Abstract
Previous studies have found an association between a single-nucleotide polymorphism 35 kb upstream of the HLA-C locus (−35 SNP), HLA-C expression, and HIV-1 set point viral loads. We show that the difference in HLA-C expression across −35 SNP genotypes can be attributed primarily to the very low expression of a single allelic product, HLA-Cw7, which is a common HLA type. We suggest that association of the −35 SNP and HIV-1 load manifests as a result of linkage disequilibrium of this polymorphism with both favorable and unfavorable HLA-C and -B alleles.
Infection with HIV-1 involves a complex interplay between virus and host. After the acute phase of infection, an equilibrium is reached between HIV-1 and the immune system, resulting in the decline of HIV-1 RNA levels to the widely variable viral set point, a known (though imperfect) predictor of time to progression to AIDS (36, 37). The early appearance of HIV-1-specific CD8 T cells and their rapid selection of escape mutants during the initial decline of viremia in primary infection imply that this T-cell response plays a central role in resolution of primary viremia and the long-term suppression of viral replication (21, 34).
The major histocompatibility complex (MHC), located on chromosome 6, contains the HLA class I and II genes, which are the most polymorphic loci in humans (9). HLA class I molecules play a significant role in the immune response against HIV-1 by presenting viral peptides to CD8 T cells (adaptive immunity) and by serving as ligands for killer cell immunoglobulin-like receptors (KIRs) expressed on natural killer (NK) cells (innate immunity). The central role of the MHC region in HIV-1 control was highlighted by a recent genomewide association study of variants that influence the control of viral set point (17, 18). In Caucasian cohorts, two single-nucleotide polymorphisms (SNPs) were identified that associated with the HIV-1 set point, and a third was associated with disease progression. All three SNPs were located in the MHC region on chromosome 6. These findings have been replicated with other independent Caucasian cohorts (10, 15, 31, 53, 55).
One minor allelic variant of an SNP (rs2395029) associated with lower viral set points is located in the HLA complex P5 (HCP5) gene and is a tag for HLA-B*5701 (17), which is associated with slow disease progression (2, 9, 25, 40, 42). The second most significant polymorphism (rs9264942), accounting for 6.5% of the variation in viral set point (18), is located 35 kb upstream of the HLA-C gene (−35 SNP), with the minor allele (C) associating with a lower set point than the major allele (T). The −35 SNP is also present in African-American populations but does not correlate with viral set point (47, 49), which suggests that the −35 SNP is a marker for another polymorphism and is not the causal SNP. Although HLA-B*5701 is in linkage disequilibrium with the −35 SNP, both associate independently with the viral load set point (17, 18).
The −35 C variant was associated with higher HLA-C mRNA expression in Epstein-Barr virus (EBV)-transformed B-cell lines, suggesting an inverse correlation between HLA-C expression and viral load (51). However, the extreme polymorphism of HLA-C and posttranslational regulation of HLA class I molecules may complicate this relationship (23, 33). The −35 SNP is known to be in strong linkage disequilibrium with HLA-C alleles (Cw*0102, -0202, -0302, -0501, -0602, -0801, -0802, -1202, -1203, and -1402 for the −35 C allele and Cw*0303, -0304, -0401, -0701, -0702, -0704, -1502, -1504, -1505, -1506, -1601, -1602, -1604, and -1701 for the −35 T allele) (17, 52), and each type may have its own intrinsic level of expression. Therefore, protein expression studies are needed to resolve the question of whether the −35 SNP is associated with increased levels of surface HLA-C. Recently, it was reported that surface expression of HLA-C varies significantly across −35 SNP genotypes and that the HLA-C alleles that are in linkage disequilibrium with the −35 C allele are expressed at higher levels than those that are in linkage disequilibrium with the −35 T allele (52). However, interpretation of this study is complicated by the known cross-reaction with HLA-E of the antibody used for the study (DT9) (7). Our results show that when this cross-reaction is taken into account, the difference in the levels of HLA-C expressed at the cell surface between −35 SNP genotypes is actually greater than previously realized. However, we also show that nearly all of this difference can be accounted for by the particularly low expression of HLA-Cw7, which is in strong linkage disequilibrium with the −35 T allele. Although HLA-Cw7 has been shown to associate with more rapid progression of disease in HIV-1-infected patients (48), it cannot account fully for association of the −35 SNP with lower HIV-1 loads and slower progression to AIDS (17).
MATERIALS AND METHODS
Subjects.
Peripheral blood mononuclear cells (PBMC) from healthy volunteers of European descent were isolated from whole blood by Ficoll-Hypaque gradient centrifugation. Written informed consent was obtained from all volunteers under appropriate university regulations. Cryopreserved PBMC from 25 HIV-1-infected subjects of European descent were obtained from the CHAVI 001 and SCOPE (22) cohorts. All experiments on HIV-infected samples were approved by the Oxford Tropical Research Ethics Committee.
−35 SNP genotyping.
Genomic DNAs from the healthy volunteers were genotyped for the −35 SNP by PCRs using a common forward primer (5′-GGGTGGTGCCAAGTATGAG-3′) and alternative allele-specific reverse primers (5′-AGAAAGTCCCACAGTGCCTA-3′ and 5′-AGAAAGTCCCACAGTGCCTG-3′). Genotyping reaction mixtures also contained control primers that amplify a constant region in the DRB1 locus (43). Thermocycling conditions were as follows: 96°C for 1 min; 5 cycles of 96°C for 25 s, 70°C for 45 s, and 72°C for 45 s; 21 cycles of 96°C for 25 s, 65°C for 50 s, and 72°C for 45 s; and 4 cycles of 96°C for 25 s, 55°C for 1 min, and 72°C for 2 min. Products were resolved in 2% agarose gels and visualized under UV with ethidium bromide. This PCR assay was validated by direct sequencing of a PCR product spanning the −35 SNP for the first 100 samples typed.
Monoclonal antibodies and flow cytometry.
MEM-E/06 and MEM-E/08 were obtained from Santa Cruz; anti-human HLA-E (3D12), anti-human HLA-A,B,C (W6/32), and a mouse IgG2b kappa isotype control (clone MPC-11) were obtained from BioLegend; CD3-PerCP (clone SK7) was obtained from Becton Dickinson; a mouse IgG2b-fluorescein isothiocyanate (FITC) (clone MOPC-141) isotype control and mouse anti-human MHC class I (W6/32)-FITC were obtained from Sigma; and rat anti-mouse kappa (clone 187.1)-FITC was obtained from Southern Biotech. Monoclonal antibody DT9 was affinity purified on protein A-Sepharose beads (Sigma-Aldrich) from a hybridoma supernatant by standard procedures and conjugated using a FluoroTag FITC conjugation kit (Sigma).
For all cell surface staining, 1 million thawed PBMC were washed once with phosphate-buffered saline (PBS; Sigma-Aldrich) and then stained with Live/Dead stain (Invitrogen) per the manufacturer's instructions. For direct antibody staining, cells were then incubated on ice for 15 min with appropriate antibodies. For indirect antibody staining, cells were incubated with primary antibodies (15 min on ice), free secondary antibody binding sites were blocked with mouse serum (Southern Biotech), and then cells were incubated with secondary antibody and anti-CD3-PerCP (15 min on ice). Cells were washed twice between each step with flow cytometry buffer (PBS containing 0.5% bovine serum albumin [BSA]). After being stained, cells were washed again, fixed with Cell Fix buffer (Becton Dickinson), and analyzed using a CyAn flow cytometer and FlowJo software (Treestar, Inc.).
Characterization of monoclonal antibodies.
LABScreen single-antigen class I Combi beads (One Lambda), each coated with a single HLA allotype (46), were incubated with DT9, 3D12, MEM-E/06, MEM-E/08, and W6/32 antibodies for 30 min at room temperature, washed 3 times with PBS, incubated at room temperature for 30 min with goat anti-mouse IgG-R-phycoerythrin (R-PE) (Invitrogen), washed 3 times as described before, and analyzed on a LABScan 100 flow analyzer.
The specificity of the monoclonal antibody panel for HLA-E was tested by binding to HLA-E on transfected 721.221 AEH cells (29), a gift from V. Braud (University of Nice).
RESULTS
Determining the specificity and applicability of HLA-C and HLA-E antibodies.
The study of individual surface HLA molecules is complicated by the scarcity of allele-specific antibodies. The monoclonal antibody DT9 (7), originally raised against MHC class I proteins from cottontop tamarins, is the only available HLA-C-specific antibody that does not extensively cross-react with HLA-A or -B alleles. However, immunoprecipitation and one-dimensional isoelectric focusing (1D-IEF) studies confirmed that it also recognizes the nonclassical HLA-E protein (6, 7). A recent study showing a correlation between surface HLA-C expression and the −35 SNP discounted cross-reaction of DT9 with HLA-E, as it was shown that the HLA-E-specific antibody MEM-E/08 stains a cell line transfected with HLA-E but does not stain PBMC (52). However, it has previously been reported that MEM-E/08 cannot be used to detect HLA-E on PBMC by flow cytometry (32). Therefore, we decided to reexamine cell surface expression of HLA-E by PBMC to better understand DT9 staining.
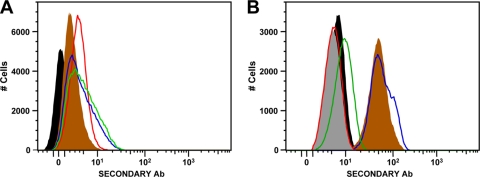
In addition to MEM-E/08, we tested two other commercially available antibodies that are nominally specific for HLA-E (MEM-E/06 and 3D12) (29, 38, 44), using 220.221 cells transfected with HLA-E. As can be seen in Fig. 1A, the levels of staining with all of these antibodies were similar to that with DT9, confirming that they all recognize HLA-E, at least as expressed by transfected cells. When we used these antibodies to stain PBMC from healthy volunteers, however, we saw marked differences in the levels of staining. Consistent with previous reports (32, 52), we did not see staining of PBMC with MEM-E/08 (Fig. 1B). In contrast, the level of staining seen with MEM-E/06 varied considerably from donor to donor, suggesting that this antibody may cross-react with other HLA proteins. Finally, we observed fairly constant levels of staining of PBMC with 3D12.
FIG. 1.
Staining of 220.221AEH cells and PBMC with DT9 and HLA-E-specific antibodies. (A) Staining of the 220.221AEH cell line, showing unstained cells (filled black area) and cells stained with MEM-E/06 (filled brown area), MEM-E/08 (red line), 3D12 (green line), and DT9 (blue line). (B) Representative staining of CD3+ lymphocytes (gating on appropriate forward and side scatter, live cells, and CD3 expression) from a healthy volunteer (HLA-A2, -B35, -B44, -Cw4, and -Cw5), showing unstained cells (filled black area) and cells stained with MEM-E/06 (filled brown area), MEM-E/08 (red line), 3D12 (green line), DT9 (blue line), and an IgG2b isotype control (filled gray area).
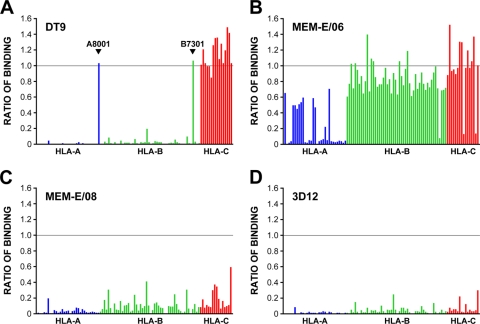
The possibility that MEM-E/06 cross-reacts with other class I proteins prompted us to test the specificity of all of the antibodies used. To do this, we employed the LABScreen system, a Luminex assay that allows detection of antibody cross-reactions with 97 common HLA class I alleles spanning HLAs A, B, and C. In addition to testing the three HLA-E antibodies, we also tested DT9. Since there can be considerable bead-to-bead differences in the amount of HLA protein present on the individual antigen beads (S. Fuggle and S. Page, personal communication), we used binding of the pan-class I antibody W6/32 (3) to normalize the results. While DT9 was highly specific for HLA-C and cross-reacted only with the rare HLA-A*8001 and -B*7301 alleles (Fig. 2A), confirming previous findings (52), MEM-E/06 did indeed show extensive cross-reactions with many HLA-A, -B, and -C proteins (Fig. 2B), as previously reported (32, 38), and there was good correlation with the levels of PBMC staining described above (data not shown). Cross-reaction of MEM-E/06 with HLA-A*0301 but not with HLA-A*0201 was confirmed by immunoprecipitation and 1D-IEF, using 220.221 cells transfected with these HLAs (data not shown). Much less cross-reaction was seen for both MEM-E/08 and 3D12 (Fig. 2C and D), and for 3D12, the level of binding seen with the beads did not translate into cross-reaction with these HLAs at the cell surface.
FIG. 2.
Characterization of monoclonal antibodies DT9, 3D12, MEM-E/06, and MEM-E/08 against 97 common HLA class I allotypes. Binding was determined for DT9 (A), MEM-E/06 (B), MEM-E/08 (C), and 3D12 (D), using beads coated with recombinant HLA-A (blue), -B (green), and -C (red) proteins, and normalized relative to the binding of W6/32 (the horizontal line corresponds to the level of binding seen with W6/32). Values shown represent the averages for three runs; the maximum standard errors of the means were 0.06 (DT9), 0.05 (MEM-E/06), 0.012 (MEM-E/08), and 0.01 (3D12). The order of the HLA allotypes tested (along the x axis) was as follows: A0101, -0201, -0203, -0206, -0301, -1101, -1102, -2301, -2402, -2403, -2501, -2601, -2901, -2902, -3001, -3002, -3101, -3201, -3301, -3303, -3401, -3402, -3601, -4301, -6601, -6602, -6801, -6802, -6901, and -7401, B8001, -0702, -0801, -1301, -1302, -1401, -1402, -1501, -1502, -1503, -1510, -1511, -1512, -1513, -1516, -1801, -2705, -2708, -3501, -3701, -3801, -3901, -4001, -4002, -4006, -4101, -4201, -4402, -4403, -4501, -4601, -4701, -4801, -4901, -5001, -5101, -5102, -5201, -5301, -5401, -5501, -5601, -5701, -5703, -5801, -5901, -6701, -7301, -7801, -8101, and -8201, and Cw0102, -0202, -0302, -0303, -0304, -0401, -0501, -0602, -0702, -0801, -1203, -1402, -1502, -1601, -1701, and -1802.
Although at present we cannot rule out cross-reaction with other nonclassical HLA proteins, it appears that 3D12 is indeed specific for HLA-E and that HLA-E is present on the surfaces of cells at levels detectable by flow cytometry. It is not clear why MEM-E/08 stains HLA-E-transfected cells and not PBMC, but one possible explanation is that this antibody is actually specific for a misfolded HLA heavy chain which may be present at increased levels on the surfaces of transfected cells.
DT9 staining of PBMC from healthy and HIV-1-infected subjects.
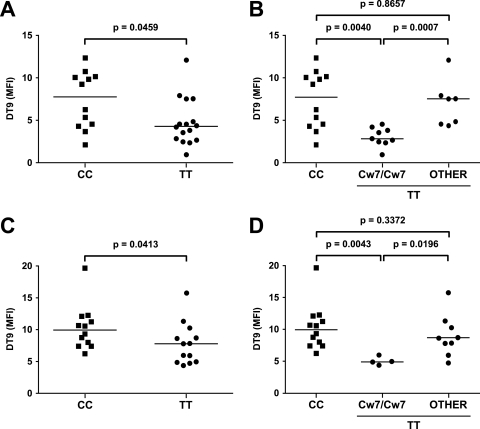
In the absence of a truly HLA-C-specific antibody, DT9 (which recognizes both HLA-C and HLA-E) was used to stain cryopreserved PBMC from 28 HIV-1-uninfected subjects selected on the basis of homozygosity of their −35 SNP genotype (both TT and CC) and the absence of the HLA alleles (A*8001 and B*7301) that cross-react with DT9. There was a significantly higher level of DT9 staining of lymphocytes (CD3-positive cells) from CC subjects than of those from TT subjects (P = 0.0459; Mann-Whitney test) (Fig. 3A). When the TT subjects were subdivided based on their HLA-C alleles, DT9 staining of TT subjects who were homozygous for HLA-Cw*07 was significantly lower than that for both CC subjects, who were all HLA-Cw*07 negative (P = 0.004), and TT subjects who were either heterozygous for or did not have HLA-Cw*07 (P = 0.0007) (Fig. 3B). There was no significant difference in the DT9 staining of PBMC from TT subjects who were not homozygous for HLA-Cw*07 and CC subjects. These findings imply that low HLA-Cw7 levels could account for the differences in HLA-C expression previously described as high levels associated with the −35 CC genotype.
FIG. 3.
Direct DT9 staining of live CD3+ T lymphocytes from −35 SNP homozygotes. PBMC from HIV-1-uninfected (A and B) and chronically HIV-1-infected (C and D) subjects were stained with DT9. DT9 median fluorescence intensities (MFI; y axis) were corrected for background autofluorescence of unstained cells. −35 SNP genotypes are indicated on the x axis. In panels B and D, “other” refers to −35 TT individuals who were not homozygous for HLA-Cw7. P values were calculated using the Mann-Whitney test.
Lymphocytes from 25 HIV-1-infected subjects were also stained with DT9. As seen before, DT9 staining of CC subjects was just significantly higher than staining of TT subjects (P = 0.0413) (Fig. 3C), and again, this difference in staining was even more striking with TT subjects who were also homozygous for HLA-Cw*07 (Fig. 3D) (P = 0.0043). Furthermore, there was no significant difference in median fluorescence intensity values comparing HIV-1-uninfected and -infected subjects, suggesting that, at least in chronically infected subjects, HIV-1 does not upregulate expression of the HLA-C protein.
Quantifying HLA-C on the cell surface.
In order to determine the contribution of HLA-C to DT9 binding, saturating amounts of DT9 and 3D12 were used to stain lymphocytes from HIV-1-uninfected subjects in indirect antibody binding assays (Fig. 4 and 5). As with direct DT9 staining, the low levels of staining observed in these indirect binding assays are consistent with cell surface levels of HLA-C protein being 10-fold less than those of HLA-A and -B proteins (50). Despite the consistently low staining with 3D12, there was a significant difference in median fluorescence intensity between unstained cells and cells stained with 3D12 (P < 0.0001; Mann-Whitney test), confirming surface expression of HLA-E (Fig. 5A), and when subjects were grouped by −35 SNP genotype, there was no significant difference in 3D12 binding (Fig. 5B). As seen before, differences in DT9 staining in this indirect staining assay were driven by the low levels of binding for TT individuals who were homozygous for HLA-Cw*07 (Fig. 5C).
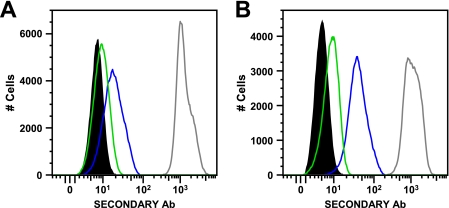
FIG. 4.
Indirect staining of lymphocytes with 3D12, DT9, and W6/32 antibodies. Representative staining results are shown for CD3+ lymphocytes (gated on appropriate forward and side scatter, live cells, and CD3 expression) from two HIV-1-uninfected subjects: a −35 TT subject (homozygous for HLA-Cw*07) (A) and a −35 CC subject (HLA-Cw*05/06) (B). Data are shown for unstained cells (filled black area), 3D12 staining (green), DT9 staining (blue), and W6/32 (pan-class I) staining (gray).
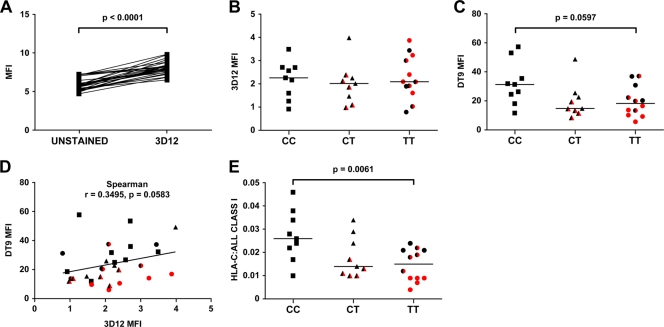
FIG. 5.
Indirect staining of lymphocytes from 30 healthy volunteers, using 3D12 and DT9. (A) Paired comparisons of MFI of unstained cells and cells stained with 3D12. (B and C) Indirect staining (with 3D12 and DT9, respectively) of live CD3+ lymphocytes, corrected for background autofluorescence of unstained cells. (D) Correlation of background-corrected DT9 and 3D12 staining. (E) Ratios of HLA-C levels (3D12 staining subtracted from DT9 staining) to total class I protein levels (W6/32 staining). For panels B to E, red and half-red symbols denote individuals homozygous and heterozygous for HLA-Cw*07, respectively. Statistical significance was calculated using the Mann-Whitney test.
There was no statistically significant correlation between 3D12 and DT9 binding (Spearman test; r = 0.3495; P = 0.0583) (Fig. 5D). These data imply that the differences seen in DT9 staining are the result of differences in HLA-C binding alone. Furthermore, the constant contribution of surface HLA-E to DT9 binding means that the difference in HLA-C surface expression between the CC and TT subjects is greater than previously recognized. To better quantify this difference, we calculated the ratio of HLA-C (3D12 staining subtracted from DT9 staining) to total class I protein on the cell surface (measured by W6/32 staining). The proportions of actual HLA-C binding to W6/32 binding ranged from 1.0 to 4.6%, 1.0 to 3.4%, and 0.4 to 2.6% for −35 CC, CT, and TT subjects, respectively. There was a significant difference in the ratios of HLA-C to W6/32 binding between −35 CC and TT subjects (Fig. 5E) (P = 0.0061; Mann-Whitney test), but again, this difference was driven exclusively by the low expression of HLA-Cw7 (P = 0.0032 for comparing −35 CC with −35 TT subjects homozygous for HLA-Cw*07). As before, there was no significant difference in the ratios of HLA-C to W6/32 binding between −35 CC and TT subjects who were not homozygous for HLA-Cw*07 (P = 0.0896). From this observation, we concluded that −35 CC subjects expressed 1.76 times more HLA-C molecules than −35 TT subjects considered as a whole but 5 times more HLA-C molecules than −35 TT subjects who were homozygous for HLA-Cw*07. Therefore, the protective −35 SNP genotype does not associate with increased expression of HLA-C. Rather, the nonprotective allele associates with the greatly reduced expression of a single HLA-C allele, the Cw*07 allele.
DISCUSSION
Previous studies of Caucasians have indicated that a single-nucleotide polymorphism 35 kb upstream of the HLA-C gene associates with control of the HIV-1 set point (17, 18) as well as with a slower progression to a CD4 count below 200 and an increased time to death (52). Transcriptional data showing that this SNP associates with differences in HLA-C expression levels and the linkage of −35 SNP alleles with HLA-C alleles (17, 52) have all been taken as evidence that this SNP is a marker for the HLA-C locus. As a result, there has been much speculation on a role for HLA-C in controlling HIV-1 infection. HIV-1 selectively downregulates surface HLA-A and -B, but not HLA-C and -E, via the action of the Nef protein (11-13, 56). The infected cells may therefore be relatively resistant to HLA-A- and -B-restricted CD8 T-cell lysis but continue to express HLA-C. While it has been shown that recognition of infected cells by HLA-C-restricted CD8 T cells is unaffected by HIV-1 Nef expression (1), these responses are relatively weak (4, 25) and are consistently associated with high levels of viremia in HIV-1-infected subjects, even when targeting the Gag protein (26). This suggests that they contribute little to viral control in vivo. HLA-C molecules also act as ligands for the inhibitory KIR2DL receptors (28, 45), so infected cells expressing high levels of HLA-C may be less vulnerable to NK cell recognition (1, 12, 13). However, it is not known whether the level of HLA-C expressed by the target cell is critical, and in addition, NK cells lacking inhibitory receptors for HLA-C and HLA-E molecules would still be able to kill HIV-1-infected T cells (5).
The prevailing model for the effects of the protective −35 C allele is that it is associated with higher cell surface levels of HLA-C protein than those with the nonprotective T allele (17, 18, 30, 48, 52). Although DT9, the best available anti-HLA-C antibody, was used to provide support for this hypothesis, this antibody is known to cross-react with HLA-E (7). It was suggested that DT9 binding to lymphocytes may be entirely specific for HLA-C, based on the observation that the nominally HLA-E-specific antibody MEM-E/08 does not stain lymphocytes (52). However, we found that the HLA-E-specific antibody 3D12, which does not cross-react significantly with any HLA-A, -B, or -C allotype, binds to lymphocytes. Thus, HLA-E is expressed at low levels on the surfaces of lymphocytes, consistent with earlier data showing that lymphocytes express HLA-E (5, 14, 41). We found that there was no significant difference in 3D12 staining when subjects were grouped according to their −35 SNP genotype. Therefore, our results show not only that the differences in DT9 staining associated with these genotypes are due to differences in HLA-C surface expression but also that the difference between HLA-C expression levels in donors with −35 CC and TT alleles is greater than previously recognized. However, our results for DT9 staining also show an important difference from those reported previously (52). Like Thomas et al. (52), we observed consistently higher levels of DT9 staining of cells from −35 CC subjects than of those from −35 TT subjects, both for healthy volunteers (P = 0.0459) and for individuals infected with HIV-1 (P = 0.0413); we also found considerable overlap in the ranges of DT9 staining levels between CC and TT subjects, reflecting a wide spread of HLA-C expression across the different alleles. However, in our data set, cells from TT subjects homozygous for HLA-Cw*07 had the least DT9 staining, by far (Fig. 3). Indeed, the difference in staining levels between CC subjects and TT subjects homozygous for HLA-Cw*07 was highly significant (P = 0.004 for healthy volunteers and P = 0.0043 for HIV-1-infected individuals). The difference in staining between TT subjects homozygous for HLA-Cw*07 and all other TT subjects was highly significant for healthy volunteers (P = 0.0007) but less significant for HIV-1-infected individuals (P = 0.0196). Importantly, there was no statistically significant difference in staining between CC subjects and TT subjects if HLA-Cw*07 homozygotes were excluded, both for the healthy volunteers and for the HIV-1-infected subjects. Note that a similar pattern of particularly low DT9 antibody binding to HLA-Cw7 was seen in the data on the staining of HIV-1-uninfected subjects presented by Thomas et al. (52).
Even when the contribution of the constant background of surface HLA-E expression to the level of DT9 staining for the healthy volunteer samples is taken into account, the picture does not change. Although the significance of the difference in expression levels increased between CC and TT subjects (P = 0.0061) (Fig. 5E), this difference was still driven entirely by the very low staining of cells from the HLA-Cw*07 homozygotes (P = 0.0032 for CC versus TT Cw*07/Cw*07 subjects and P = 0.0896 for CC versus TT non-Cw*07/Cw*07 subjects). Overall, we estimate that levels of surface HLA-C expression were approximately 1.76-fold greater for PBMC from CC subjects than for cells from TT subjects, but this difference was almost all accounted for by the approximately 5-fold-decreased expression of HLA-C in −35 TT donors who were also homozygous for HLA-Cw*07 compared with that in −35 CC subjects. Therefore, we concluded that the −35 SNP is actually most strongly associated with intrinsically low expression of HLA-Cw7 rather than with high expression of several −35 C-associated HLA-C allotypes. We cannot exclude, however, the possibility that other HLA-C alleles may also have different intrinsic levels of expression that could contribute to the overall levels of surface staining.
Despite the evidence that HLA-C-restricted CD8 T cells contribute little to the control of HIV-1 at the population level, it is still possible that significantly reduced expression of HLA-Cw7 could have significant effects on an individual level. Low expression of HLA-Cw7 could result in less effective presentation of peptides to CD8 T cells and in less control of viral replication. Low expression could also mean positive selection of fewer HLA-Cw7-restricted T cells in the thymus. In addition, by analogy with the suggestion that much of the benefit of protective HLA-B alleles arises from the presentation of particularly effective peptides (27), it is possible that HLA-Cw7 might bind to less-favorable peptides in HIV-1. Consistent with these possibilities, Fellay et al. showed that of all the HLA-C alleles, HLA-Cw*07 is significantly associated with the highest mean viral load set point (17). However, the exclusion of subjects homozygous for HLA-Cw*07 does not abolish the association of the −35 SNP with control of HIV-1 infection. This suggests that lower expression of HLA-Cw7 (a particularly common allele in people of European descent [26.0 to 66.2%] [39]) contributes only a part of the protective effect marked by the −35 SNP. The International HIV Controllers Study recently implicated seven HLA class I alleles with control of HIV-1 and showed that HLA-Cw*07 is a risk allele associated with high viremia and more rapid disease progression (48).
Indeed, it is becoming increasingly clear that genomewide association studies of human diseases can be hampered by the complexity of linkage disequilibrium patterns in the MHC region. These studies capture the majority of common variants in the genome that explain only a small proportion of heritability, while low-frequency polymorphisms, each conferring an intermediate increase in risk, can explain a significant proportion of the genetic susceptibility to common diseases (19, 20). Recently, Dickson et al. argued that rare variants can create synthetic association signals in genomewide association studies by occurring more often in association with one of the alleles of an SNP, which would therefore synthetically confer an increased risk for disease (16). We suggest, therefore, that the −35 SNP effect is an example of just such a synthetic association, arising as a compound effect of linkage disequilibrium. Specifically, HLA-C alleles associated with rapid progression tend to be in linkage disequilibrium with the −35 T allele, while protective alleles such as HLA-B*57 and -B*27 are in linkage disequilibrium with the protective −35 C allele. Studies on immune responses in HIV-1 have shown that HLA-B allotypes most frequently induce detectable T-cell responses and that these are generally of greater magnitude than responses restricted by either HLA-A or HLA-C. Therefore, HLA-B-restricted CD8 T-cell responses play a greater role in the immune response against HIV-1 infection (4, 25, 26). It is very well established that pairs of HLA-B and HLA-C alleles are in linkage disequilibrium and that particular haplotypes can be either protective (B*14-Cw*0802 in Caucasians and B*3910-Cw*1203 and B*8101-Cw*0401 in Africans) or harmful (B*35-Cw*0401 in Caucasians) in the context of HIV-1 disease progression (8, 17, 30, 48). HLA-Cw*07 is in strong linkage disequilibrium with HLA-B*07 and -B*08, and the latter is known to have an unfavorable impact on HIV-1 disease progression (9, 24, 35, 42, 48, 54). Although particular HLA alleles, particularly those of the HLA-B locus, can have a strong individual impact, overall HIV-1 control is likely to be influenced by the combination of all HLA alleles present (30).
In conclusion, it is extremely unlikely that the association of the −35 SNP with control of HIV-1 infection is a direct result of the SNP itself, and the role of increased expression of HLA-C allotypes associated with the −35 C allele (18, 52) is uncertain. Our data imply that it is the particularly low expression of the −35 T-associated HLA-Cw*07 allele that contributes substantially to the low HLA-C expression associated with the −35 T allele, and possibly to the relatively high risk of disease progression.
Acknowledgments
This work was supported by the Center for HIV/AIDS Vaccine Immunology (CHAVI), by NIAID grant A1067854, and by the Wellcome Trust (grant 081799/Z/06/Z to T.W.C.). Additional support came from the Medical Research Center Human Immunology Unit, the UK National Institute for Health Research, and the Oxford Biomedical Research Center.
We are grateful to the healthy volunteers and patients who provided samples for study. We are grateful to David Goldstein for review and discussion of the manuscript and to Marco Colonna for helpful discussions on HLA-C- and HLA-E-specific monoclonal antibodies. We also thank Susan Fuggle and Suzanne Page at the Oxford Transplant Centre for assistance with LABScreen single-antigen bead staining.
Footnotes
Published ahead of print on 19 January 2011.
REFERENCES
- 1.Adnan, S., et al. 2006. Nef interference with HIV-1-specific CTL antiviral activity is epitope specific. Blood 108:3414-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altfeld, M., et al. 2003. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 17:2581-2591. [DOI] [PubMed] [Google Scholar]
- 3.Barnstable, C. J., et al. 1978. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens—new tools for genetic analysis. Cell 14:9-20. [DOI] [PubMed] [Google Scholar]
- 4.Bihl, F., et al. 2006. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J. Immunol. 176:4094-4101. [DOI] [PubMed] [Google Scholar]
- 5.Bonaparte, M. I., and E. Barker. 2004. Killing of human immunodeficiency virus-infected primary T-cell blasts by autologous natural killer cells is dependent on the ability of the virus to alter the expression of major histocompatibility complex class I molecules. Blood 104:2087-2094. [DOI] [PubMed] [Google Scholar]
- 6.Braud, V. M., et al. 1998. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391:795-799. [DOI] [PubMed] [Google Scholar]
- 7.Braud, V. M., D. S. Allan, D. Wilson, and A. J. McMichael. 1998. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr. Biol. 8:1-10. [DOI] [PubMed] [Google Scholar]
- 8.Carrington, M., et al. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748-1752. [DOI] [PubMed] [Google Scholar]
- 9.Carrington, M., and S. J. O'Brien. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54:535-551. [DOI] [PubMed] [Google Scholar]
- 10.Catano, G., et al. 2008. HIV-1 disease-influencing effects associated with ZNRD1, HCP5 and HLA-C alleles are attributable mainly to either HLA-A10 or HLA-B*57 alleles. PLoS One 3:e3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen, G. B., et al. 1999. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10:661-671. [DOI] [PubMed] [Google Scholar]
- 12.Collins, K. L., and D. Baltimore. 1999. HIV's evasion of the cellular immune response. Immunol. Rev. 168:65-74. [DOI] [PubMed] [Google Scholar]
- 13.Collins, K. L., B. K. Chen, S. A. Kalams, B. D. Walker, and D. Baltimore. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397-401. [DOI] [PubMed] [Google Scholar]
- 14.Coupel, S., et al. 2007. Expression and release of soluble HLA-E is an immunoregulatory feature of endothelial cell activation. Blood 109:2806-2814. [DOI] [PubMed] [Google Scholar]
- 15.Dalmasso, C., et al. 2008. Distinct genetic loci control plasma HIV-RNA and cellular HIV-DNA levels in HIV-1 infection: the ANRS Genome Wide Association 01 study. PLoS One 3:e3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickson, S. P., K. Wang, I. Krantz, H. Hakonarson, and D. B. Goldstein. Rare variants create synthetic genome-wide associations. PLoS Biol. 8:e1000294. [DOI] [PMC free article] [PubMed]
- 17.Fellay, J., et al. 2009. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 5:e1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fellay, J., et al. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frazer, K. A., S. S. Murray, N. J. Schork, and E. J. Topol. 2009. Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 10:241-251. [DOI] [PubMed] [Google Scholar]
- 20.Goldstein, D. B. 2009. Common genetic variation and human traits. N. Engl. J. Med. 360:1696-1698. [DOI] [PubMed] [Google Scholar]
- 21.Goulder, P. J., and D. I. Watkins. 2008. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 8:619-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunt, P. W., et al. 2003. T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. J. Infect. Dis. 187:1534-1543. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, D. R. 2000. Differential expression of human major histocompatibility class I loci: HLA-A, -B, and -C. Hum. Immunol. 61:389-396. [DOI] [PubMed] [Google Scholar]
- 24.Kaslow, R. A., et al. 1990. A1, Cw7, B8, DR3 HLA antigen combination associated with rapid decline of T-helper lymphocytes in HIV-1 infection. A report from the Multicenter AIDS Cohort Study. Lancet 335:927-930. [DOI] [PubMed] [Google Scholar]
- 25.Kiepiela, P., et al. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769-775. [DOI] [PubMed] [Google Scholar]
- 26.Kiepiela, P., et al. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46-53. [DOI] [PubMed] [Google Scholar]
- 27.Kosmrlj, A., et al. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature 465:350-354. [DOI] [PMC free article] [PubMed]
- 28.Lanier, L. L. 2005. NK cell recognition. Annu. Rev. Immunol. 23:225-274. [DOI] [PubMed] [Google Scholar]
- 29.Lee, N., D. R. Goodlett, A. Ishitani, H. Marquardt, and D. E. Geraghty. 1998. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J. Immunol. 160:4951-4960. [PubMed] [Google Scholar]
- 30.Leslie, A., et al. 2010. Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J. Virol. 84:9879-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Limou, S., et al. 2009. Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02). J. Infect. Dis. 199:419-426. [DOI] [PubMed] [Google Scholar]
- 32.Lo Monaco, E., et al. 2008. HLA-E: strong association with beta2-microglobulin and surface expression in the absence of HLA class I signal sequence-derived peptides. J. Immunol. 181:5442-5450. [DOI] [PubMed] [Google Scholar]
- 33.McCutcheon, J. A., J. Gumperz, K. D. Smith, C. T. Lutz, and P. Parham. 1995. Low HLA-C expression at cell surfaces correlates with increased turnover of heavy chain mRNA. J. Exp. Med. 181:2085-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMichael, A. J., P. Borrow, G. D. Tomaras, N. Goonetilleke, and B. F. Haynes. The immune response during acute HIV-1 infection: clues for vaccine development. Nat. Rev. Immunol. 10:11-23. [DOI] [PMC free article] [PubMed]
- 35.McNeil, A. J., et al. 1996. Association of HLA types A1-B8-DR3 and B27 with rapid and slow progression of HIV disease. QJM 89:177-185. [DOI] [PubMed] [Google Scholar]
- 36.Mellors, J. W., et al. 2007. Prognostic value of HIV-1 RNA, CD4 cell count, and CD4 cell count slope for progression to AIDS and death in untreated HIV-1 infection. JAMA 297:2349-2350. [DOI] [PubMed] [Google Scholar]
- 37.Mellors, J. W., et al. 1997. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann. Intern. Med. 126:946-954. [DOI] [PubMed] [Google Scholar]
- 38.Menier, C., et al. 2003. Characterization of monoclonal antibodies recognizing HLA-G or HLA-E: new tools to analyze the expression of nonclassical HLA class I molecules. Hum. Immunol. 64:315-326. [DOI] [PubMed] [Google Scholar]
- 39.Middleton, D., L. Menchaca, H. Rood, and R. Komerofsky. 2003. New allele frequency database: http://www.allelefrequencies.net. Tissue Antigens 61:403-407. [DOI] [PubMed] [Google Scholar]
- 40.Migueles, S. A., et al. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. U. S. A. 97:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nattermann, J., et al. 2005. HIV-1 infection leads to increased HLA-E expression resulting in impaired function of natural killer cells. Antivir. Ther. 10:95-107. [DOI] [PubMed] [Google Scholar]
- 42.O'Brien, S. J., X. Gao, and M. Carrington. 2001. HLA and AIDS: a cautionary tale. Trends Mol. Med. 7:379-381. [DOI] [PubMed] [Google Scholar]
- 43.Olerup, O., and H. Zetterquist. 1992. HLA-DR typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours: an alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens 39:225-235. [DOI] [PubMed] [Google Scholar]
- 44.Palmisano, G. L., et al. 2005. HLA-E surface expression is independent of the availability of HLA class I signal sequence-derived peptides in human tumor cell lines. Hum. Immunol. 66:1-12. [DOI] [PubMed] [Google Scholar]
- 45.Parham, P. 2005. MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 5:201-214. [DOI] [PubMed] [Google Scholar]
- 46.Pei, R., J. H. Lee, N. J. Shih, M. Chen, and P. I. Terasaki. 2003. Single human leukocyte antigen flow cytometry beads for accurate identification of human leukocyte antigen antibody specificities. Transplantation 75:43-49. [DOI] [PubMed] [Google Scholar]
- 47.Pelak, K., et al. Host determinants of HIV-1 control in African Americans. J. Infect. Dis. 201:1141-1149. [DOI] [PMC free article] [PubMed]
- 48.Pereyra, F., et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shrestha, S., et al. 2009. Host genetics and HIV-1 viral load set-point in African-Americans. AIDS 23:673-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snary, D., C. J. Barnstable, W. F. Bodmer, and M. J. Crumpton. 1977. Molecular structure of human histocompatibility antigens: the HLA-C series. Eur. J. Immunol. 7:580-585. [DOI] [PubMed] [Google Scholar]
- 51.Stranger, B. E., et al. 2005. Genome-wide associations of gene expression variation in humans. PLoS Genet. 1:e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas, R., et al. 2009. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat. Genet. 41:1290-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trachtenberg, E., et al. 2009. The HLA-B/-C haplotype block contains major determinants for host control of HIV. Genes Immun. 10:673-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turnbull, E. L., et al. 2006. HIV-1 epitope-specific CD8+ T cell responses strongly associated with delayed disease progression cross-recognize epitope variants efficiently. J. Immunol. 176:6130-6146. [DOI] [PubMed] [Google Scholar]
- 55.van Manen, D., et al. 2009. Association of HLA-C and HCP5 gene regions with the clinical course of HIV-1 infection. AIDS 23:19-28. [DOI] [PubMed] [Google Scholar]
- 56.Williams, M., et al. 2002. Direct binding of human immunodeficiency virus type 1 Nef to the major histocompatibility complex class I (MHC-I) cytoplasmic tail disrupts MHC-I trafficking. J. Virol. 76:12173-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]