Abstract
BHK cells remain resistant to xenotropic murine retrovirus-related virus (XMRV) or gibbon ape leukemia virus (GALV) infection, even when their respective receptors, Xpr1 or PiT1, are expressed. We set out to determine the stage at which viral infection is blocked and whether this block is mediated by a dominant-negative factor or the absence of a requisite ancillary factor. BHK cells bind neither XMRV nor GALV envelope proteins. BHK cells expressing the appropriate receptors bind XMRV or GALV envelope proteins. BHK cells can be infected by NZB-XMV(New Zealand Black mouse xenotropic murine virus)-enveloped vectors, expressing an envelope derived from a xenotropic retrovirus that, like XMRV, employs Xpr1 as a receptor, and also by vectors bearing the envelope of 10A1 murine leukemia virus (MLV), a murine retrovirus that can use PiT1 as a receptor. The retroviral vectors used in these analyses differ solely in their viral envelope proteins, suggesting that the block to XMRV and GALV infection is mediated at the level of envelope-receptor interactions. N-linked glycosylation of the receptors was not found to mediate resistance of receptor-expressing BHK cells to GALV or XMRV, as shown by tunicamycin treatment and mutation of the specific glycosylation site of the PiT1 receptor. Hybrid cells produced by fusing BHKXpr1 or BHKPiT1 to XMRV- or GALV-resistant cells, respectively, can mediate efficient XMRV or GALV infection. These findings indicate that BHK cells lack a factor that is required for infection by primate xenotropic viruses. This factor is not required for viruses that use the same receptors but were directly isolated from mice.
The prototypic xenotropic retrovirus NZB-XMV (New Zealand Black mouse xenotropic murine virus) was recovered from New Zealand Black mouse embryo fibroblasts as well as from adult NZB mouse tissues and was originally described as unique among other murine leukemia viruses (MLVs) in that it fails to propagate in the mouse cells from which it was recovered but grows in a wide range of cells derived from heterologous species, including human cells (19). Since then it has been shown that XMVs use Xpr1 as a receptor (3, 26, 36), and certain murine cells are resistant because XMVs fail to employ the murine Xpr1 ortholog expressed on laboratory mice and European Mus musculus domesticus-derived cells but can use the Xpr1 ortholog found on other feral mouse cells (35).
Gibbon ape leukemia virus (GALV) represents a xenotropic virus isolated from primates (gibbon apes) (16) that, like XMVs, can infect a wide variety of cells from heterologous species but is unable to infect laboratory mouse-derived cells. In contrast to XMVs, GALV fails to infect most cells derived from feral mice, including Mus dunni cells (18). GALV also shows a significant potential for penetrance into other mammalian species, as evidenced by the isolation of conspecific viruses from a woolly monkey (WMV) (29) and koalas (KoRV) (15). The recent recovery of a xenotropic murine leukemia virus-related virus (XMRV) from human prostate cancer tissues, peripheral blood mononuclear cells (PBMCs) of chronic fatigue syndrome patients, and the respiratory tract of immunocompromised patients (14, 20, 32) raises the concern of a threat to public health from cross-species transmission of XMVs. It is with this in mind that we sought to investigate differences in the infectivity properties of XMVs isolated from primates from those originally isolated from mice. As previously reported, XMVs isolated from mice have a broader host range than XMRV (35) or GALV (22). We initially posited that the expanded host range of murine XMVs was attributable to their ability to use a wider repertoire of receptor orthologs. NZB-XMV and XMRV both infect human cells by employing the human Xpr1 receptor. 10A1 MLV and GALV utilize human PiT1, a transporter of inorganic phosphate (Pi) also referred to as SLC20A1, as a receptor. Expression of Xpr1 or PiT1 in resistant hamster CHOK1 cells renders them susceptible to XMRV (6) and GALV (22), respectively. In contrast, hamster BHK cells are susceptible to NZB-XMV and 10A1 MLV but resistant to XMRV and GALV, and expression of human Xpr1 or PiT1 does not render BHK cells susceptible to either XMRV or GALV. Thus, in BHK cells, expression of a functional receptor is not sufficient to confer susceptibility to these primate viral isolates. Studies of the spread of xenotropic viruses can provide important information enabling the identification of risk factors that may facilitate their spread to humans. Therefore, determination of important cellular factors, in addition to the viral receptor, that are involved in the entry of XMRV and GALV into target cells is imperative.
MATERIALS AND METHODS
Cell lines.
BHK hamster kidney cells (ATCC CCL 10), 293T human embryonic kidney cells (ATCC CCL 11268), Mus dunni tail fibroblast (MDTF) cells (18), and cells of the Chinese hamster E36 line (provided by Christine Kozak, NIAID, Bethesda, MD) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Chinese hamster ovary cells of the CHOK1 line (ATCC CCL 61) were grown in alpha-MEM medium containing the same supplements as those provided in cells grown in DMEM. Cells of CHOK1 derivatives pgsA745 and pgsA745XTII (8) were obtained from Frank W. Ruscetti (SAIC, Frederick, MD) and grown under the same conditions as CHOK1 cells. MDTF cells expressing different receptors were constructed by transducing these cells with retroviral vectors using vesicular stomatitis virus glycoprotein (VSV-G) envelope and a pLNSX-derived genome expressing the appropriate receptor cDNA, as previously described (10). Hygromycin-resistant MDTF (MDTF-hygro) cells were obtained by transducing MDTF cells with VSV-G-enveloped pLHCX genome. Puromycin-resistant CHOK1 (CHOK1-puro) cells were made similarly by using transduced pBABE-puro as a selectable marker. After transduction for 48 h, MDTF-Hygro and CHOK1-puro cells were selected with 800 mg/ml hygromycin and 6 mg/ml puromycin, respectively, for 2 weeks.
Production of retroviral vectors and infection.
The calcium phosphate precipitation method (Promega, Madison, WI) was used to transfect 293T cells by using a 3-plasmid system encoding envelope, MLV Gag/Pol, and retroviral genomes on individual plasmids as previously described (10). The viral supernatant was collected 48 and 72 h after transfection, pooled, and then passed through a 0.45-μm-pore-size filter (Millipore, Bedford, MA) before the supernatant was applied to target cells. Target cells were seeded in a 24-well plate at a density of 4 × 104 cells per well the day before infection. Infection with viruses using different envelopes, except VSV-G, was performed with 10 μg/ml Polybrene. When the β-galactosidase (LacZ) gene was used as the indicator gene, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining was carried out 48 h after exposing cells to viral supernatant.
Plasmid construction.
XMRV envelope-expressing plasmid pHCMVenv VP62 was generously provided by Kathryn S. Jones (SAIC, Frederick, MD). The NZB-XMV envelope-expressing plasmid pFBsalf-XMVenv and human Xpr1-expressing plasmid pLNSX-Xpr1 were obtained from Chetankumar S. Tailor (Toronto, Ontario, Canada). Xpr1 was tagged with a double-hemagglutinin (HA) epitope (YPYDVPDYA), as previously described for PiT1 (5). Xpr1 was tagged at the amino terminus with the FLAG epitopes obtained from the p3×FLAG-myc-CMV-25 expression vector (Sigma, St. Louis, MO). Briefly, the start codon of Xpr1 was replaced with a HindIII restriction site by PCR mutagenesis using sense primer 5′-GGAAACGGCAAGCTTAAGTTCGCC-3′, and a BamHI site was introduced into C-terminal end of Xpr1 by antisense primer 5′-GGAGACTAAATAGGATCCTTTATTTTATCG-3′. The PCR product was subcloned into p3×FLAG-myc-CMV-25 vector by replacing the sequences between the HindIII and BamHI sites. The resultant p3×FLAG-Xpr1-CMV25 plasmid has three tandem FLAG epitopes fused with the N-terminal end of Xpr1 within the same open reading frame. For the construction of FLAG-tagged Xpr1 in a retroviral vector, an AvrII site was introduced into the N-terminal FLAGXpr1 sequences by using sense primer 5′-GTTGACGCAAATGGGCCCTAGGCGTGTACG-3′ and the complementary antisense primer. The PCR product was cloned into a TA vector with the TOPO-TA cloning kit (Invitrogen, Carlsbad, CA). After AvrII and BstXI digestion, the FLAGXpr1 fragment was ligated into the original pLNSX-Xpr1 vector, replacing the Xpr1 segment within these restriction sites. The resultant pLNSX-FLAGXpr1 sequence was confirmed. The XMRV envelope surface unit (SU) glycoprotein was tagged with a double-HA tag by the same strategy used for tagging Xpr1. The HindIII sense primer 5′-GGTCTATATAAGCAAAGCTTTCTGGCTAAC-3′ and SacI antisense primer 5′-CCTTTGGCCAGAGCTCAACCAGGACACAG-3′ were used to introduce HindIII and SacI restriction sites (underlined) at the 5′ and 3′ ends of XMRV envelope SU PCR product, respectively. These two restriction sites were then used to subclone XMRV envelope SU into pCS2-HA vector.
Binding assay and flow cytometry analysis.
pCS2 plasmids expressing HA-tagged soluble XMRV SU or V5-tagged GALV SU (11) were transfected into 293T cells. Forty-eight hours posttransfection, the supernatants were used to perform the binding assay. Target BHK cells expressing various receptor cDNAs were detached from tissue culture flasks with Cellstripper cell dissociation solution (Mediatech, Reston, VA); 5 × 105 cells for each receptor cell line were suspended in 0.5 ml of supernatant containing soluble HA-tagged XMRV SU or V5-tagged soluble GALV SU and incubated at 37°C for 45 min with shaking. Cells were washed twice with Hanks buffered saline solution (HBSS) containing 1% fetal bovine serum and then incubated for 30 min at room temperature in the presence of 1 μg of anti-HA-Alexa Fluor 488 (Invitrogen) or anti-V5-fluorescein isothiocyanate (FITC) (Invitrogen) monoclonal antibody in HBSS; cells were then washed with HBSS and analyzed by flow cytometry. The accessibility of N and C termini of Xpr1 on the membrane of nonpermeabilized cells was detected with monoclonal anti-FLAG M2-FITC conjugate antibody (Sigma) for FLAG-Xpr1 expressed in BHK or E36 cells and monoclonal antibody anti-HA-Alexa Fluor 488 for Xpr1HA expressed in BHK or E36 cells. The level of heparan sulfate proteoglycans (HSPGs) on CHOK1Xpr1 cells was detected by using FITC-conjugated heparan sulfate mouse anti-human monoclonal antibody (LifeSpan Biosciences, Seattle, WA).
Cysteine residue labeling.
The construction of pLNSX plasmids expressing different mutant PiT1HA proteins with single cysteine substitutions has been previously described (12). Transduction of BHK cells with VSV envelope vectors expressing different cysteine mutant PiT1HA proteins and cysteine residue labeling with 250 μM 3-(N-maleimidyl propionyl)biocytin (biotin maleimide, or B-mal) (Sigma) were performed as previously described (12). The samples were stored at −20°C before Western blot analysis.
Western blotting.
Lysates from B-mal-labeled cells were heated to 95°C for 5 min before loading on Tris-glycine-SDS-PAGE gels (Invitrogen). HA-labeled proteins were detected by incubation with anti-HA monoclonal antibody HA.11 (Covance, Princeton, NJ) at 1:1,000. After incubation with the second antibody, goat anti-mouse IgG conjugated to horseradish peroxidase (HRP) (Pierce, Rockford, IL) (1:25,000), signals were detected with Immobilon Western chemiluminescent HRP substrate (Millipore, Bedford, MA), followed by exposure and analysis with a CoolSnap HQ2 camera and Ivision software (Biovision Technologies, Exton, PA). Western blot analysis for PiT1 expression in BHK and MDTF cells using anti-HA monoclonal antibody HA.11 was performed as described above. Xpr1 expression in BHK and E36 cells was detected with anti-FLAG M2 FLAG-HRP monoclonal antibody for Xpr1 protein with a FLAG tag at the amino terminus (Sigma) or with anti-HA monoclonal antibody HA.11 for Xpr1 protein with an HA tag at the carboxy terminus.
Cell fusion.
Hybrid cells were generated by polyethylene glycol (PEG)-mediated cell fusion. For the fusion of BHKPiT1 cells with MDTF cells, 1 × 107 cells of both the BHKPiT1 and MDTF-hygro lines were mixed, 1 ml prewarmed 50% PEG 1500 (Sigma) was then added, and the cells were stirred for 3 min. Next, 3 ml of prewarmed serum-free DMEM was added to the mixture, and the cells were stirred for another 3 min. Finally, 10 ml of serum-free DMEM was added slowly to the mixture, and the fused cells were left at room temperature for 5 min. The cells were washed and plated in 10 10-cm plates. Hybrid cells were obtained after selection with 800 μg/ml of both hygromycin and G418. Fusion of BHKXpr1 cells with CHOK1 cells was carried out in a similar manner. CHOK1-puro cells that have pBABE-puro retroviral genome integrated were fused with BHKXpr1 cells by using PEG 1500, and the fused cells were selected with 6 μg/ml puromycin and 800 μg/ml G418 for 2 weeks when control CHOK1 or BHK cells were killed by drug selection.
Heparinase treatment.
CHOK1Xpr cells were treated with 5 U/ml heparinase II (Sigma) for 1 h at 30°C. Cells were then washed with HBSS and used in subsequent experiments.
RESULTS
XMRV or GALV envelope components do not permit entry in resistant cells.
We exposed cell lines derived from Chinese hamster E36, CHOK1, or MDTF cells to various gammaretroviral vectors. All vectors contain the same MLV-based β-galactosidase-encoding genome and MLV gag/pol genes. The only variable among the vectors was the envelope component. The titer of NZB-XMV vectors on CHOK1 or E36 cells was approximately 104 focus-forming units (FFU)/ml. Expression of the human ortholog of the xenotropic receptor Xpr1 enhanced the susceptibility of E36 cells to NZB-XMV infection by 2 orders of magnitude (Table 1). Even though XMRV, like NZB-XMV, employs Xpr1 as a receptor, E36 cells exposed to XMRV-enveloped vectors are resistant to infection (Table 1). Expression of Xpr1 rendered E36 cells susceptible to infection by XMRV (Table 1). Neither GALV nor 10A1 MLV can infect CHOK1 cells. 10A1 MLV and GALV differ in their abilities to infect murine MDTF cells (18). The murine retroviruses NZB-XMV and 10A1 MLV infected MDTF cells, as did XMRV, but MDTF cells were resistant to GALV, as shown in Table 1. MDTF cells that express the GALV receptor PiT1 were rendered susceptible to GALV-enveloped vectors (Table 1).
TABLE 1.
Host range is determined by viral envelope
| Target cells | Titer (FFU/ml) ofa: |
|||
|---|---|---|---|---|
| NZB-XMV | XMRV | 10A1 MLV | GALV | |
| E36 | (1.3 ± 0.3) × 104 | <5 | (6.3 ± 0.4) × 106 | (3.1 ± 0.5) × 106 |
| E36Xpr1 | (1.1 ± 0.2) × 106 | (5.6 ± 0.2) × 105 | NDb | ND |
| CHOK1 | (3.4 ± 0.5) × 102 | <5 | <5 | <5 |
| CHOK1Xpr1 | (4.5 ± 2.0) × 104 | (7.6 ± 2.8) × 103 | ND | ND |
| MDTF | (5.1 ± 0.4) × 106 | (2.9 ± 0.5) × 105 | (3.5 ± 0.5) × 106 | <5 |
| MDTFPiT1 | ND | ND | ND | (5 ± 0.5) × 106 |
Titer was determined by the mean number of FFU ± standard deviation. The average of at least three independent experiments was calculated.
ND, not determined.
Neither XMRV nor GALV can infect BHK cells even when appropriate functional receptors are expressed.
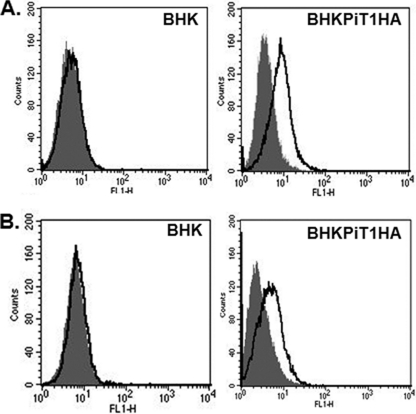
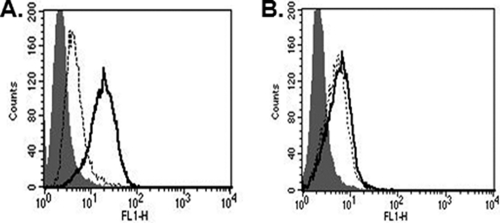
Both BHK cells and BHK cells expressing the GALV receptor PiT1 are resistant to GALV. As mentioned above, 10A1 MLV can employ the same receptor as GALV (PiT1) to infect cells, and 10A1 vectors, in contrast to GALV, can infect BHK cells (Table 2). BHK and MDTF cells display different properties of susceptibility to GALV: MDTF cells expressing PiT1 become susceptible to GALV, whereas BHK cells expressing PiT1 remain resistant (Table 2). The expression of PiT1 on BHK cells was validated by using PiT1 containing a carboxy-terminal HA epitope tag. Because the carboxy terminus of PiT1 is extracellular, it can be detected on the surface of cells by flow cytometry and an HA antibody (12) (Fig. 1A).
TABLE 2.
Inability of XMRV nor GALV to infect BHK cells, even when appropriate functional receptors are expressed
| Target cells | Titer (FFU/ml) ofa: |
||||
|---|---|---|---|---|---|
| NZB-XMV | 10A1 MLV | A-MLV | XMRV | GALV | |
| BHK | (1.6 ± 0.5) × 103 | (2.3 ± 0.4) × 103 | <5 | <5 | <5 |
| BHKFLAGXpr1 | (3.9 ± 0.8) × 104 | NDb | <5 | <5 | <5 |
| BHKPiT1HA | ND | ND | <5 | ND | <5 |
| BHKPiT2HA | ND | ND | (6.6 ± 0.5) × 104 | ND | ND |
| BHKC1FHA | ND | ND | (6.0 ± 0.2) × 104 | ND | <5 |
| BHKN96CHA | ND | ND | ND | ND | <5 |
Titer was determined by the mean number of FFU ± standard deviation. The average of at least three independent experiments was calculated.
ND, not determined.
FIG. 1.
(A) PiT1 protein is expressed on the BHK cell surface. The expression of HA on the cell surface of BHK cells expressing PiT1HA was detected by flow cytometry after staining with Alexa Fluor 488-conjugated anti-HA antibody for 1 h (areas under bold line). Isotype controls are in shaded areas. (B) BHKPiT1HA can bind GALV envelope protein. V5-tagged GALV SU was used to bind BHK or BHKPiT1HA cells as described in Materials and Methods. FITC-conjugated anti-V5 antibody was used to detect GALV SU binding on the cell surface, as represented by the areas under the bold line; shaded areas represent isotype controls.
The amphotropic murine retrovirus (A-MLV) employs PiT2 (also referred to as SLC20A2) as a receptor. PiT2, like PiT1 (SLC20A1), normally functions as a transporter of inorganic phosphate (Pi), and the two proteins share greater than 60% residue identity (22). Nonetheless, A-MLV and GALV are restricted to utilization of their respective receptors to infect human cells. BHK cells are resistant to A-MLV, but BHK cells expressing PiT2 are susceptible to infection by A-MLV vectors (Table 2). We have previously reported the construction of a chimeric receptor designated C1F that is composed of a PiT1 backbone but contains the first extracellular loop domain of PiT2 (13). C1F functions as a dual receptor for GALV and A-MLV (13). To determine whether BHK cells expressing this dual receptor could confer susceptibility to GALV or A-MLV, we expressed a carboxy-terminal HA-tagged C1F (C1FHA) in BHK cells. BHK cells expressing C1FHA are rendered susceptible to A-MLV, suggesting that C1FHA is expressed on BHK and functions as a gammaretroviral receptor; however, the resistance of BHKC1FHA to GALV infection indicated that this receptor like PiT1 is not sufficient to confer GALV infectivity to BHK cells (Table 2).
The block to XMRV or GALV infection of BHKXpr1 or BHKPiT1 is not mediated by N-linked glycosylation.
PiT1 had been previously reported to be a glycoprotein. The asparagine residue at position 96 is the single site of glycosylation (10). All the other asparagine residues present in the context of a canonical sequence for N-linked glycosylation have been shown to be intracellular (10). To test the influence of N-linked glycosylation-dependent modification of PiT1 on its receptor function in BHK cells, the glycosylation site, located in the first extracellular loop of PiT1HA, was eliminated by substitution of a cysteine for this asparagine residue by site-directed mutagenesis. The resultant mutant, PiT1N96CHA, functions efficiently as a receptor for GALV infection when the cDNA is expressed in MDTF cells (12). We expressed PiT1N96CHA in BHK cells and exposed these cells to GALV vectors. As shown in Table 2, BHKPiT1N96CHA remained resistant to GALV, indicating that N-linked glycosylation of PiT1 does not account for the resistance of BHK cells to GALV.
The Xpr1 protein has seven potential N-linked glycosylation sites. Although glycosylation-mediated inhibition of gammaretrovirus receptors expressed on hamster cells has been implicated in restricting infection by several gammaretroviral vectors (33), glycosylation modification of the Xpr1 protein expressed on hamster cells has not been implicated as conferring resistance to XMVs or XMRV (35). We tested whether inhibition of glycosylation of BHKXpr1 cells circumvented resistance to XMRV. BHKXpr1 cells incubated in medium containing 0.15 μg/ml tunicamycin for 24 h remained resistant to XMRV vectors but not NZB-XMV vectors (data not shown). Thus, N-linked glycosylation does not appear to regulate the susceptibility of BHKXpr1 cells to XMRV.
The retroviral receptors expressed on BHK cells bind XMRV or GALV envelope proteins.
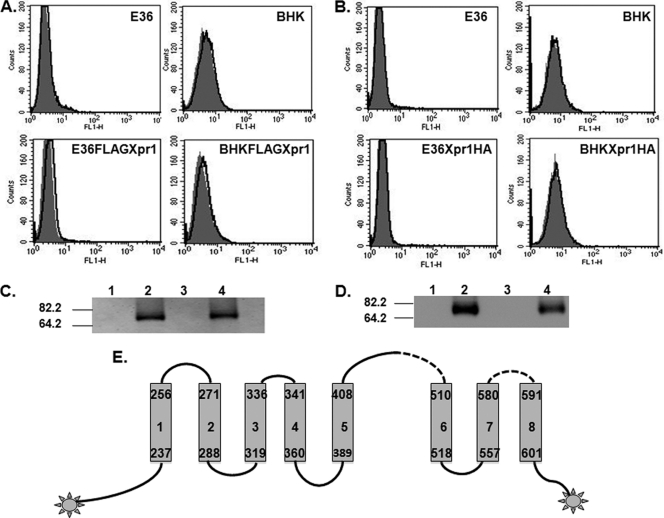
We have previously described the employment of truncated GALV SU as a method for assessing the ability of GALV to bind wild-type and various mutant PiT1 receptors (9, 12). Using GALV SU with a V5 epitope tag at the C terminus, we found BHK cells expressing PiT1 bound to GALV SU (Fig. 1B). To explore why the human receptor Xpr1 does not allow entry of XMRV into BHK cells, we fused, in frame, a double-HA-epitope tag to the C terminus of human Xpr1, expressed the tagged receptor in BHK cells, and undertook flow cytometry analysis of the BHK cells expressing Xpr1HA. The HA tag was not detected on the surface of either E36 cells or BHK cells expressing Xpr1HA, suggesting that the C terminus of Xpr1 is intracellular (Fig. 2A). We next introduced a triple-FLAG tag at the N terminus of Xpr1. Flow cytometry analysis of BHK and E36 cells expressing FLAGXpr1 failed to detect the FLAG epitope, indicating that the orientation of N- and C-terminal ends of Xpr1 in BHK cells is the same as that in E36 cells and that both ends of the Xpr1 protein are within the cell (Fig. 2B). Even though flow cytometry analysis failed to detect the presence of N- or C-terminally tagged Xpr1, Western blot analysis confirmed the expression of FLAGXpr1 and Xpr1HA in BHK and E36 cells transduced with pLNSX-FLAGXpr1 and Xpr1HA separately (Fig. 2C and D). Using a constrained prediction wherein the N and C termini of the protein are designated as intracellular and the predicted receptor binding domain (35) as extracellular in concert with HMMTOP (http://www.enzim.hu/hmmtop/) (30, 31), we conceived a topological model of Xpr1 shown in Fig. 2E. This model differs from the experimentally validated topology of PiT1 (12) and the proposed structure of PiT2 (23) in that it contains only 8 rather than 12 transmembrane domains and lacks the large cytoplasmic loop region predicted for the PiT receptors.
FIG. 2.
The N and C termini of Xpr1 are located inside the cell membrane. BHK and E36 cells expressing Xpr1 with FLAG tag at the N terminus (FLAGXpr1) or HA tag at the C terminus (Xpr1HA) were established as described in Materials and Methods. (A) The expression of FLAG on the cell surface was detected by flow cytometry analysis after staining with FITC-conjugated anti-FLAG antibody for 1 h (areas under bold line). Shaded areas represent isotype controls. (B) Same as in panel A, but with anti-HA-Alexa Fluor 488 to detect Xpr1HA expression on the surface of BHKXpr1HA or E36Xpr1HA cells (areas under the bold line). (C) Western blots for FLAGXpr1 expression in E36, E36FLAGXpr1, BHK, and BHKFLAGXpr1 cells (lanes 1 to 4, respectively) were performed with anti-FLAG-HRP antibody. Molecular mass markers in kilodaltons are to the left. The same cell number equivalents of whole-cell lysates from each cell line were loaded. (D) As described for panel C, except that anti-HA monoclonal antibody was used to detect Xpr1HA expression in BHK, BHKXpr1HA, E36, and E36-Xpr1HA cells (lanes 1 to 4, respectively). (E) Predicted topology of Xpr1. Shown is a topological model for human Xpr1 obtained by the HMMTOP method with constrained prediction: the N and C termini of the protein are designated as intracellular, as experimentally validated by labeling with FLAG and HA tags, and the predicted receptor binding domains (35) are extracellular. The transmembrane regions are represented by the gray bars and numbered 1 through 8. The lines on top of the designated transmembrane regions correspond to extracellular regions, with dotted lines representing the proposed receptor binding domains.
We constructed an XMRV SU protein fused to a double-HA tag and used this reagent to determine the ability of XMRV SU to bind BHK cells expressing Xpr1. XMRV SU was not able to bind BHK cells, but the expression of Xpr1 in BHK cells was sufficient to confer XMRV SU binding to BHKXpr1 cells (Fig. 3).
FIG. 3.
Expression of Xpr1 in BHK cells confers XMRV envelope protein binding. The binding of XMRV envelope to BHK or BHKXpr1 cells was determined with HA-tagged XMRV SU. HA antibody conjugated with Alexa Fluor 488 was used to detect XMRV binding on cell surface (areas under bold line). Shaded areas represent isotype controls.
PiT1 protein expressed on the membrane of BHK cells has a similar configuration as when it is expressed in MDTF cells.
Even though PiT1 is expressed on the cell surface of BHK cells and binds GALV envelope protein, it remains possible that the conformation of the PiT1 protein as expressed on the surface of BHK cells is compromised and thus cannot contribute to postbinding entry events. The topology of a functional human PiT1 receptor in BHK cells was therefore determined as in previous studies by the substituted cysteine accessibility method (SCAM) scanning mutagenesis technique (12). SCAM is a powerful method that has been successfully employed in establishing the topology of a large number of multiple membrane-spanning proteins because it minimally perturbs the structure and function of the assessed protein (4). In brief, each of the 13 cysteine residues present in PiT1HA was replaced with an alanine residue to produce a cysteineless protein (PiT1-13HA). PiT1-13HA fully retained its GALV receptor function (12). The PiT1-13HA plasmid was subsequently used as a template for mutagenesis, in which an individual cysteine residue was substituted for a specific residue of interest at selected positions in the protein (12).
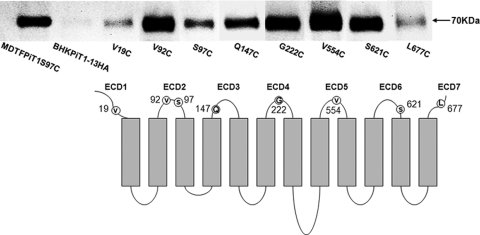
To validate that the topology of PiT1 expressed on BHK cells is similar to that expressed on MDTF cells, we individually expressed PiT1-13HA mutants containing a single cysteine residue in each of the regions previously determined to constitute an extracellular region (ECR). As shown in Fig. 4, residues 19 from ECR1, 92 and 97 from ECR2, 147 from ECR3, 222 from ECR4, 554 from ECR5, 621 from ECR6, and 677 from ECR7 were each reacted with B-mal, recovered by avidin-coated beads, and then detected with an HA antibody on a Western blot (Fig. 4). BHK cells expressing a cysteineless form of PiT1 (BHKPiT1-13HA), as expected, failed to react with B-mal, and no signal was detected with an HA antibody on a Western blot. Figure 4 shows that all the representative residues from each of the ECRs are located outside the BHK cell membrane, indicating that PiT1 expressed in BHK cells assumes a topology similar to that observed in MDTF cells. Therefore, the resistance of BHKPiT1 to GALV infection is not due to an overt conformational reconfiguration of PiT1 on the surface of BHK cells.
FIG. 4.
PiT1 protein expressed on the surface of BHK cells shares a similar configuration to PiT1 expressed on susceptible MDTFPiT1 cells. The top lane shows a representative Western blot of BHK cell lysates expressing PiT1-13HA mutants containing the individual cysteine substitutions V19C, S97C, Q147C, G222C, V554C, S621C, and L677C reacted with B-mal. BHK cells expressing the functional but cysteineless PiT1-13HA mutant were used as a negative control for B-mal reactivity. MDTF cells expressing PiT1-13HA mutant S97C were used as a positive control. The same cell number equivalents of whole-cell lysates from each cell line were loaded. The bottom lane shows the topological model for SLC20A1 (PiT1) expressed in MDTF cells validated by SCAM experimentally (10). The gray bars represent transmembrane regions. The extracellular regions are numbered 1 through 7. Individual residues presented in Western blots are localized in each ECR outside the cell membrane of MDTF cells.
Hybrid cells formed between cells lacking functional receptors and BHK cells expressing Xpr1 or PiT1 are susceptible to the appropriate primate retroviral vectors.
Our observations showed (i) retroviral vectors that differ exclusively in the composition of their envelope protein (e.g., A-MLV, 10A1 MLV, and GALV or NZB-XMV and XMRV) vary in their ability to infect BHK cells, demonstrating that the XMRV/GALV envelope component restricts entry; (ii) BHK cells expressing the appropriate receptor bind XMRV or GALV SU proteins, showing that the block to infection is mediated at a postbinding stage of entry; and (iii) MDTF and CHOK1 cells are resistant to GALV or XMRV, but expression of their respective receptors is sufficient to render them susceptible to infection. These collective observations suggest the block to infection is not attributable to a dominant-negative cellular factor that targets capsid or blocks integration and/or expression of the integrated vector. Therefore, BHK cells may lack a factor present in MDTF or CHOK1 cells, and this factor is requisite for infection by the assessed primate retroviruses.
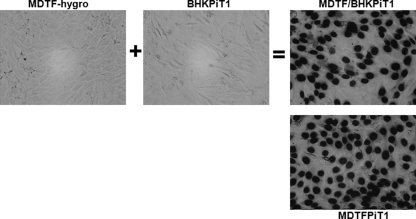
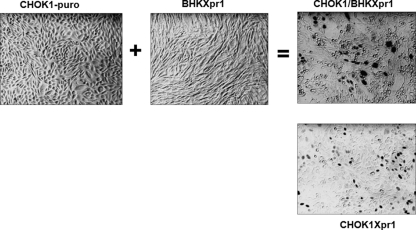
To further explore the mechanism for resistance of BHKPiT1 cells to GALV infection, we conducted a cell fusion assay between BHKPiT1 and MDTF cells. G418-resistant BHKPiT1 cells were fused with hygromycin-resistant MDTF cells and then selected for dual drug resistance. Although BHKPiT1 and MDTF cells are individually resistant to GALV infection, hybrid cells formed by fusing MDTF and BHKPiT1 were efficiently infected by GALV (Fig. 5). Our findings suggest that MDTF cells contain an ancillary factor that is absent in BHKPiT1 cells and that the absence of this ancillary factor determines the resistance of BHKPiT1 cells to GALV infection. In contrast to BHK cells, CHOK1 cells can be rendered susceptible to XMRV infection following Xpr1 expression (Table 1). To investigate whether the mechanism for the resistance of BHKXpr1 to XMRV infection is also due to the lack of an ancillary factor, XMRV-resistant CHOK1 cells and BHKXpr1 cells were fused together in a similar manner to MDTF/BHKPiT1 fusion. The CHOK1/BHKXpr1 hybrid cells can be infected by XMRV at a titer similar to that in CHOK1Xpr1 cells (4 × 103 FFU/ml) (Fig. 6). Therefore, the underlying mechanism of the resistance of BHKXpr1 to XMRV infection is similar to that effecting resistance of BHKPiT1 to GALV infection—that is, the lack of an ancillary factor.
FIG. 5.
Hybrids formed between nonpermissive MDTF and BHKPiT1 cells are susceptible to GALV infection. Hygromycin-resistant MDTF (MDTF-hygro) cells were generated by transducing MDTF cells with retroviral vector pLHCX, and BHKPiT1 and MDTFPiT1 cells were constructed with pLNSX-PiT1, which confers G418 resistance. The MDTF/BHKPiT1 hybrid was produced by coselection with hygromycin and G418 after fusion of MDTF-hygro with BHKPiT1 cells. MDTF-hygro, BHKPiT1, MDTF/BHKPiT1 hybrid, and MDTFPiT1 cells were exposed to GALV-enveloped retroviral vectors expressing nuclear localized β-galactosidase. Micrographs were taken with a 40× objective on a phase-contrast microscope.
FIG. 6.
Hybrids formed between nonpermissive CHOK1 and BHKXpr1 cells are susceptible to XMRV infection. Retroviral vector pBABE-puro was used to make puromycin-resistant CHOK1 (CHOK1-puro) cells. BHK or CHOK1 cells exposed to pLNSX-Xpr1 were used to generate the G418-resistant BHKXpr1 and CHOK1Xpr1 cells. After fusion of CHOK1-puro with BHKXpr1, puromycin and G418 were used for double selection of the CHOK1/BHKXpr1 hybrid. CHOK1-puro, BHKXpr1, CHOK1/BHKXpr1, and CHOK1Xpr1 cells were infected with XMRV-enveloped retroviral vector expressing nuclear localized β-galactosidase. Micrographs were obtained with a 20× objective.
DISCUSSION
Even though XMRV and NZB-XMV or GALV and 10A1 employ a common receptor to infect human cells, they differ in their abilities to infect hamster BHK cells expressing their respective human functional viral receptors. NZB-XMV and 10A1 MLV, but not XMRV and GALV, can infect BHK cells. BHK cells expressing the human Xpr1 or PiT1 receptor are efficiently infected by NZB-XMV and 10A1 MLV, but these cells remain resistant to XMRV and GALV. Several groups, including ours, have found that retrovirus or vector entry can be inhibited by N-linked glycosylation of cellular proteins, and this effect is cell and virus specific (27, 33, 34). This does not appear to be the case for BHK cells as tunicamycin treatment of BHK cells expressing Xpr1 or PiT1 did not ameliorate resistance to XMRV or GALV. Furthermore, BHK cells expressing a functional PiT1 that is glycosylation impaired did not circumvent BHK resistance to GALV (Table 2). Topology studies of PiT1 in BHK cells by SCAM analysis showed that the configuration of PiT1 is similar to that present on GALV-susceptible MDTFPiT1 cells. Thus, any obvious change in configuration is not responsible for the resistance of BHKPiT1 to GALV (Fig. 5).
Several studies report a crucial role for cell-associated HSPGs in the entry of human immunodeficiency virus type 1 (HIV-1) into permissive cells (2). It has also been demonstrated that the ability of the ecotropic MLV isolate PVC211 to infect brain capillary endothelial cells occurs via increased heparin binding activity of BCEC-tropic viruses in the initial step of infection (16). We were unable to determine any associative role between the absence of HSPGs correlating with the restriction of BHK to XMRV or GALV infection. Diminished expression of HSPGs after treatment of CHOK1Xpr1 cells with heparinase did not reduce XMRV binding significantly (Fig. 7). pgsA745 is a CHOK1 hamster cell line deficient in xylosyltransferases (XTI and XTII). This deficiency results in diminished initiation of the biosynthesis of both HSPGs and chondroitin sulfate proteoglycans (8). pgsA745 cells expressing Xpr1 (pgsA745-Xpr1) can be infected by XMRV at a similar titer to pgsA745-Xpr1 cells expressing XTII xylosyltransferases (Table 3). This observation is consistent with published findings that heparin sulfate proteoglycans are abundantly expressed on the surface of BHK cells (5, 17).
FIG. 7.
Binding of XMRV is heparin independent. (A) After CHOK1Xpr1 cells were treated with (areas under dotted line) or without (areas under bold line) heparinase II, the expression of HSPGs on the cell surface was detected by flow cytometry analysis after staining with anti-human heparan sulfate monoclonal antibody-FITC conjugate for 1 h. (B) Binding of XMRV envelope to CHOK1Xpr1 cells was determined with HA-tagged XMRV SU, similar to in Fig. 3. Areas under the dotted line show XMRV binding to CHOK1Xpr1 after treatment with heparinase II. Areas under the bold line represent XMRV binding to CHOK1Xpr1 without heparinase II treatment. Shaded areas represent isotype controls.
TABLE 3.
Infection with XMRV is heparin independent
| Target cells | XMRV titer (FFU/ml)a |
|---|---|
| pgsA745 | <5 |
| pgsA745XTII | <5 |
| pgsA745-Xpr1 | (1.2 ± 0.2) × 104 |
| pgsA745XTII-Xpr1 | (1.1 ± 0.3) × 104 |
Titer was determined by the mean number of FFU ± standard deviation. The average of at least three independent experiments was calculated.
Resistance of BHK cells expressing Xpr1 or PiT1 to XMRV or GALV is a postbinding event, and the viral component responsible for restricting viral entry is the envelope protein as substitution of NZB-XMV for XMRV or 10A1 MLV for GALV envelopes circumvents the block to vector infection. Thus, the restriction of XMRV or GALV to infection of BHK receptor-expressing cells appears to be envelope mediated and to occur at an early stage of viral entry. In some cases, restriction to infection by retroviruses that occurs at a postbinding stage of infection is a consequence of host cell factors that exert an inhibitory effect in a dominant-negative manner. For example, hybrid cells formed between cells dependent on the immunosuppressive drug cyclosporine (CsA) and CsA-independent cells indicated that the CsA-dependent HIV-1 infection is due to dominant cellular restriction, and their presence supports the existence of a cell-specific factor capable of restricting HIV-1 at an early postentry step by a CypA-dependent mechanism (24). Similarly, Fv1 (Friend virus susceptibility factor 1) restricts retroviral infection at a postbinding stage prior to integration and formation of proviral structures (25). Fv1-mediated restriction is dominant in hybrid cells formed between cells infected with the appropriate virus but lacking this restrictive factor and Fv1-expressing cells (28). In contrast, hybrid cells formed between cells expressing factors that restrict viral entry and cells lacking these factors restore virus entry. Most notably this was found to occur when HIV-1 nonpermissive NIH 3T3 cells expressing CD4 were fused to HeLa cells lacking CD4. These studies demonstrated that NIH 3T3 cells, although capable of binding HIV, were blocked at a postentry stage of infection, and this block was complemented by fusion to HeLa cells (7). Later this ancillary factor was determined to be the HIV receptor cofactor CCR5 or CXCR4. In addition, analysis of hybrid cells formed between CD4-expressing MDTF and human 293T cells suggested that MDTF cells lack a cofactor that is required for efficient virus assembly and release (21). Thus, the fusion of resistant cells that express the primary viral receptor with resistant cells that provide a complementary factor has provided a means of identifying components in addition to the viral receptor that are required for efficient infection by retroviruses.
Our findings suggest that MDTF cells and CHOK1 cells can provide an unknown factor that is necessary for GALV and XMRV infection of BHKPiT1 and BHKXpr1 cells, respectively. XMRV and GALV may interact with different orthologs of the same factor present in cells derived from different species, or these viruses may require distinct cofactors. Both suppositions are consistent with our findings, as is the possibility that other gammaretroviruses, in addition to GALV and XMRV, require ancillary factors. The latter possibility is supported by the published observation that feline leukemia virus subgroup T (FeLV-T) requires a factor in addition to its primary receptor, PiT1, to infect cells (1). The identification of this factor is a critical next step in enabling the identification of risk factors that may facilitate virus spread to humans.
Acknowledgments
We thank Karen B. Farrell for expert technical assistance and advice; Jim Nagle and Debbie Kauffmann at the NINDS Sequencing Facility (National Institutes of Health) for sequencing; Laura Li, Jill Russ, David Kim, and Mickeyas Alemayehu for experimental help and comments; and Arlette Kouwenhoven for critical reading of the manuscript.
Footnotes
Published ahead of print on 26 January 2011.
REFERENCES
- 1.Anderson, M. M., A. S. Lauring, C. C. Burns, and J. Overbaugh. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828-1830. [DOI] [PubMed] [Google Scholar]
- 2.Argyris, E. G., et al. 2003. Human immunodeficiency virus type 1 enters primary human brain microvascular endothelial cells by a mechanism involving cell surface proteoglycans independent of lipid rafts. J. Virol. 77:12140-12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battini, J. L., J. E. Rasko, and A. D. Miller. 1999. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc. Natl. Acad. Sci. U. S. A. 96:1385-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogdanov, M., W. Zhang, J. Xie, and W. Dowhan. 2005. Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM): application to lipid-specific membrane protein topogenesis. Methods 36:148-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bretscher, M. S. 1985. Heparan sulphate proteoglycans and their polypeptide chains from BHK cells. EMBO J. 4:1941-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong, B., et al. 2007. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc. Natl. Acad. Sci. U. S. A. 104:1655-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dragic, T., P. Charneau, F. Clavel, and M. Alizon. 1992. Complementation of murine cells for human immunodeficiency virus envelope/CD4-mediated fusion in human/murine heterokaryons. J. Virol. 66:4794-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esko, J. D., T. E. Stewart, and W. H. Taylor. 1985. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 82:3197-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrell, K. B., and M. V. Eiden. 2005. Dissection of gammaretroviral receptor function by using type III phosphate transporters as models. J. Virol. 79:9332-9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrell, K. B., J. L. Russ, R. K. Murthy, and M. V. Eiden. 2002. Reassessing the role of region A in Pit1-mediated viral entry. J. Virol. 76:7683-7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrell, K. B., Y. T. Ting, and M. V. Eiden. 2002. Fusion-defective gibbon ape leukemia virus vectors can be rescued by homologous but not heterologous soluble envelope proteins. J. Virol. 76:4267-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrell, K. B., G. E. Tusnady, and M. V. Eiden. 2009. New structural arrangement of the extracellular regions of the phosphate transporter SLC20A1, the receptor for gibbon ape leukemia virus. J. Biol. Chem. 284:29979-29987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldman, S. A., K. B. Farrell, R. K. Murthy, J. L. Russ, and M. V. Eiden. 2004. Identification of an extracellular domain within the human PiT2 receptor that is required for amphotropic murine leukemia virus binding. J. Virol. 78:595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer, N., et al. 2010. Xenotropic murine leukemia virus-related gammaretrovirus in respiratory tract. Emerg. Infect. Dis. 16:1000-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanger, J. J., L. D. Bromham, J. J. McKee, T. M. O'Brien, and W. F. Robinson. 2000. The nucleotide sequence of koala (Phascolarctos cinereus) retrovirus: a novel type C endogenous virus related to gibbon ape leukemia virus. J. Virol. 74:4264-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jinno-Oue, A., M. Oue, and S. K. Ruscetti. 2001. A unique heparin-binding domain in the envelope protein of the neuropathogenic PVC-211 murine leukemia virus may contribute to its brain capillary endothelial cell tropism. J. Virol. 75:12439-12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karreman, C., S. Karreman, and H. Hauser. 1996. Retroviral infection of Syrian hamster BHK cells depends on age and susceptibility toward sialidase. Virology 220:46-50. [DOI] [PubMed] [Google Scholar]
- 18.Lander, M. R., and S. K. Chattopadhyay. 1984. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ectropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J. Virol. 52:695-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy, J. A. 1973. Xenotropic viruses: murine leukemia viruses associated with NIH Swiss, NZB, and other mouse strains. Science 182:1151-1153. [DOI] [PubMed] [Google Scholar]
- 20.Lombardi, V. C., et al. 2009. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science 326:585-589. [DOI] [PubMed] [Google Scholar]
- 21.Mariani, R., et al. 2001. Mouse-human heterokaryons support efficient human immunodeficiency virus type 1 assembly. J. Virol. 75:3141-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Overbaugh, J., A. D. Miller, and M. V. Eiden. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol. Mol. Biol. Rev. 65:371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salaun, C., P. Rodrigues, and J. M. Heard. 2001. Transmembrane topology of PiT-2, a phosphate transporter-retrovirus receptor. J. Virol. 75:5584-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song, C., and C. Aiken. 2007. Analysis of human cell heterokaryons demonstrates that target cell restriction of cyclosporine-resistant human immunodeficiency virus type 1 mutants is genetically dominant. J. Virol. 81:11946-11956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoye, J. P. 1998. Fv1, the mouse retrovirus resistance gene. Rev. Sci. Tech. 17:269-277. [DOI] [PubMed] [Google Scholar]
- 26.Tailor, C. S., A. Nouri, C. G. Lee, C. Kozak, and D. Kabat. 1999. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc. Natl. Acad. Sci. U. S. A. 96:927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tavoloni, N., and A. Rudenholz. 1997. Variable transduction efficiency of murine leukemia retroviral vector on mammalian cells: role of cellular glycosylation. Virology 229:49-56. [DOI] [PubMed] [Google Scholar]
- 28.Tennant, R. W., F. E. Myer, and L. McGrath. 1974. Effect of the Fv-1 gene on leukemia virus in mouse cell heterokaryons. Int. J. Cancer 14:504-513. [DOI] [PubMed] [Google Scholar]
- 29.Theilen, G. H., D. Gould, M. Fowler, and D. L. Dungworth. 1971. C-type virus in tumor tissue of a woolly monkey [Lagothix spp.] with fibrosarcoma. J. Natl. Cancer Inst. 47:881-885. [PubMed] [Google Scholar]
- 30.Tusnady, G. E., L. Kalmar, H. Hegyi, P. Tompa, and I. Simon. 2008. TOPDOM: database of domains and motifs with conservative location in transmembrane proteins. Bioinformatics 24:1469-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tusnady, G. E., L. Kalmar, and I. Simon. 2008. TOPDB: topology data bank of transmembrane proteins. Nucleic Acids Res. 36:D234-D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urisman, A., et al. 2006. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2:e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Wilson, C. A., and M. V. Eiden. 1991. Viral and cellular factors governing hamster cell infection by murine and gibbon ape leukemia viruses. J. Virol. 65:5975-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan, Y., Y. T. Jung, T. Wu, and C. A. Kozak. 2008. Role of receptor polymorphism and glycosylation in syncytium induction and host range variation of ecotropic mouse gammaretroviruses. Retrovirology 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan, Y., Q. Liu, and C. A. Kozak. 2009. Six host range variants of the xenotropic/polytropic gammaretroviruses define determinants for entry in the XPR1 cell surface receptor. Retrovirology 6:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, Y. L., et al. 1999. Receptors for polytropic and xenotropic mouse leukaemia viruses encoded by a single gene at Rmc1. Nat. Genet. 21:216-219. [DOI] [PubMed] [Google Scholar]