ or, more rarely, blunt DNA end, transposition of SB occurs via a 3′ overhang (36). Because key INSTI-vDNA contacts occur via the penultimate LTR C/G base pair (10, 11), it seems possible that the lack of pairing bases at the reactive transposon end might also contribute to INSTI resistance. Based on the relative activities of low-micromolar preclinical compounds, the Tc/mariner Mos1 transposase has been proposed as a surrogate to identify HIV-1 IN inhibitors (3). Our results indicate potential limitations to utilizing Tc/mariner elements to identify highly efficacious drugs.
The inability to degrade preintegrative retrotransposon DNA can increase susceptibility to autoimmune dysfunction (44), and inhibition by RAL transiently increases unintegrated LTR circular DNA forms during acute HIV-1 infection (12). The rate of onset of autoimmune disease has, moreover, been observed to increase in susceptible mice treated with RAL (1). As we have demonstrated differential susceptibilities of retroviruses and retrotransposons to inhibition by RAL versus EVG (Tables 2 and 4), novel INSTIs should be evaluated individually for inhibitory activities against potentially medically relevant (1, 25) non-HIV elements.
Acknowledgments
We are indebted to the following colleagues for their generous donations of reagents: H. Kazazian for L1RP-EGFP(puro) and pL1RP(JM111)-EGFP(puro), J. Takeda for pDE1 and pQ14CAG, P. Mead for pT2βGFP, Z. Izsvák for pSB100X, W. Johnson and E. Hunter for pSARM-EGFP, and T. Hatziioannou for the HIV-2ROD reporter virus. RAL and AZT were obtained from the NIH AIDS Research and Reference Reagent Program.
This work was supported by U.S. NIH grant AI039394 (A.E.), the Mitsubishi Pharma Foundation (Y.K.), the Japanese Association for Infectious Diseases (Y.K.), the Japanese Society of Chemotherapy (Y.K.), and the Harvard University Center for AIDS Research, an NIH-funded program (P30AI060354) that is supported by the following NIH institutes and centers: NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, and NCCAM.
Expressed scientific views and commercial reagent sources in no way reflect opinions or endorsement by funding agencies.
Footnotes
Published ahead of print on 26 January 2011.
REFERENCES
- 1.Beck-Engeser, G. B., D. Eilat, T. Harrer, H. M. Jäck, and M. Wabl. 2009. Early onset of autoimmune disease by the retroviral integrase inhibitor raltegravir. Proc. Natl. Acad. Sci. U. S. A. 106:20865-20870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeke, J. D., and J. P. Stoye. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements p. 343-435. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- 3.Bouchet, N., et al. 2009. First Mariner Mos1 transposase inhibitors. Mini Rev. Med. Chem. 9:431-439. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Z., et al. 2000. X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-293)—an initial glance of the viral DNA binding platform. J. Mol. Biol. 296:521-533. [DOI] [PubMed] [Google Scholar]
- 5.Cooper, D. A., et al. 2008. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N. Engl. J. Med. 359:355-365. [DOI] [PubMed] [Google Scholar]
- 6.Delelis, O., et al. 2009. The G140S mutation in HIV integrases from raltegravir-resistant patients rescues catalytic defect due to the resistance Q148H mutation. Nucleic Acids Res. 37:1193-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dyda, F., et al. 1994. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266:1981-1986. [DOI] [PubMed] [Google Scholar]
- 8.Engelman, A. 2010. Reverse transcription and integration, p. 129-159. In R. Kurth and N. Bannert (ed.), Retroviruses: molecular biology, genomics and pathogenesis. Caister Academic Press, Norfolk, United Kingdom.
- 9.Fransen, S., et al. 2009. Loss of raltegravir susceptibility by human immunodeficiency virus type 1 is conferred via multiple nonoverlapping genetic pathways. J. Virol. 83:11440-11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hare, S., S. S. Gupta, E. Valkov, A. Engelman, and P. Cherepanov. 2010. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464:232-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hare, S., et al. 2010. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc. Natl. Acad. Sci. U. S. A. 107:20057-20062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hazuda, D. J., et al. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 13.Horie, K., et al. 2007. Retrotransposons influence the mouse transcriptome: implication for the divergence of genetic traits. Genetics 176:815-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Izsvák, Z., M. K. Chuah, T. Vandendriessche, and Z. Ivics. 2009. Efficient stable gene transfer into human cells by the Sleeping Beauty transposon vectors. Methods 49:287-297. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins, T. M., D. Esposito, A. Engelman, and R. Craigie. 1997. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 16:6849-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi, M., et al. 2008. Selection of diverse and clinically relevant integrase inhibitor-resistant human immunodeficiency virus type 1 mutants. Antiviral Res. 80:213-222. [DOI] [PubMed] [Google Scholar]
- 17.Koh, Y., H. Haim, and A. Engelman. 2011. Identification and characterization of persistent intracellular human immunodeficiency virus type 1 strand transfer inhibitor activity. Antimicrob. Agents Chemother. 55:42-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koh, Y., et al. 2003. Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob. Agents Chemother. 47:3123-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krishnan, L., et al. 2010. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc. Natl. Acad. Sci. U. S. A. 107:15910-15915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnan, L., et al. 2010. The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. J. Virol. 84:397-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis, M. G., et al. 2010. Response of a simian immunodeficiency virus (SIVmac251) to raltegravir: a basis for a new treatment for simian AIDS and an animal model for studying lentiviral persistence during antiretroviral therapy. Retrovirology 7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu, R., et al. 2004. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J. Virol. 78:12735-12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mátés, L., et al. 2009. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 41:753-761. [DOI] [PubMed] [Google Scholar]
- 24.McColl, D. J., and X. Chen. 2010. Strand transfer inhibitors of HIV-1 integrase: bringing IN a new era of antiretroviral therapy. Antiviral Res. 85:101-118. [DOI] [PubMed] [Google Scholar]
- 25.Melek, M., et al. 2002. Effect of HIV integrase inhibitors on the RAG1/2 recombinase. Proc. Natl. Acad. Sci. U. S. A. 99:134-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Métifiot, M., et al. 2010. Biochemical and pharmacological analyses of HIV-1 integrase flexible loop mutants resistant to raltegravir. Biochemistry 49:3715-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Métifiot, M., C. Marchand, K. Maddali, and Y. Pommier. 2010. Resistance to integrase inhibitors. Viruses 2:1347-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitsuya, H., et al. 1985. 3′-Azido-3′-deoxythymidine (BW A509U): an antiviral agent that inhibits the infectivity and cytopathic effect of human T-lymphotropic virus type III/lymphadenopathy-associated virus in vitro. Proc. Natl. Acad. Sci. U. S. A. 82:7096-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moebes, A., et al. 1997. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 71:7305-7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman, R. M., et al. 2006. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5alpha. Proc. Natl. Acad. Sci. U. S. A. 103:19134-19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.North, T. W., G. L. North, and N. C. Pedersen. 1989. Feline immunodeficiency virus, a model for reverse transcriptase-targeted chemotherapy for acquired immune deficiency syndrome. Antimicrob. Agents Chemother. 33:915-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowotny, M. 2009. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 10:144-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ostertag, E. M., E. T. Prak, R. J. DeBerardinis, J. V. Moran, and H. H. Kazazian. 2000. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 28:1418-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paprotka, T., et al. 2010. Inhibition of xenotropic murine leukemia virus-related virus by APOBEC3 proteins and antiviral drugs. J. Virol. 84:5719-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pauwels, R., et al. 1990. Potent and selective inhibition of HIV-1 replication in vitro by a novel series of TIBO derivatives. Nature 343:470-474. [DOI] [PubMed] [Google Scholar]
- 36.Plasterk, R. H., and H. G. van Luenen. 2002. The Tc1/mariner family of transposable elements, p. 519-532. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 37.Richardson, J. M., S. D. Colloms, D. J. Finnegan, and M. D. Walkinshaw. 2009. Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote. Cell 138:1096-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roquebert, B., et al. 2008. HIV-2 integrase gene polymorphism and phenotypic susceptibility of HIV-2 clinical isolates to the integrase inhibitors raltegravir and elvitegravir in vitro. J. Antimicrob. Chemother. 62:914-920. [DOI] [PubMed] [Google Scholar]
- 39.Ruprecht, R. M., L. G. O'Brien, L. D. Rossoni, and S. Nusinoff-Lehrman. 1986. Suppression of mouse viraemia and retroviral disease by 3′-azido-3′-deoxythymidine. Nature 323:467-469. [DOI] [PubMed] [Google Scholar]
- 40.Shimura, K., et al. 2008. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137). J. Virol. 82:764-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimura, K., and E. N. Kodama. 2009. Elvitegravir: a new HIV integrase inhibitor. Antivir. Chem. Chemother. 20:79-85. [DOI] [PubMed] [Google Scholar]
- 42.Singh, I. R., J. E. Gorzynski, D. Drobysheva, L. Bassit, and R. F. Schinazi. 2010. Raltegravir is a potent inhibitor of XMRV, a virus implicated in prostate cancer and chronic fatigue syndrome. PLoS One 5:e9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, R. A., G. S. Gottlieb, and A. D. Miller. 2010. Susceptibility of the human retrovirus XMRV to antiretroviral inhibitors. Retrovirology 7:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stetson, D. B., J. S. Ko, T. Heidmann, and R. Medzhitov. 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Summa, V., et al. 2008. Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J. Med. Chem. 51:5843-5855. [DOI] [PubMed] [Google Scholar]
- 46.Tobin, G. J., W. H. Ennis, D. J. Clanton, and M. A. Gonda. 1996. Inhibition of bovine immunodeficiency virus by anti-HIV-1 compounds in a cell culture-based assay. Antiviral Res. 33:21-31. [DOI] [PubMed] [Google Scholar]
- 47.Tsai, C. C., et al. 1995. Prevention of SIV infection in macaques by (R)-9-(2-phosphonylmethoxypropyl)adenine. Science 270:1197-1199. [DOI] [PubMed] [Google Scholar]
- 48.Valkov, E., et al. 2009. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 37:243-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weichenrieder, O., K. Repanas, and A. Perrakis. 2004. Crystal structure of the targeting endonuclease of the human LINE-1 retrotransposon. Structure 12:975-986. [DOI] [PubMed] [Google Scholar]
- 50.Witvrouw, M., et al. 1999. Activity of non-nucleoside reverse transcriptase inhibitors against HIV-2 and SIV. AIDS 13:1477-1483. [DOI] [PubMed] [Google Scholar]
- 51.Yan, N., P. Cherepanov, J. E. Daigle, A. Engelman, and J. Lieberman. 2009. The SET complex acts as a barrier to autointegration of HIV-1. PLoS Pathog. 5:e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang, Z.-N., T. C. Mueser, F. D. Bushman, and C. C. Hyde. 2000. Crystal structure of an active two-domain derivative of Rous sarcoma virus integrase. J. Mol. Biol. 296:535-548. [DOI] [PubMed] [Google Scholar]
- 53.Yergeau, D. A., et al. 2009. Transgenesis in Xenopus using the Sleeping Beauty transposon system. Dev. Dyn. 238:1727-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, F., T. Hatziioannou, D. Perez-Caballero, D. Derse, and P. D. Bieniasz. 2006. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology 353:396-409. [DOI] [PubMed] [Google Scholar]