Abstract
A common polymorphism at codon 72 in the p53 tumor suppressor gene encodes either proline (P72) or arginine (R72). Several groups have reported that in cultured cells, this polymorphism influences p53's transcriptional, senescence, and apoptotic functions. However, the impact of this polymorphism within the context of a living organism is poorly understood. We generated knock-in mice with the P72 and R72 variants and analyzed the tissues of these mice for apoptosis and transcription. In the thymus, we find that the P72 variant induces increased apoptosis following ionizing radiation, along with increased transactivation of a subset of p53 target genes, which includes murine Caspase 4 (also called Caspase 11), which we show is a direct p53 target gene. Interestingly, the majority of genes in this subset have roles in inflammation, and their promoters contain NF-κB binding sites. We show that caspase 4/11 requires both p53 and NF-κB for full induction after DNA damage and that the P72 variant shows increased interaction with p65 RelA, a subunit of NF-κB. Consistent with this, we show that P72 mice have a markedly enhanced response to inflammatory challenge compared to that of R72 mice. Our data indicate that the codon 72 polymorphism impacts p53's role in inflammation.
Within the p53 tumor suppressor gene exists a common polymorphism at codon 72, encoding either proline or arginine (P72 or R72, respectively). That these two variants might possess altered biological function was first suggested by the finding of a linear relationship between geographic latitude and the frequency of these variants, suggesting that there may be selection for the P72 allele in environments subject to high ultraviolet light or warmer winter temperatures (2, 40, 41). Using genetically engineered inducible cell lines as well as human tumor cell lines homozygous for each variant, we and others have shown that these two forms of p53 differ in their ability to induce growth arrest and apoptosis. Specifically, the P72 variant possesses an increased ability to transactivate p21 and induce growth arrest (5, 36, 43, 46), while the R72 variant demonstrates superior mitochondrial localization in tumor cell lines (8). However, differences in the function of these variants within an intact organism are unclear. To date, there have been a large number of epidemiological studies investigating the impact of this polymorphism on cancer risk. At present, the combined studies suggest that there may be a minor association between the P72 allele and increased cancer risk (10, 55). The high degree of genetic variability in human populations, combined with a dearth of information regarding potential tissue-specific effects of the codon 72 polymorphism on p53 function, suggests that it is critical that a mouse model be created in order to analyze these variants.
Like p53, NF-κB is a stress-inducible transcription factor that plays a central role in proliferation and apoptosis. It also plays a key role in the regulation of immunity and inflammation. Given the pivotal role of both proteins in tumorigenesis, it is not surprising that there is considerable cross talk between them. Most of the available evidence indicates that these two transcription factors function antagonistically. For example, NF-κB transcriptionally induces the negative regulator of p53, MDM2 (21, 45). p53 inhibits the ability of NF-κB to transactivate NF-κB-responsive promoters (15, 17), and p53 and NF-κB compete for binding to p300 on target promoters (33, 50, 52). Restoring p53 function inhibits NF-κB (26), and loss of p53 is associated with increased NF-κB activity (22, 23). Conversely, under certain circumstances, p53 and NF-κB can cooperate with each other, such as in the transactivation of genes that contain both p53 and NF-κB response elements in their promoters, like the Skp2 gene (1). p53 and NF-κB likewise cooperate to transactivate certain target genes in cells exposed to hydroxyurea (38). Finally, in certain cells, p53 requires NF-κB in order to efficiently induce apoptosis (35). Clearly, depending upon circumstances, the impact of these proteins on each other's function can be either antagonistic or cooperative.
A clear role for p53 in the control of innate immunity has emerged in recent years. The transcription of the p53 gene is controlled by type I interferon (IFN) signaling (44), and the induction of p53 participates in the host defense against viral infection (6, 28). p53 also interacts with interferon regulatory factor 9 (IRF9) to enhance IFN signaling (6). Additionally, p53 regulates the transcription of several cytokines and chemokines involved in innate immunity; this activity of p53 is believed to contribute to the ability of the immune system to eliminate senescent cells (56). The role of p53 in the control of innate immunity was recently found to be evolutionarily conserved: specifically, a mutation in the nucleolar protein Nol-6 leads to ribosomal stress-induced p53 activation in Caenorhabditis elegans, which results in increased innate immune function (11).
We have focused on elucidating the impact of the codon 72 polymorphism on p53 function. While previous studies support the premise that these two variants have altered functions, the majority of these studies were cell line based and utilized overexpression of p53 alleles. We recently described a humanized p53 knock-in (Hupki) mouse in which exons 4 to 9, encoding the human proline-rich and DNA binding domains of p53, replace those of the mouse. Because codon 72 in mouse encodes alanine, it was important to study the codon 72 polymorphism in a humanized version of p53. Importantly, Hupki p53 is fully functional and tumor suppressive in the mouse (24, 34, 54). In the present study, we compare the biological functions of the P72 and R72 variants in inbred mice and show that these variants demonstrate significant differences in apoptotic function. Moreover, these studies led us to find for the first time that the codon 72 polymorphism markedly affects the ability of p53 to interact and cooperate with NF-κB in the transactivation of genes involved in immunity and inflammation.
MATERIALS AND METHODS
Mouse studies and treatment.
P72 and R72 mice were generated on a mixed C57BL/6-129 (C57/129) background. These mice were backcrossed to C57BL/6 mice (Jackson Laboratories) for seven generations. P72 and R72 mice were then crossed to each other for three generations; single nucleotide polymorphism (SNP) genotyping analysis of markers polymorphic between the C57BL/6 and 129 backgrounds (performed by the Jackson Laboratories) indicates that these mice can be considered C57BL/6 in genotype. All studies with mice complied with all federal and institutional guidelines. For irradiation experiments, mice were exposed to a cesium-137 gamma source (Fox Chase Cancer Center Irradiation Facility). For lipopolysaccharide (LPS) treatment, P72 and R72 mice were injected intraperitoneally with 20 mg/kg LPS (Escherichia coli 0111; B4 Calbiochem), and survival was tracked daily. A total of 4 h after injection, the thymuses from mice were harvested and analyzed by immunohistochemistry, or instead, thymocytes were purified, and RNA was used for quantitative reverse transcription-PCR (QPCR). For tumor studies, Eμ-myc and p53+/− mice were obtained from Jackson Laboratories (C57BL/6 background).
Cell culture, drug treatments, and Western analysis.
Primary mouse embryo fibroblast (MEF) lines were derived from 13.5-day-old embryos as described previously (54); only cells from passages 0 to 4 were used for these studies. H1299 cells with Tet-inducible P72 and R72 were treated with 0.75 μg/ml doxycycline for 6 h to induce p53. Normal human fibroblast lines 6113 (Pro/Pro) and 5386 (Arg/Arg) were obtained from the Coriell Institute for Medical Research and cultured in Dulbecco modified Eagle medium (DMEM), 15% fetal bovine serum (FBS), 1% Pen/Strep, and 1% l-glutamine. Adriamycin (Sigma) at a concentration of 0.5 μg/ml was used. Etoposide (Sigma) at a concentration of 100 μM was used. BAY-11-7082 (Cayman Chemicals) at 1 μM was used. Western analysis was performed as described previously (8). Membranes were blocked and probed with antibodies for 1:500 p53 505 (Novocastra), 1:10,000 actin AC15 (Sigma), 1:200 each Mdm2 Ab1 and Ab2 (Calbiochem), 1:100 p21 Ab 4 (Calbiochem), 1:1,000 Ras (BD Biosciences), 1:500 cleaved caspase 3 (Cell Signaling), 1:200 p53 FL393 goat (Santa Cruz), 1:1,000 cleaved lamin A (Cell Signaling), 1:200 cleaved caspase 11 M20 (Santa Cruz), 1:1,000 tubulin (Sigma), 1:1,000 p65 ab7970 (Abcam), 1:1,000 p105/p50 ab7971 (Abcam), 1:1,000 p53 Ab 6 (Calbiochem), and 1:500 p53 ser15 (Cell Signaling). Horseradish peroxidase-conjugated secondaries (Jackson Immunochemicals) were used at a dilution of 1:10,000. Blots were exposed to ECL (Amersham).
RNA isolation, QPCR, and microarray.
RNA was isolated from MEFs and cell lines using RNeasy (Qiagen), and RNA from thymocytes was isolated using Trizol (Invitrogen). For microarray analysis, RNA was amplified and labeled using the Agilent Quick Amp labeling kit. A total of 1.65 μg of Cy3-labeled cRNA targets was hybridized onto Agilent 4×44k whole-genome arrays for 17 h at 65 degrees and washed according to the Agilent protocol. Hybridized slides were scanned at a 5-μm resolution on an Agilent scanner, and fluorescence intensities of hybridization signals were extracted using Agilent Feature Extraction software. Raw expression data obtained from Agilent microarrays were background corrected and quantile normalized across the experimental conditions (3). The LIMMA (Linear Models for Microarray Data) methodology outlined in reference 42 was applied to the log2-transformed expression data to identify differentially expressed genes in each comparison. The LIMMA module in the Open Source R/Bioconductor package (13) was utilized in the computations. Differentially expressed genes were identified based on statistical significance as well as biological significance. Statistical significance was measured by the false discovery rate (FDR) to account for multiple testing; genes showing an FDR of less than 5% were considered statistically significant. QPCR was performed in the Fox Chase Cancer Center QPCR Facility, using ABI primer sets purchased for the indicated genes.
siRNA transfection.
A total of 50 μM of small interfering RNA (siRNA) (negative control; p65 and p50 from DharmaFECT) was transfected using DharmaFECT 1 for 20 h. After an 8-hour recovery period, cells were treated with etoposide (100 μM) for 16 to 24 h.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed using the SimpleChIP enzymatic chromatin IP kit, as per the protocols provided (Cell Signaling), using the following primer sets: p21-For, 5′GGTGGGGACTAGCTTTCTGG3′; p21-Rev, 5′TCCACCACCCTGCACTGA3′; Noxa-For, 5′GGGGTTGAGCAGGACTCGT3′; Noxa-Rev, 5′GAGCGAAGTGGAGCAGGTCT3′; Casp4-p53-1-For, 5′AAGTTGTATTTGTCAGCTTAGGTCCA3′; Casp4-p53-1-Rev, 5′ATGATCAGACGCTTGTCGTTTTTA3′; Casp4-p53-2-For, 5′CCACCTTGCTGTCTATACCAGATACT3′; Casp4-p53-2-Rev, 5′ATTAAAAGACAGTGTCCCAGAGAAGA3′; Casp4-NF-κB-For, 5′ACTTTCTGAGCAGCTCTTTCAACA3′; and Casp4-NF-κB-Rev, 5′GCCATGAGAAAAAGCCTCAGTT3′. The IGX1A negative-control primer set was purchased from Qiagen.
Immunohistochemistry.
Tissues were harvested and fixed in 10% phosphate-buffered formaldehyde (F79-4; Fischer Scientific) for 48 h and embedded in paraffin. Following deparaffinization, antigen retrieval was performed with citrate buffer at pH 6 for 10 min in a microwave oven. Endogenous peroxidases were quenched with 0.3% hydrogen peroxide in methanol. Sections were incubated overnight with the primary antibody, washed the next day with phosphate-buffered saline (PBS), incubated with biotinylated secondary antibodies (Vector Labs), stained with the Vectastain Elite ABC kit (Vector Labs), developed with a DAB kit (Vector Labs), and lightly counterstained with Meyer's hematoxylin. Negative controls were stained without primary antibody. Specimens were documented photographically using a Nikon Optiphot microscope equipped with an Optronics charge-coupled-device (CCD) camera. Antisera used included p53 CM5 (Novocastra), cleaved caspase 11 M20 (Santa Cruz), p53 ser15 (Cell Signaling), cleaved caspase 3 (Cell Signaling), and cleaved lamin A (Cell Signaling).
Immunoprecipitation and Western blotting.
A total of 250 to 3,000 μg of whole-cell lysate was incubated with 1 μg of antibody overnight. Protein G-agarose beads were added for 1 h, followed by washes and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The antibodies used were the same as those described above for Western blotting, with the addition of p300 N-15 (Santa Cruz). Equal amounts of IgG were used as negative controls. Horseradish peroxidase-conjugated light-chain-specific secondary antibody was used (Jackson Immunochemicals).
Statistical analysis.
Data were analyzed by two-sided unpaired Student's t test using a GraphPad software package. For the animal cancer studies, data were analyzed with the one-sided Wilcoxon 2 sample test. For the LPS studies, data were analyzed using the log rank test.
Microarray data accession numbers.
Microarray data have been submitted to the GEO database and have been assigned accession no. GSE26851.
RESULTS
Increased induction of p21 in P72 Hupki MEFs.
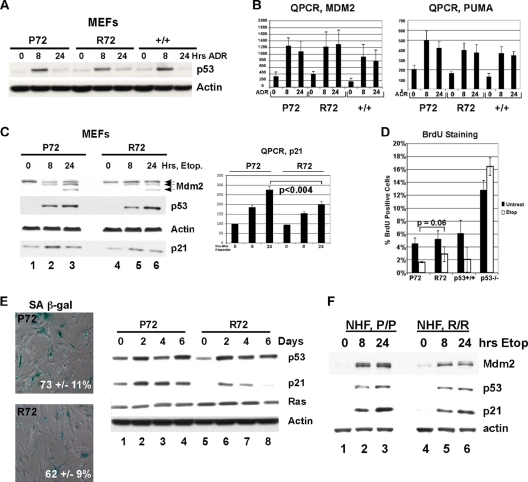
Humanized p53 knock-in (Hupki) mice carrying the P72 and R72 alleles were generated in a mixed C57/129 background (34), backcrossed for multiple generations to a C57BL/6 background, and then crossed to each other to generate the mice analyzed in this study (see Materials and Methods for details). For simplicity, these will be referred to as P72 and R72 mice. To initially compare the expression and function of P72 and R72 variants, primary mouse embryo fibroblast (MEF) cultures were generated for each variant and analyzed for p53 stabilization and the transcriptional response to genotoxic stress. As depicted in Fig. 1, the levels of stabilization of p53 following treatment with adriamycin in P72, R72, and wild-type C57BL/6 (+/+) primary MEF cultures were comparable (Fig. 1A). Similarly, quantitative reverse transcription-PCR (QPCR) indicated that the transactivation of two p53 target genes, Mdm2 and Puma (Bbc3), was indistinguishable between these two variants and murine wild-type p53 (Fig. 1B).
FIG. 1.
The P72 variant demonstrates an increased ability to transactivate p21 and induce senescence. (A) Western analysis of whole-cell lysates from primary mouse embryo fibroblasts (MEFs) isolated from P72, R72, and wild-type (+/+) mice treated with 0.5 μg/ml of adriamycin (ADR) for 8 or 24 h and probed with antisera for p53 and actin. (B) Quantitative reverse transcription-PCR (QPCR) to obtain the levels of Mdm2 and Puma in mouse embryo fibroblasts (MEFs) treated with 0.5 μg/ml adriamycin. The results shown are averaged values from two independent MEF cultures of each genotype. (C, left) Western analysis of primary MEFs treated with 100 μM etoposide (Etop) for 8 or 24 h and probed with the indicated antibodies. (Right) Quantitative reverse transcription-PCR (QPCR) analysis to obtain the level of p21 normalized to that of the control. Error bars depict standard errors. Statistical significance was calculated using the Student's t test. (D) Percentage of bromodeoxyuridine (BrdU)-positive cells from primary P72, R72, +/+, and −/− MEFs treated with etoposide (Etop; 100 μM) after 24 h. Error bars depict standard errors. (E, left) Senescence-associated β-galactosidase (SA β-gal) staining of primary P72 and R72 MEF cultures infected with oncogenic Ha-Ras retrovirus, selected for 4 days for Ha-Ras expression, and assayed after the times indicated; the data depicted are representative of 4 days postselection. (Right) Western analysis of the levels of p53, p21, Ras, and control (actin) in Ha-Ras-infected MEFs of the genotypes shown. (F) Western analysis of lysates from normal human fibroblasts (NHFs) that are homozygous P72 or R72 treated with 100 μM etoposide for 8 and 24 h and probed for Mdm2, p53, p21, and actin.
In response to the DNA-damaging agent etoposide, the stabilization of the P72 and R72 proteins and the induction of the p53 target gene Mdm2 were similar between these variants in primary MEF cultures (Fig. 1C). There was, however, a moderate but consistent increase in the induction of the cyclin-dependent kinase inhibitor p21 (Cdkn1a) in P72 MEFs, as determined by Western analysis and QPCR (Fig. 1C); this difference was by approximately 1.5-fold and was evident in multiple P72 and R72 MEF lines (A. K. Frank, unpublished results). These data were notable because in human cell lines, a similar increased ability of the P72 variant to transactivate p21 has been noted (5, 36, 46). To investigate further, we compared the abilities of the P72 and R72 variants to induce cell cycle arrest and Ras-mediated senescence, both of which require p21. P72 MEFs showed a moderate but consistent increase in cell cycle arrest following etoposide treatment (Fig. 1D) and gamma irradiation (data not shown), as assessed by the percentage of cells positive for bromodeoxyuridine (BrdU). We also noted increased p21 levels in P72 MEFs following infection with oncogenic Ha-ras, along with increased numbers of cells positive for senescence-associated β-galactosidase (Fig. 1E). Similarly, we noted a consistently increased ability of P72 to induce p21 in normal human fibroblasts homozygous for P72 and R72 (Fig. 1F). Overall, these data were notable because they recapitulate what has been found for the human codon 72 variants in normal fibroblasts (5, 36) and therefore support the use of Hupki mice as a valid model in which to investigate functional differences between codon 72 variants.
Increased apoptosis in the P72 thymus.
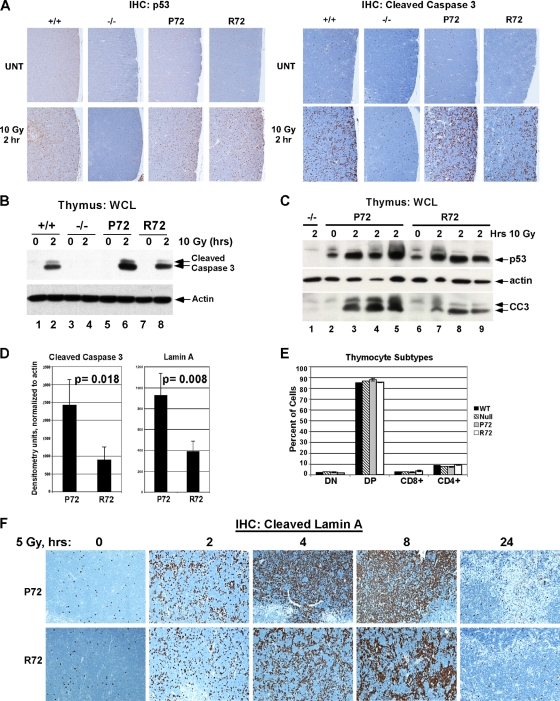
P72 and R72 mice were next subjected to gamma irradiation, and the thymuses of these mice were analyzed by immunohistochemistry using antisera for p53, cleaved caspase 3, and caspase-cleaved lamin A to measure apoptosis. Immunostaining for p53 in the thymuses of P72, R72, and wild-type (+/+) mice showed no difference in p53 accumulation at 2 h following 10 Gy of gamma radiation (Fig. 2 A, left). There were, however, marked differences in apoptosis, with approximately a 2-fold increased number of apoptotic cells in the P72 thymus compared to that in the R72 thymus (Fig. 2A, right); this difference was also evident by Western analysis of cleaved caspase 3 (Fig. 2B). Interestingly, this was not the case in other tissues, as there was consistently greater apoptosis in the R72 small intestine and equal apoptosis seen between these variants in the spleen (data not shown). To account for possible differences due to biological variation, we next analyzed three P72 and R72 sibling littermates side by side for their levels of apoptosis after gamma irradiation using Western analysis of cleaved caspase 3 and cleaved lamin A. Again these data indicated between 2- and 2.5-fold increased apoptosis levels in the thymuses of P72 mice (Fig. 2C and D). These data could not be explained by differences in the differentiation of the thymocytes in these mice, as P72 and R72 mice showed identical profiles of thymocyte development (Fig. 2E). We next performed a time course experiment following a lower dose of gamma irradiation (5 Gy). This analysis also showed increased apoptosis in the P72 thymus at all time points, as assessed by immunohistochemical (IHC) analysis of cleaved lamin A (Fig. 2F), with the most marked differences between P72 and R72 mice at 4 and 8 h postradiation.
FIG. 2.
Increased apoptosis in P72 thymocytes following ionizing radiation. (A) Wild-type (+/+), p53-null (−/−), P72, and R72 mice were irradiated with 10 Gy, and after 2 h, thymuses were collected and subjected to immunohistochemical (IHC) analysis of p53 (left) and cleaved (activated) caspase 3 (right). UNT, untreated. (B) Western analysis of thymic extracts from P72 and R72 mice and p53-null mice (−/−) that were irradiated with 10 Gy, harvested after 2 h, and probed for p53, actin, and cleaved caspase 3 (CC3). The lower-molecular-weight band is the fully active form of caspase 3. WCL, whole-cell lysate. (C) As in panel B, except that sibling littermates were analyzed. (D) The averaged densitometry results for Western analyses for cleaved caspase 3 shown in panel B and cleaved lamin A in irradiated thymocytes normalized to actin. The data depicted are averaged values from 3 mice per genotype. Statistical significance was calculated using the Student's t test. (E) Thymocytes were isolated from P72, R72, wild-type, and p53-null mice and subjected to fluorescence-activated cell sorter (FACS) analysis of the cell surface markers CD8 and CD4. The percentages of cells that were double negative (DN), double positive (DP), or singly positive (CD8+ and CD4+) were graphed. Three mice with each genotype were analyzed. (F) Immunohistochemical analysis of cleaved lamin A obtained from thymuses from P72 and R72 mice that were irradiated with 5 Gy and harvested after the times indicated.
Increased induction of a subset of p53 target genes in the P72 thymus.
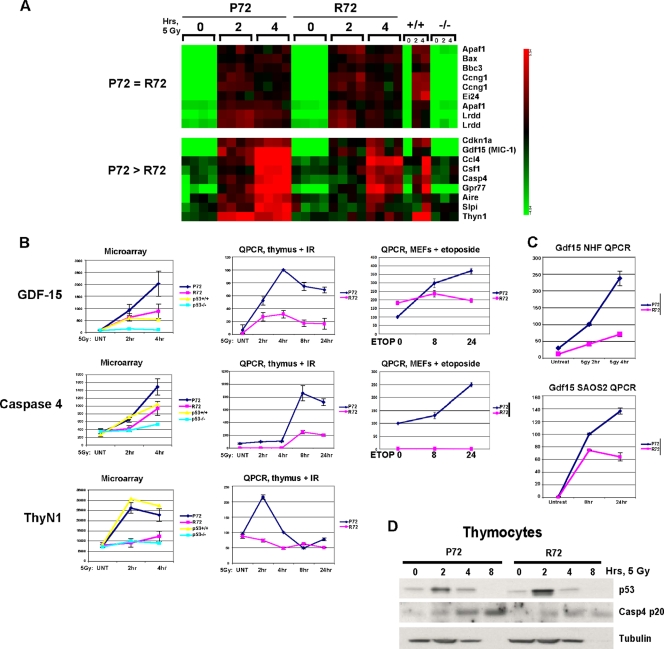
The p53-dependent apoptotic pathway possesses both transcription-dependent and -independent arms (4). To determine the underlying basis for increased apoptosis in the P72 thymus, we analyzed thymocytes from gamma-irradiated P72 and R72 mice for mitochondrial localization of p53 and for the transactivation of p53 target genes with known roles in cell death (Puma [Bbc3], Noxa [Pmaip1], Bax, and Pidd [Lrdd]). Surprisingly, no differences in the localization of these variants to mitochondria, or in the transactivation of these target genes, were detected (data not shown). These findings prompted us to consider the possibility that a previously unidentified p53 target gene(s) might play a role in the increased apoptosis found in P72 thymocytes. With this hypothesis in mind, we performed microarray analysis on thymocytes isolated from P72 and R72 mice that were subjected to 5 Gy of gamma radiation and isolated after 0, 2, and 4 h. Four independent biological replicates of each experiment were conducted, using two mice per time point along with p53+/+ and p53−/− mice as controls. A heat map depicting known p53 target genes, including Apaf1, Bax, Puma (Bbc3), and Pidd (Lrdd), is depicted in Fig. 3 A; this heat map indicates no differences in the transactivation of proapoptotic p53 target genes following gamma radiation in P72, R72, and +/+ mice (Fig. 3A). Indeed, of the more than 100 genes that were upregulated in a p53-dependent manner following gamma radiation, the overwhelming majority displayed no differences between the P72 and R72 thymocytes (data not shown). There was, however, a small subset of genes that were upregulated in a p53-dependent manner and which were transactivated to higher levels in P72 thymocytes. These genes included the known p53 target genes p21 (Cdkn1a), Gdf15 (31), Csf-1, and Cxcl1 (56). Other preferential P72 targets included Ccl4, Caspase 4 (originally called Caspase 11 in mouse and most likely the orthologue of human Caspase 5), Gpr77, Aire, Slpi, and ThyN1 (Fig. 3A). Quantitation of the data from the microarray analysis of the genes Gdf15, ThyN1, and Caspase 4/11, as well as confirmatory QPCR analysis of thymocytes and primary MEFs, is depicted in Fig. 3B. Overall, we noted a 2- to 4-fold increase in the levels of these genes in P72 cells relative to those in R72 cells (Fig. 3B). Of these three genes, only Gdf15 was expressed in normal human fibroblasts; QPCR analysis confirmed that there is increased expression of Gdf15 in P72 normal human fibroblasts (NHFs) and in inducible P72 cells compared to that in R72 NHFs (Fig. 3C). Western analysis of caspase 4/11 confirmed that this protein is induced by gamma radiation to higher levels in P72 thymocytes than in R72 thymocytes (Fig. 3D). Caspase 4/11 functions in innate immunity by processing caspase 1, which processes and activates interleukin-1β; alternatively, caspase 4/11 can behave as an initiator caspase by directly activating caspase 3 (19, 20). We confirmed that the increased apoptosis in P72 thymocytes may be caused by enhanced induction of caspase 4/11, as we found that expression of full-length caspase 4/11 in transfected cells is sufficient to induce apoptosis (data not shown) (19, 20).
FIG. 3.
Increased expression of Gdf15, Casp4, and ThyN1 in the P72 thymus after irradiation. (A) Portions of the heat map generated from microarray analysis of thymocytes purified from P72, R72, wild-type (+/+), and p53-null (−/−) mice following treatment with 5 Gy and harvested after 2 or 4 h. Each square represents one of four independent biological replicates, using 2 mice per genotype. (B, left) Graphic representation of the generated microarray data shown in panel A, showing values for Gdf-15, caspase 4/11, and ThyN1. (Middle) QPCR analysis of a time course experiment with an independent set of thymocyte RNA samples for P72 and R72 thymocytes following treatment with 5 Gy, depicting Gdf-15, caspase 4/11, and ThyN1. IR, ionizing radiation. (Right) QPCR of RNA isolated from P72 and R72 mouse embryo fibroblasts (MEFs) treated with 100 μM etoposide for 0, 8, or 24 h. The results depicted are averaged values from four independent QPCR samples performed in duplicate; the error bars depict standard errors. (C, top) QPCR of the Gdf15 level in normal human fibroblasts (NHFs) homozygous for either P72 or R72 that were irradiated with 5 Gy and harvested after 2 and 4 h. (Bottom) QPCR of the Gdf15 level in Saos-2 cells containing a temperature-sensitive inducible form of P72 or R72 shifted to the permissive temperature (wild-type p53) for 8 and 24 h. (D) Western analysis of thymocytes purified from P72 and R72 mice following irradiation with 5 Gy and harvested after the time points indicated for p53, caspase 4/11 (Casp4 p20), and tubulin; the loss of tubulin at 8 h likely represents substantial cell death at this time point.
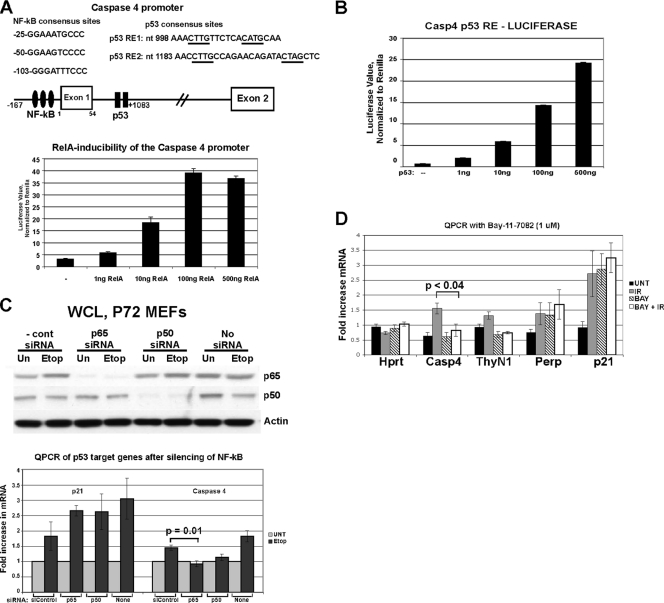
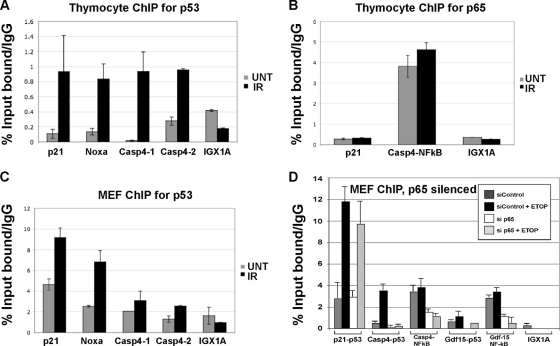
A list of all genes identified by microarray analysis as being induced to higher levels in P72 thymocytes is presented in Table 1. In this table, the ratio of P72/R72 induction of each gene is presented at 2 and 4 h after gamma radiation and ranges from 1.4- to 2.9-fold. Several of these genes (Gdf15, Rnd3, Csf-1, and Cxcl1) are known to be upregulated by p53 (29, 31, 56, 57). Interestingly, 10 out of 12 of these genes are known or postulated NF-κB target genes (see, for example, http://bioinfo.lifl.fr/NF-KB/), suggesting the involvement of this transcription factor. Consistent with this, IPA pathway analysis of this subset of genes indicated that NF-κB represented a common node of regulation (data not shown). We cloned the 5′ regulatory sequences of the murine Caspase 4/11 promoter and identified three consensus NF-κB binding sites in the upstream regulatory sequences and two closely linked consensus binding sites for p53 in the first intron (Fig. 4 A, diagram). In a reporter construct controlled by this promoter, we found that both p65 RelA and p53 were able to substantially activate reporter activity (Fig. 4A and B), suggesting that this promoter is positively regulated by both NF-κB and p53. We next tested the possibility that p53 cooperates with NF-κB in the DNA damage-mediated induction of caspase 4/11. Notably, we found that transfection of primary MEFs with siRNA of the NF-κB subunit p65 RelA or p50 markedly inhibited the ability of p53 to induce caspase 4/11, but not p21, following etoposide treatment (Fig. 4C). Similarly, we found that the NF-κB inhibitor BAY-11-7082 could effectively inhibit the ability of p53 in thymocytes to transactivate Caspase 4/11 but not p21 or Perp (Fig. 4D). The combined data support the premise that p53 and NF-κB cooperate in the regulation of Caspase 4/11 in MEFs and thymocytes.
TABLE 1.
List of genes demonstrating increased transactivation in P72 thymocytes following gamma radiation
| Gene | Full name | Increase in transactivation (fold) aftera: |
Activity |
Immunity/inflammation | ||
|---|---|---|---|---|---|---|
| 2 h | 4 h | NF-κB target | p53 | |||
| Slpi | Secretory leukocyte peptidase inhibitor | 1.6 | 2.6 | Unknown | − | Yes |
| Gdf15 | Macrophage inhibitory cytokine-1 | 1.5 | 2.4 | + | + | Yes |
| ThyN1 | Thymocyte nuclear protein 1 | 3 | 1.9 | Unknown | − | |
| Gpr77 | G protein coupled receptor 77 | 1.4 | 1.8 | + | − | Yes |
| Aire | Autoimmune polyendocrinopathy | 1.5 | 1.7 | + | − | |
| Jun | Jun oncogene | 1.3 | 1.6 | + | − | Yes |
| Casp4 | Caspase 4/11 | 1.6 | 1.6 | + | − | Yes |
| Ccl4 | Chemokine ligand 4 | 1.4 | 1.6 | + | − | Yes |
| Rnd3 | Rho family GTPase 3 | 1.7 | 1.5 | + | + | Yes |
| CSF1 | Macrophage colony-stimulating factor 1 | 1 | 1.5 | + | + | Yes |
| Cxcl1 | Chemokine ligand 1 | 1 | 1.4 | + | + | Yes |
The fold increase in transactivation in P72 cells compared to that detected in R72 cells is depicted after 2 and 4 h.
FIG. 4.
Induction of caspase 4/11 after DNA damage is impaired following silencing or inactivation NF-κB. (A, top) Schematic of the Caspase 4/11 promoter, depicting consensus NF-κB binding sites (filled ovals) and p53 binding sites (filled rectangles). The corresponding sequences of these sites and their locations relative to the start site of transcription are shown. (Bottom) Luciferase assays on lysates from H1299 cells transfected with a luciferase construct containing the full-length caspase 4 promoter (nucleotides −167 to +1083) along with increasing amounts of p65 RelA. The graph represents two independent transfections, each analyzed in triplicate. (B) Luciferase assay of lysates from H1299 cells transfected with a luciferase construct containing the p53 response element from caspase 4 (nucleotides +998 to 1083) and increasing amounts of p53. (C, top) Western analysis of primary mouse embryo fibroblasts (MEFs) transfected with a nontargeting control siRNA (siControl) or siRNA from p65 RelA (p65), p50, or left untreated (no siRNA). Following transfection, cells were treated with 100 μM etoposide (Etop) for 20 h or left untreated (Un). (Bottom) QPCR determination of the levels of the p53 target genes p21 and Caspase 4/11 in the siRNA-treated cells. The results shown were obtained from 2 independent experiments each done in duplicate; error bars depict standard errors. Statistical significance was calculated using Student's t test. (D) QPCR for caspase 4/11, normalized to the Hprt control. Thymocytes isolated from P72 mice were pretreated with 1 μM Bay-11-7082 for 1 h or left untreated, subjected to 5 Gy, and harvested after 2 h. QPCR was performed for Hprt, Casp4, ThyN1, Perp, and p21. The results depicted represent three independent experiments analyzed in duplicate. Error bars show standard errors of measurement. Statistical significance was calculated using Student's t test.
p53 and p65 RelA bind to the Caspase 4 promoter.
We next performed chromatin immunoprecipitation (ChIP) to determine whether p53 and NF-κB bind to the Caspase 4/11 promoter. In P72 thymocytes treated with gamma radiation, we were able to reproducibly detect a fragment of the Caspase 4/11 promoter containing the closely linked p53 binding site(s) immunoprecipitating with antisera to p53 but not the negative-control gene IGX1A (Fig. 5 A). Similarly, NF-κB binding was readily detectable at the closely linked consensus NF-κB sites (Fig. 5B). In MEFs treated with etoposide, we were likewise able to perform ChIP of the p53 binding site of Caspase 4/11 with p53 antisera (Fig. 5C). These data prompted us to evaluate the impact of the silencing of p65 RelA on the ability of p53 to bind to this promoter. Notably, we found that treatment of MEFs with siRNA to p65 RelA markedly inhibited the ability of p53 to chromatin immunoprecipitate the promoters of caspase 4/11 and Gdf-15 but not p21 (Fig. 5D). These data indicate that NF-κB is required for p53 binding to these promoters. We noted in these experiments that both the P72 and R72 proteins had comparable abilities in chromatin immunoprecipitating the Caspase 4/11 promoter in thymocytes and MEFs (A. K. Frank, unpublished results). These data prompted us to analyze the interaction between the codon 72 variants of p53 with NF-κB.
FIG. 5.
p53 binds to the Caspase 4/11 promoter in an NF-κB-dependent manner. (A) Chromatin immunoprecipitation using p53 antisera (antibody fl-393G) of P72 thymocytes treated with 5 Gy and harvested after 4 h. Immunoprecipitated DNA was analyzed by QPCR for the consensus p53 binding sites in Caspase 4/11 as well as for known p53 binding sites in p21 and Noxa and in the negative control IGX1A. All ChIP data were obtained from two independent experiments performed in duplicate, normalized to IgG control. The error bars depict standard errors. (B) Chromatin immunoprecipitation using antisera to p65 RelA (p65) for the p53 binding site in p21 as well as for the set of three closely linked NF-κB binding sites in Caspase 4/11. (C) Chromatin immunoprecipitation using p53 antisera of P72 MEFs treated with 100 μM etoposide (Etop) for 16 h or left untreated (UNT). Immunoprecipitated DNA was analyzed by QPCR for the consensus p53 binding sites in Caspase 4/11 as well as for known p53 binding sites in p21 and Noxa. (D) Chromatin immunoprecipitation using p53 and p65 RelA antisera of P72 MEFs treated with 100 μM etoposide for 16 h or left untreated (UNT), following 24 h transfection with siControl or siRNA to p65 RelA. Immunoprecipitated DNA was analyzed by QPCR for the consensus p53 and NF-κB binding sites in Caspase 4/11 and Gdf-15 as well as for the known p53 binding site in p21.
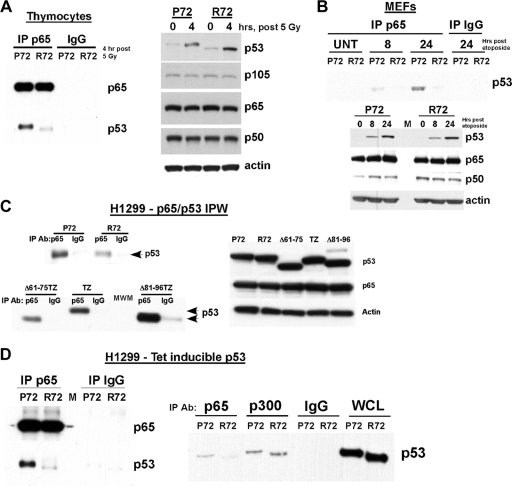
Increased association of P72 with the p65 subunit of NF-κB.
p53 and the p65 RelA subunit of NF-κB directly interact (15, 17). We isolated thymocytes from irradiated P72 and R72 mice and immunoprecipitated p65 RelA, followed by immunoblotting for associated p53. As shown in Fig. 6 A, p53 was clearly immunoprecipitated with p65 RelA in P72 thymocytes; however, there was evidence showing minimal R72 protein in these p65 immunoprecipitates, despite comparable total levels of p53. Similarly, in a time course experiment of etoposide-treated P72 MEFs, a p65 RelA/p53 complex was detectable at 8 h posttreatment and was enriched at 24 h (Fig. 6B). Again, however, there was minimal interaction between the R72 variant and p65 RelA. As both p53 and p65 can “shuttle” between the nucleus and the cytoplasm, it was formally possible that the impaired interaction between p65 and R72 was the result of differences in the nuclear/cytosolic localization of these p53 variants. However, cell fractionation experiments demonstrated no differences in the nuclear/cytosolic localization of P72 and R72 variants in etoposide-treated MEFs (data not shown). In transfected cells, we found that the R72 variant possessed a decreased ability to interact with p65 RelA and that deletion of the proline-rich domain of p53 likewise impaired the ability of p53 to interact with p65 RelA (Fig. 6C). We next tested the influence of the codon 72 polymorphism on the ability of human p53 to interact with p65. As depicted in Fig. 6D, we consistently found that the R72 variant had an impaired ability to interact with p65 RelA (Fig. 6D, left); in contrast, both variants showed identical abilities to immunoprecipitate with p300 (Fig. 6D, right). The combined data support the premise that the codon 72 polymorphism of p53 influences its ability to interact and cooperate with NF-κB.
FIG. 6.
Impaired interaction of the R72 variant of p53 with p65 RelA. (A, left) Immunoprecipitation-Western analysis of thymocytes from P72 and R72 mice harvested 4 h after 5 Gy; the immunoprecipitating (IP) antibody is denoted, and the blot was probed for p53 and p65 RelA. Equal exposures of p65 RelA and p53 are shown. (Right) Western analysis of thymocyte lysates for p53, the NF-κB subunits p105, p65, and p50, and the actin control. The data depicted are representative of three independent experiments. (B, top) Immunoprecipitation-Western analysis of P72 and R72 MEFs treated with 100 μM etoposide for 8 and 24 h or untreated (UNT); the immunoprecipitating (IP) antibody is denoted, and the blot was probed for p53. (Bottom) Western analysis of MEF lysates for p53, the NF-κB subunits p65 and p50, and the actin control. (C, left) A total of 250 μg of lysates from H1299 cells transfected for 24 h with plasmids containing P72, R72, Δ61-75TZ (tetramerization zipper [TZ] with deletion of positions 61 to 75), TZ, and Δ81-96TZ p53 variants were immunoprecipitated (IP) with an antibody (Ab) to p65 RelA and IgG. The TZ mutant lacks the oligomerization domain of p53, which reduces spurious interactions. IPW, immunoprecipitation-Western analysis. (Right) Western analysis of whole-cell lysates from H1299 (p53-null) cells transfected with the p53 constructs denoted and probed using antisera specific for p53, p65, and actin. (D) Immunoprecipitation-Western analysis of H1299 cells containing tetracycline-inducible versions of P72 and R72 following treatment with doxycycline for 6 h; the immunoprecipitating antibody is denoted, and the blot was probed for p53 and p65 RelA. Equal exposures of p65 RelA and p53 are shown. WCL, whole-cell lysate.
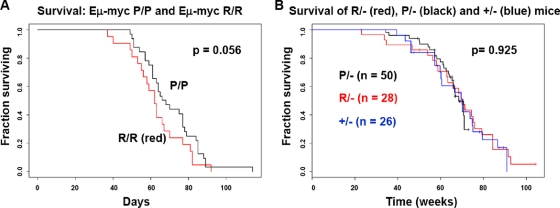
In order to assess the potential impact of the codon 72 polymorphism on cancer incidence, we crossed P72 and R72 mice with the Eμ-myc mouse, which develops B cell lymphoma; the development of this tumor is known to be limited by p53 (32, 37). We also crossed P72 and R72 mice with p53+/− mice to generate P72/− and R72/− mice; these would be expected to develop T cell lymphoma and sarcoma, predominantly (7). As depicted in Fig. 7 A, in the Eμ-myc background there was a modest increase in survival imparted by the P72 allele; this may be due to the increased ability of P72 to induce senescence, which is known to control the development of tumors in Eμ-myc mice (32). We noted, however, no difference in the survival (Fig. 7B) or tumor spectrum (Table 2) between P72/−, R72/−, and +/− mice. Therefore, consistent with the current literature on human populations and the findings of others (55, 58), we noted only the minor impact of the codon 72 polymorphism on cancer risk, at least in these models.
FIG. 7.
Limited impact of the codon 72 polymorphism of p53 on cancer incidence. (A) Kaplan-Meier analysis of survival data obtained from P72 and R72 mice crossed with Eμ-myc transgenic mice (C57BL/6 background). Statistical significance was assessed using the log rank test. (B) Kaplan-Meier analysis of survival data from P72 and R72 mice crossed with p53+/− mice to generate P/−, R/−, and +/− mice (C57BL/6 background). Statistical significance was assessed using the log rank test.
TABLE 2.
Tumor spectrum in P/−, R/−, and +/− mice
| Type of tumor | No. of tumors in indicated mice |
||
|---|---|---|---|
| P/− (n = 23) | R/− (n = 23) | +/− (n = 16) | |
| Lymphoma | 12 | 12 | 10 |
| Spindle cell sarcoma | 4a | 3a | 0 |
| Osteosarcoma | 3a | 4b | 3b |
| Hemangiosarcoma | 0 | 1 | 0 |
| Histiocytic sarcoma | 1 | 0 | 1 |
| Sarcoma (undetermined origin) | 1 | 0 | 1 |
| Mammary adenocarcinoma | 0 | 1a | 1a |
| Squamous cell carcinoma | 2b | 2b | 0 |
One mouse had multiple tumors.
Two mice had multiple tumors.
Enhanced response of P72 mice to LPS challenge.
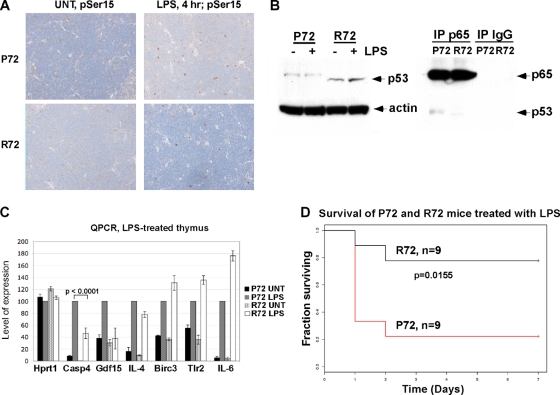
Our finding that the codon 72 polymorphism influences the ability of p53 to cooperate with NF-κB in the transactivation of a subset of proinflammatory p53 target genes prompted us to test the hypothesis that this polymorphism might have an impact on the inflammatory response. To test this hypothesis, we treated P72 and R72 mice with the endotoxin lipopolysaccharide (LPS), which stimulates a strong inflammatory response. Importantly, susceptibility to LPS challenge is known to be mediated by caspase 4/11, and knockout mice for this caspase show resistance to toxicity by LPS (51). We found that LPS treatment induces an accumulation of p53 protein that is phosphorylated on serine 15 (a common marker for activated p53) (Fig. 8 A). Additionally, a p65 RelA/p53 complex was detectable in LPS-treated P72 thymocytes (Fig. 8B). Analysis of RNA isolated from the thymocytes of LPS-treated mice indicated that Caspase 4/11 and Gdf-15 demonstrated markedly increased transactivation in P72 mice. In contrast, there was no difference in the transactivation of NF-κB target genes that lack p53 binding sites, including Birc3 and Tlr2 (Fig. 8C). Notably, the response to LPS challenge differed significantly for the two p53 genotypes, and whereas the majority of P72 mice succumbed to the septic shock induced by LPS within 2 days, the majority of R72 mice survived this treatment (Fig. 7D). This difference was highly significant (P = 0.0155; log rank test). These data indicate that the codon 72 polymorphism of p53 significantly affects the inflammatory response to LPS challenge in mice.
FIG. 8.
P72 mice demonstrate an enhanced response to lipopolysaccharide (LPS) challenge. (A) Thymuses were isolated from P72 and R72 mice that were subjected to intraperitoneal injection of 20 mg/kg of LPS or left untreated (UNT); after 4 h, thymuses were harvested, fixed, and stained for p53 phosphorylated on serine 15 using phosphospecific antibody (pSer15). In both P72 and R72 thymuses, approximately 4% of cells stain positively for p53 phosphorylated on serine 15 after 4 h. (B, left) Western analysis of whole-cell lysates from mice treated with 20 mg/kg of lipopolysaccharide (LPS) for 4 h. (Right) Immunoprecipitation-Western analysis of the p65 RelA-p53 interaction in LPS-treated thymocytes. (C) QPCR analysis of control (Hprt1), candidate p53 target genes (Casp4 and Gdf15), and the NF-κB target genes IL-4, Birc3, Tlr2, and IL-6 in thymocytes isolated from mice treated with LPS after 4 h. The results shown are averaged values from 2 independent experiments, each done in duplicate; error bars depict standard errors. (D) Kaplan-Meier plot of the survival of P72 and R72 mice injected with 20 mg/kg of LPS. Statistical significance was calculated using the Wilcoxon 2 sample test.
DISCUSSION
We describe the first detailed functional analysis of the Hupki mouse model of the p53 codon 72 polymorphism. We find that the P72 variant is associated with increased transactivation of the cyclin-dependent kinase inhibitor p21, along with an increased ability to induce growth arrest and senescence in MEFs; these data mirror the findings on human codon 72 variants of others (5, 36). We find that the transforming growth factor β (TGF-β) superfamily member Gdf15, which is a known p53 target gene, shows increased transactivation in P72 Hupki cells, normal human fibroblasts homozygous for P72, and inducible cell lines containing P72. We used microarray analysis to highlight the NF-κB pathway as being differentially impacted by the codon 72 polymorphism, leading us to the finding that the P72 variant shows increased interaction with p65 RelA in both mouse and human cells. That these three pieces of data are concordant between Hupki and human p53 lends credence to the premise that human polymorphisms can be effectively modeled in the mouse, with relevant biological discoveries as the outcome.
Our data indicate that the codon 72 polymorphism of p53 influences the p53-mediated inflammatory response. The role of p53 in innate immunity and the inflammatory response is now well established (6, 28, 44) and, importantly, is evolutionarily conserved (11). Whereas the impact of the codon 72 polymorphism on cancer risk appears to be minor, there are compelling examples in the literature of an effect of this polymorphism on diseases associated with inflammation. For example, studies of human ulcerative colitis (UC) indicate that there is a significant association of the P72 allele with UC (47), with the clinical course and duration of UC (49), and with the risk of UC-associated colorectal cancer (9). Likewise, significant associations between this polymorphism and the incidence and severity of type II diabetes (12) and rheumatoid arthritis (25) have been noted, two diseases whose severity is associated with increased inflammation. Infection and chronic inflammation are known to contribute to increased cancer risk, and this may explain part of the increased cancer risk seen previously for the P72 variant. These findings suggest that when analyzing the potential impact of this polymorphism on cancer risk, it may be most informative to analyze inflammation-associated cancers.
P72 is the ancestral allele carried by the p53 gene, and it is believed that the R72 allele arose some 30,000 to 50,000 years ago (16). It is unclear why the codon 72 polymorphism displays a geographical bias in the distribution of alleles, with P72 apparently selected for at the equator and R72 selected for in more northern latitudes. One possibility might be that the increased innate immune function associated with the P72 allele is selected for near the equator because immune challenge is greater there. In support of this notion, the P72 variant is generally associated with longevity, even following noncancerous illness (30, 48). The strong selection for the R72 allele in northern latitudes remains to be explained. Levine and colleagues have reported that the R72 allele demonstrates a 2-fold increased ability to transactivate LIF, a cytokine necessary for embryo implantation, thus raising the possibility that the selection for the R72 allele involves an impact on fecundity (14, 18). Along these lines, while the ability to fight infection is aided by a robust innate immune response, reproductive success requires a more tolerant immune response so that the fetus is not affected. In support of this, high inflammatory cytokine profiles are associated with decreased fecundity, and a reduced innate immune response is associated with increased fecundity (53). An alternative hypothesis is that the abilities of p53 and NF-κB to play opposing roles in the control of metabolism explains the geographic distribution of these alleles (22, 27).
One reason to model the p53 polymorphism in mouse was because of the possibility that the influence of this polymorphism on apoptosis might be tissue specific. Cell line-based studies suggest that the R72 allele has superior proapoptotic function in human tumor cell lines (8). Recently, a study by Zhu and colleagues described another mouse model for the codon 72 polymorphism; this study indicates that the R72 variant induces increased apoptosis in MEFs and in the small intestines of mice following ionizing radiation (58). We also have found that the R72 variant is associated with increased apoptosis in these tissues (data not shown), along with decreased apoptosis in the thymus. Such tissue-specific influences of this polymorphism on apoptosis may explain why human studies have been inconclusive regarding the role of this polymorphism on cancer risk.
The present study represents the first unbiased analysis of differences in transcriptional potential between P72 and R72 variants. Somewhat surprisingly, while others have found that the R72 variant shows increased transactivation of proapoptotic genes like Perp, Puma (Bbc3), and Noxa (Pmaip1) (16, 58), we find no evidence for increased transactivation of these genes in R72 thymocytes, MEFs, or normal human fibroblasts (this study; M. E. Murphy, unpublished data). The reasons for these discrepant findings are not presently clear. One explanation may be that these studies were done in cells that expressed supraphysiological levels of p53, while our Hupki mice maintain normal levels and regulation of this protein. In our model, we find that only a small subset of p53 target genes, the majority of which appear to contain binding sites for both p53 and NF-κB, show increased transactivation by P72. Interestingly, we find that NF-κB is required for the ability of p53 to bind to the promoters of caspase 4/11 and Gdf-15, suggesting that these two transcription factors may function cooperatively in the transactivation of this subset of target genes. Similar findings have been described previously for p53 and the p52 subunit of NF-κB (39). It remains to be determined if the cooperation between these two transcription factors is stress specific or cell type specific.
There are important clinical implications for this work. The codon 72 polymorphism shows a significant ethnic bias in North American, with the P72 allele significantly more prevalent in African-Americans than in Caucasian-Americans (41). Therefore, study of the codon 72 polymorphism of p53 has the potential to aid in our efforts to understand health disparities in African-Americans. For example, it is not understood why African-American women typically have a poorer prognosis for breast cancer or why African-American men have an increased incidence of multiple myeloma. Additionally, African-Americans show increased an incidence and severity of certain diseases associated with inflammation, including type II diabetes, heart disease, and obesity. It is anticipated that the Hupki model will be an invaluable preclinical tool to address such issues.
Acknowledgments
We thank Donna George and Steven McMahon for critical reading of the manuscript and members of the Murphy lab for valuable input. We thank Glenn Rall, Dave Wiest, and Rugang Zhang for guidance with the MEF, thymocyte, and senescence assays, respectively. We thank Steven McMahon (Kimmel Cancer Center, Thomas Jefferson University) for providing Tet-inducible P72 and R72 H1299 cells. We thank David Johnson for communication of results prior to publication. We are grateful to Emmanuelle Nicolas for QPCR expertise, and we acknowledge the support of the Laboratory Animal Facility, DNA Microarray, Biostatistics, Immunohistochemistry, and Genotyping Facilities at Fox Chase Cancer Center.
We acknowledge funding received from the National Institutes of Health. We acknowledge core budget support received from the German Cancer Research Center. This project was funded in part by a grant from the Pennsylvania Department of Health. We declare that no conflicts of interest exist.
The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions found in this paper.
A.K.F designed the research, performed experiments, analyzed data, and cowrote the paper. J.I.-J.L., T.N., and M.H. designed the research, performed experiments, and analyzed data. K.D. and Y.Z. provided analysis of microarray data. A.K.-S. provided analysis of immunohistochemistry. M.E.M. designed the research, analyzed data, and cowrote the paper.
Footnotes
Published ahead of print on 18 January 2011.
REFERENCES
- 1.Barre, B., and N. D. Perkins. 2010. The Skp2 promoter integrates signaling through the NF-kappaB, p53, and Akt/GSK3beta pathways to regulate autophagy and apoptosis. Mol. Cell 38:524-538. [DOI] [PubMed] [Google Scholar]
- 2.Beckman, G., et al. 1994. Is p53 polymorphism maintained by natural selection? Hum. Hered. 44:266-270. [DOI] [PubMed] [Google Scholar]
- 3.Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185-193. [DOI] [PubMed] [Google Scholar]
- 4.Chipuk, J. E., and D. R. Green. 2006. Dissecting p53-dependent apoptosis. Cell Death Differ. 13:994-1002. [DOI] [PubMed] [Google Scholar]
- 5.den Reijer, P. M., A. B. Maier, R. G. Westendorp, and D. van Heemst. 2008. Influence of the TP53 codon 72 polymorphism on the cellular responses to X-irradiation in fibroblasts from nonagenarians. Mech. Ageing Dev. 129:175-182. [DOI] [PubMed] [Google Scholar]
- 6.Dharel, N., et al. 2008. Potential contribution of tumor suppressor p53 in the host defense against hepatitis C virus. Hepatology 47:1136-1149. [DOI] [PubMed] [Google Scholar]
- 7.Donehower, L. A., et al. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356:215-221. [DOI] [PubMed] [Google Scholar]
- 8.Dumont, P., J. I. Leu, A. C. Della Pietra III, D. L. George, and M. Murphy. 2003. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 33:357-365. [DOI] [PubMed] [Google Scholar]
- 9.Eren, F., et al. 2010. R72P polymorphism of TP53 in ulcerative colitis patients is associated with the incidence of colectomy, use of steroids and the presence of a positive family history. Pathol. Oncol. Res. 16:503-508. [DOI] [PubMed] [Google Scholar]
- 10.Fang, S., et al. 2010. Effects of MDM2, MDM4 and TP53 codon 72 polymorphisms on cancer risk in a cohort study of carriers of TP53 germline mutations. PLoS One 5:e10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuhrman, L. E., A. K. Goel, J. Smith, K. V. Shianna, and A. Aballay. 2009. Nucleolar proteins suppress Caenorhabditis elegans innate immunity by inhibiting p53/CEP-1. PLoS Genet. 5:e1000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaulton, K. J., et al. 2008. Comprehensive association study of type 2 diabetes and related quantitative traits with 222 candidate genes. Diabetes 57:3136-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gentleman, R. C., et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu, W., Z. Feng, A. K. Teresky, and A. J. Levine. 2007. p53 regulates maternal reproduction through LIF. Nature 450:721-724. [DOI] [PubMed] [Google Scholar]
- 15.Ikeda, A., et al. 2000. p300/CBP-dependent and -independent transcriptional interference between NF-kappaB RelA and p53. Biochem. Biophys. Res. Commun. 272:375-379. [DOI] [PubMed] [Google Scholar]
- 16.Jeong, B. S., W. Hu, V. Belyi, R. Rabadan, and A. J. Levine. 2010. Differential levels of transcription of p53-regulated genes by the arginine/proline polymorphism: p53 with arginine at codon 72 favors apoptosis. FASEB J. 24:1347-1353. [DOI] [PubMed] [Google Scholar]
- 17.Jeong, S. J., M. Radonovich, J. N. Brady, and C. A. Pise-Masison. 2004. HTLV-I Tax induces a novel interaction between p65/RelA and p53 that results in inhibition of p53 transcriptional activity. Blood 104:1490-1497. [DOI] [PubMed] [Google Scholar]
- 18.Kang, H. J., et al. 2009. Single-nucleotide polymorphisms in the p53 pathway regulate fertility in humans. Proc. Natl. Acad. Sci. U. S. A. 106:9761-9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang, S. J., et al. 2000. Dual role of caspase-11 in mediating activation of caspase-1 and caspase-3 under pathological conditions. J. Cell Biol. 149:613-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang, S. J., S. Wang, K. Kuida, and J. Yuan. 2002. Distinct downstream pathways of caspase-11 in regulating apoptosis and cytokine maturation during septic shock response. Cell Death Differ. 9:1115-1125. [DOI] [PubMed] [Google Scholar]
- 21.Kashatus, D., P. Cogswell, and A. S. Baldwin. 2006. Expression of the Bcl-3 proto-oncogene suppresses p53 activation. Genes Dev. 20:225-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawauchi, K., K. Araki, K. Tobiume, and N. Tanaka. 2008. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat. Cell Biol. 10:611-618. [DOI] [PubMed] [Google Scholar]
- 23.Komarova, E. A., et al. 2005. p53 is a suppressor of inflammatory response in mice. FASEB J. 19:1030-1032. [DOI] [PubMed] [Google Scholar]
- 24.Luo, J. L., et al. 2001. Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene 20:320-328. [DOI] [PubMed] [Google Scholar]
- 25.Macchioni, P., et al. 2007. The codon 72 polymorphic variants of p53 in Italian rheumatoid arthritis patients. Clin. Exp. Rheumatol. 25:416-421. [PubMed] [Google Scholar]
- 26.Meylan, E., et al. 2009. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature 462:104-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minamino, T., et al. 2009. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat. Med. 15:1082-1087. [DOI] [PubMed] [Google Scholar]
- 28.Munoz-Fontela, C., et al. 2008. Transcriptional role of p53 in interferon-mediated antiviral immunity. J. Exp. Med. 205:1929-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ongusaha, P. P., et al. 2006. RhoE is a pro-survival p53 target gene that inhibits ROCK I-mediated apoptosis in response to genotoxic stress. Curr. Biol. 16:2466-2472. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Orsted, D. D., S. E. Bojesen, A. Tybjaerg-Hansen, and B. G. Nordestgaard. 2007. Tumor suppressor p53 Arg72Pro polymorphism and longevity, cancer survival, and risk of cancer in the general population. J. Exp. Med. 204:1295-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osada, M., et al. 2007. A p53-type response element in the GDF15 promoter confers high specificity for p53 activation. Biochem. Biophys. Res. Commun. 354:913-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Post, S. M., et al. 2010. p53-dependent senescence delays Emu-myc-induced B-cell lymphomagenesis. Oncogene 29:1260-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravi, R., et al. 1998. p53-mediated repression of nuclear factor-kappaB RelA via the transcriptional integrator p300. Cancer Res. 58:4531-4536. [PubMed] [Google Scholar]
- 34.Reinbold, M., et al. 2008. Common tumour p53 mutations in immortalized cells from Hupki mice heterozygous at codon 72. Oncogene 27:2788-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan, K. M., M. K. Ernst, N. R. Rice, and K. H. Vousden. 2000. Role of NF-kappaB in p53-mediated programmed cell death. Nature 404:892-897. [DOI] [PubMed] [Google Scholar]
- 36.Salvioli, S., et al. 2005. p53 codon 72 alleles influence the response to anticancer drugs in cells from aged people by regulating the cell cycle inhibitor p21WAF1. Cell Cycle 4:1264-1271. [DOI] [PubMed] [Google Scholar]
- 37.Schmitt, C. A., R. R. Wallace-Brodeur, C. T. Rosenthal, M. E. McCurrach, and S. W. Lowe. 2000. DNA damage responses and chemosensitivity in the E mu-myc mouse lymphoma model. Cold Spring Harb. Symp. Quant. Biol. 65:499-510. [DOI] [PubMed] [Google Scholar]
- 38.Schneider, G., et al. 2010. Cross talk between stimulated NF-kappaB and the tumor suppressor p53. Oncogene 29:2795-2806. [DOI] [PubMed] [Google Scholar]
- 39.Schumm, K., S. Rocha, J. Caamano, and N. D. Perkins. 2006. Regulation of p53 tumour suppressor target gene expression by the p52 NF-kappaB subunit. EMBO J. 25:4820-4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi, H., et al. 2009. Winter temperature and UV are tightly linked to genetic changes in the p53 tumor suppressor pathway in Eastern Asia. Am. J. Hum. Genet. 84:534-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sjalander, A., R. Birgander, A. Kivela, and G. Beckman. 1995. p53 polymorphisms and haplotypes in different ethnic groups. Hum. Hered. 45:144-149. [DOI] [PubMed] [Google Scholar]
- 42.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. [DOI] [PubMed] [Google Scholar]
- 43.Sullivan, A., et al. 2004. Polymorphism in wild-type p53 modulates response to chemotherapy in vitro and in vivo. Oncogene 23:3328-3337. [DOI] [PubMed] [Google Scholar]
- 44.Takaoka, A., et al. 2003. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature 424:516-523. [DOI] [PubMed] [Google Scholar]
- 45.Tergaonkar, V., M. Pando, O. Vafa, G. Wahl, and I. Verma. 2002. p53 stabilization is decreased upon NFkappaB activation: a role for NFkappaB in acquisition of resistance to chemotherapy. Cancer Cell 1:493-503. [DOI] [PubMed] [Google Scholar]
- 46.Thomas, M., et al. 1999. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol. Cell. Biol. 19:1092-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaji, S., Z. Salehi, and K. Aminian. 2010. Association of p53 codon 72 genetic polymorphism with the risk of ulcerative colitis in northern Iran. Int. J. Colorectal Dis. 26:235-238. [DOI] [PubMed] [Google Scholar]
- 48.van Heemst, D., et al. 2005. Variation in the human TP53 gene affects old age survival and cancer mortality. Exp. Gerontol. 40:11-15. [DOI] [PubMed] [Google Scholar]
- 49.Vietri, M. T., et al. 2007. p53 codon 72 polymorphism in patients affected with ulcerative colitis. J. Gastroenterol. 42:456-460. [DOI] [PubMed] [Google Scholar]
- 50.Wadgaonkar, R., et al. 1999. CREB-binding protein is a nuclear integrator of nuclear factor-kappaB and p53 signaling. J. Biol. Chem. 274:1879-1882. [DOI] [PubMed] [Google Scholar]
- 51.Wang, S., et al. 1998. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell 92:501-509. [DOI] [PubMed] [Google Scholar]
- 52.Webster, G. A., and N. D. Perkins. 1999. Transcriptional cross talk between NF-kappaB and p53. Mol. Cell. Biol. 19:3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westendorp, R. G. 2004. Are we becoming less disposable? EMBO Rep. 5:2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whibley, C., et al. 2010. Wild-type and Hupki (human p53 knock-in) murine embryonic fibroblasts: p53/ARF pathway disruption in spontaneous escape from senescence. J. Biol. Chem. 285:11326-11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whibley, C., P. D. Pharoah, and M. Hollstein. 2009. p53 polymorphisms: cancer implications. Nat. Rev. Cancer 9:95-107. [DOI] [PubMed] [Google Scholar]
- 56.Xue, W., et al. 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445:656-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang, H., Z. Filipovic, D. Brown, S. N. Breit, and L. T. Vassilev. 2003. Macrophage inhibitory cytokine-1: a novel biomarker for p53 pathway activation. Mol. Cancer Ther. 2:1023-1029. [PubMed] [Google Scholar]
- 58.Zhu, F., et al. 2010. Mouse models for the p53 R72P polymorphism mimic human phenotypes. Cancer Res. 70:5851-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]