Abstract
Spt2 is a chromatin component with roles in transcription and posttranscriptional regulation. Recently, we found that Spt2 travels with RNA polymerase II (RNAP II), is involved in elongation, and plays important roles in chromatin modulations associated with this process. In this work, we dissect the function of Spt2 in the repression of SER3. This gene is repressed by a transcription interference mechanism involving the transcription of an adjacent intergenic region, SRG1, that leads to the production of a noncoding RNA (ncRNA). We find that Spt2 and Spt6 are required for the repression of SER3 by SRG1 transcription. Intriguingly, we demonstrate that these effects are not mediated through modulations of the SRG1 transcription rate. Instead, we show that the SRG1 region overlapping the SER3 promoter is occluded by randomly positioned nucleosomes that are deposited behind RNAP II transcribing SRG1 and that their deposition is dependent on the presence of Spt2. Our data indicate that Spt2 is required for the major chromatin deposition pathway that uses old histones to refold nucleosomes in the wake of RNAP II at the SRG1-SER3 locus. Altogether, these observations suggest a new mechanism of repression by ncRNA transcription involving a repressive nucleosomal structure produced by an Spt2-dependent pathway following RNAP II passage.
In eukaryotes, modulation of the chromatin structure is a key aspect of important processes involving DNA. The basic unit of chromatin structure is the nucleosome, which consists of 146 bp of DNA wrapped around an octamer of histones (20). In addition to histones, many nonhistone proteins play important roles in regulating chromatin structure and chromatin-related processes (41). In Saccharomyces cerevisiae, one such nonhistone chromatin component is the HMG-like protein Spt2/Sin1, which was first identified genetically, by analysis of secondary mutations that suppress Ty and δ insertion mutations (spt2 for “suppressor of Ty 2”), in the HIS4 promoter (43).The sequence identity of Spt2 to HMG proteins is weak. However, like these proteins, it binds DNA nonspecifically and has significant affinity to four-way junction DNA, a structure similar to that found at the entrance/exit point of DNA from a nucleosome (25, 46). Several studies indicated that this factor could play a negative role in transcription initiation. Indeed, Spt2 was identified in a second screen for mutations that suppress the loss of the Swi/Snf chromatin remodeling complex and was called SIN1 (39). In addition, mutations in SPT2 suppress defects caused by mutations of SAGA histone acetyltransferase complex components (27, 31) and by deletion mutations in the RNA polymerase II (RNAP II) largest subunit gene (RPB1) (28). Finally, spt2 mutations have been shown to derepress the heat shock SSA3 gene (2).
While these mutant phenotypes suggested a negative role for Spt2 in transcription initiation, our recent work showed that it has an important function in transcription elongation (24). We demonstrated that Spt2 plays a significant role in the maintenance of proper chromatin structure over transcribed regions of the two active genes PMA1 and GAL1. Similarly to other elongation factors, it is required for the repression of transcription from the FLO8 cryptic promoter within its coding region (24). This factor is also involved in 3′-end processing of RNA and has been shown to specifically affect the polyadenylation of mRNA (11). Finally, in addition to its role in transcription, the spt2Δ mutation enhances recombination where transcription is active and causes defects in chromosome segregation and stability (16, 24, 38). Collectively, these observations suggest that Spt2p protects genome integrity at transcriptionally active regions, presumably by maintaining chromatin structure.
Global analyses of Spt2 localization showed that it is generally associated with coding regions of actively transcribed genes (24). However, a few exceptions have been observed, and among these infrequent Spt2 localizations is the SRG1-SER3 intergenic region. Interestingly, the SER3 gene is regulated by a transcription interference mechanism involving the transcription of a noncoding RNA (ncRNA) (21), produced from the intergenic region (SRG1) where Spt2 is localized. Noncoding RNAs were shown to be major players in gene expression regulation. They are produced by transcription across entire genomes, including intergenic regions, and regulate gene expression by different mechanisms, including RNA interference (RNAi)-mediated pathways of gene repression (10). However, production of some ncRNAs, as is the case for the ncRNA SRG1, regulates gene expression in cis. Indeed, recent observations indicated that, rather than the ncRNA product itself, it was the act of transcription and its associated processes that were important for the regulation of adjacent genes. Moreover, a few studies showed that the chromatin modulations associated with transcription of ncDNA play a major regulatory role. The activation of the Schizosaccharomyces pombe fbp1+ gene requires displacement by ncRNA transcription of key nucleosomes at the fbp1+ promoter (12). Similarly, antisense transcription enhances PHO5 activation by facilitating nucleosome displacement from the PHO5 promoter (40). Other studies have highlighted the roles of specific chromatin marks induced by the transcription of ncRNAs in the regulation of adjacent genes (reviewed in reference 10). Specifically, the histone posttranslational modifications directed to GAL1 and GAL10 regulatory regions by transcription of the ucut Gal ncRNA were shown to directly affect the expression of these genes (13, 30).
In the SRG1-SER3 system, the act of transcribing SRG1, rather than the ncRNA product, mediates regulation of the SER3 gene (21). The current model posits that in serine-rich medium SRG1 transcription interferes with the downstream SER3 promoter, thereby blocking SER3 expression and unnecessary serine biosynthesis (see Fig. 1 A) (23, 37). Although the specific molecular mechanism of transcription interference is unknown, several models were proposed, including promoter occlusion by the transcription machinery, collisions between RNAP II complexes, and promoter competition (37). In the case of the SRG1-SER3 regulatory system, the current preferred model postulates that the passage of RNA polymerase II transcribing SRG1 ncDNA through the SER3 promoter inhibits the assembly of the preinitiation complex and represses SER3 transcription (21). Importantly, in the absence of SRG1 ncRNA, Spt2 is completely delocalized from the intergenic region, suggesting a tight association between the regulation by SRG1 ncRNA and Spt2 targeting to the intergenic DNA (24). Moreover, deletion of the SPT2 gene affects dramatically the transcription of SER3, apparently by disturbing the regulation by transcription interference associated with SRG1 ncRNA production (24).
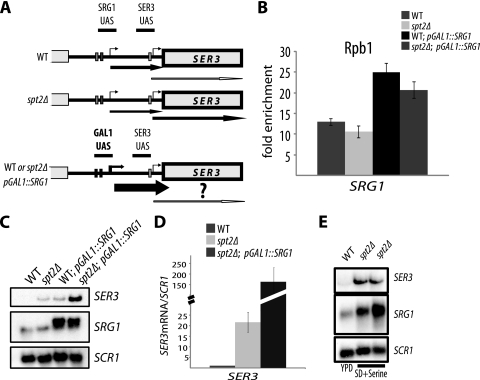
FIG. 1.
Deletion of the SPT2 gene affects SER3 regulation and reduces ncRNA SRG1 levels. (A) Diagram explaining the complex regulation of SER3 by the intergenic transcription of the ncDNA SRG1. In the presence of high levels of serine in the medium, ncDNA SRG1 is actively transcribed and SER3 is inhibited by transcription interference. In the absence of serine, SRG1 ncDNA transcription activity is reduced and SER3 is then induced. (B) spt2Δ mutation affects the transcript levels of SER3 and ncDNA SRG1. Wild-type (WT) (YAN1034) and spt2Δ (YAN1035) strains were grown in YPD at 30°C. Total RNA was extracted and analyzed by Northern blotting with a probe against SER3 and SRG1. SCR1 served as a loading control. (C and D) Quantification of the SRG1 ncRNA and SER3 mRNA by RT-qPCR. Total RNA analyzed by Northern blotting as shown in panel B was used to produce cDNA and quantified by qPCR. The relative level is the ratio of the indicated RNA to the SCR1 transcript level. All values shown are the average results with standard errors from three independent experiments. *, P < 0.05. (E) Association of RNAP II with ncDNA SRG1 is partially dependent on Spt2. Yeast cells from the wild-type (YAN1034) or spt2Δ (YAN1035) strains were grown in YPD medium to mid-log phase and then cross-linked with 1% formaldehyde. Chromatin immunoprecipitations were performed using the 8WG16 antibody against Rpb1. The fold enrichment is the ratio of the percent immunoprecipitation (%IP) of the ncDNA SRG1 region to the %IP of the nontranscribed control region (NoORF). (F) Run-on assay showing that deletion of SPT2 results in a decrease of transcription activity at intergenic ncDNA SRG1. Diagram showing the different probes used in the run-on assay conducted on the SRG1-SER3 locus. Radiolabeled RNA extracted from wild-type (YAN1034) or spt2Δ (YAN1035) strains was hybridized on membranes containing immobilized probes spanning the SRG1-SER3 locus. The ACT1 probe was used as a control.
In this work, we analyzed the mechanism by which the elongation factor Spt2 modulates expression of the SER3 gene by ncRNA SRG1 transcription. We found that the change in ncRNA SRG1 transcription in the spt2Δ background cannot explain the loss of SER3 repression. Importantly, we show that Spt2 is involved in the shaping of a particular nucleosomal structure at the SRG1-SER3 locus. Our data indicate that nucleosomes are deposited specifically within the ncDNA SRG1 region, corresponding to the SER3 promoter regulatory elements, following the passage of RNAP II. Interestingly, this deposition is severely impaired by the spt2Δ mutation, suggesting that the loss of nucleosomes at this region could be the consequence of disrupted nucleosome deposition in the wake of transcription. Therefore, in contrast to previous observations and models involving nucleosome displacement, histone posttranslational modifications, and promoter occlusion by RNAP II, our data suggest a model in which specific nucleosomes, deposited by a transcription-dependent mechanism involving Spt2, play a central role in the repression by ncRNA transcription.
MATERIALS AND METHODS
S. cerevisiae strains, media, and genetic methods.
All S. cerevisiae strains (see Table ST1 at http://www.crc.ulaval.ca/nourani/supplemental_data) are isogenic to a GAL2 derivative of S288C. Strains (44) were constructed by standard methods, either by crosses or by transformation. The HIS3-pGAL1::SRG1 and HIS3-pGAL1::FMP27 alleles were constructed by replacing the promoter of the corresponding gene with the HIS3-pGAL1 cassette (1, 19). SPT15-3HA, marked with HIS3, was generated by integrating DNA encoding three copies of the HA epitope at the 3′ end of the gene (19). For experiments involving galactose induction, cells were grown to an optical density at 600 nm (OD600) of 0.5 in YP (1% yeast extract, 2% peptone) supplemented with 2% raffinose (YPraf). The cells were then centrifuged, resuspended in YP medium containing 2% galactose (YPgal), and grown for 2 h before being harvested. In the experiments involving glucose repression, cells were grown to an OD600 of 0.5 in YPgal; glucose was then added to reach a concentration of 4% in the medium. Efficient G1 arrest (at least 95%) of cells was achieved by adding 500 ng/ml of α-factor for 2 to 3 h. Sequences of all oligonucleotides used in strain constructions, Northern blotting, reverse transcription (RT)-PCR, run-on transcription, nucleosome scanning assays, and chromatin immunoprecipitation (ChIP) assays are available upon request.
Chromatin immunoprecipitation experiments.
Chromatin immunoprecipitation experiments were performed as previously described (22). For the immunoprecipitation of Spt2-13Myc, TBP-3HA, and Gal4, we used, respectively, the antibodies 9E10 anti-Myc (1 μl per immunoprecipitation; Covance), HA.11 anti-HA (1.5 μl per immunoprecipitation; Covance), and Gal4 (DBD):sc-577 (1 μl per immunoprecipitation; Santa Cruz Biotechnology). Immunoprecipitation of Rpb1 was performed using the 8WG16 anti-CTD antibody (2 μl per immunoprecipitation; Covance). The histone H3 immunoprecipitation was done using rabbit anti-H3 antibody (0.2 μl per immunoprecipitation; Abcam). The PCR amplification was performed with 1% of the precipitated material and 0.05% of the input DNA using the LightCycler 480 Sybr green I master kit from Roche.
RNA analyses.
Total RNA was isolated using the hot-phenol method (35). In Northern blot analyses, 20 to 40 μg of RNA were separated on a 1% agarose formaldehyde-MOPS gel and transferred to a nylon membrane. The SRG1, SER3, and SCR1 probes were amplified by PCR and radiolabeled by random priming. The SRG1, SER3, and SCR1 probes were also quantified by RT-quantitative PCR (qPCR). For that process, cDNAs were generated using the Invitrogene M-MLV reverse transcriptase kit and their levels were measured by real-time PCR using LightCycler 480 Sybr green I master kit purchased from Roche. The run-on experiments were conducted as described in reference 21.
Nucleosome scanning assay.
Nucleosome scanning experiments were performed using a method adapted from those previously described (3, 17, 42). Cells were grown to 2 × 107 to 3 × 107 cells/ml and treated with formaldehyde (2% final concentration) for 30 min at 30°C and then glycine (125 mM final concentration) for 10 min at room temperature. Formaldehyde-treated cells (1.2 × 109) were harvested by centrifugation, washed with Tris-buffered saline, and then incubated in ZDB buffer (50 mM Tris Cl, pH 7.5, 1 M sorbitol, 10 mM β-mercaptoethanol) containing 1.5 mg Zymolase 20T at 30°C for 30 min on a rocker platform. Spheroplasts were pelleted by low-speed centrifugation, gently washed with NP buffer (1 M sorbitol, 50 mM NaCl, 10 mM Tris Cl, pH 7.4, 5 mM MgCl2, 1 mM CaCl2, 0.075% NP-40, 1 mM β-mercaptoethanol, and 500 μM spermidine), and resuspended in 1.8 ml NP buffer. Samples were divided into six 300-μl aliquots that were then digested with 0, 1, 2.5, 5, 10, and 20 units of micrococcal nuclease (MNase) (Nuclease S7; Roche) for 45 min at 37°C. Digestions were stopped with 75 μl Stop buffer (5% SDS, 50 mM EDTA) and treated with 100 μg proteinase K for 12 to 16 h at 65°C. DNA was extracted by phenol-chloroform using PLG-H tubes (5 Prime) and incubated with 50 μg RNase A for 1 h at 37°C. DNA was reextracted with phenol-chloroform, precipitated with an equal volume of isopropanol, washed with 100% ethanol, and resuspended in 100 μl TE. MNase digestions were evaluated by two methods. First, one-fifth of digested DNA was separated by gel electrophoresis. Second, previously characterized GAL1 promoter sequences (3, 8, 18), one within a positioned nucleosome (GAL1 NB) and a second in an adjacent region (GAL1 NUB) that is rapidly digested by MNase, were amplified by qPCR from MNase-treated and untreated samples. The MNase concentration that resulted in mostly mononucleosome-sized DNA with a GAL1 NUB/NB ratio of <15% was subjected to further qPCR using tiled SER3 primer pairs that amplify 38 unique SER3 sequences that range from 90 to 114 bp in size, with an average overlap of 69 bp between sequences. For each SER3 primer set, the amount of protected template was calculated as a ratio between amounts of MNase-digested and undigested samples and then normalized to the amount of protected GAL1 NB template. All nucleosome scanning assays were done in triplicate using at least two independent strains as indicated in the figure legends. The qPCR data were obtained using an ABI 7300 real-time PCR system with Sybr green reagents (Fermentas). All calculations were performed using Pfaffl methodology for relative quantitation of real-time PCR (29).
RESULTS
Spt2 is required for normal SER3 repression and affects ncRNA SRG1 production.
The SER3 gene is regulated by a complex mechanism involving noncoding RNA SRG1 (ncRNA SRG1) transcription (21). As indicated in Fig. 1A, in the presence of serine in the medium, the SER3 gene is inhibited by the active transcription of the SRG1 gene. This gene produces a noncoding RNA and its transcription interferes with that of SER3. In the absence of serine from the medium, the SRG1 gene is repressed and SER3 is in turn rapidly activated (23). Spt2 is recruited to the ncDNA SRG1, and its association to this location is tightly linked to the presence of the ncRNA SRG1 (24). We decided to further study the role of the elongation factor Spt2 in the complex regulation of SER3. Since SER3 regulation is tightly associated with the transcription of the ncRNA SRG1, we wanted to know the effect of SPT2 deletion on the levels of SER3 mRNA and SRG1 ncRNA. For that, we extracted total RNA from a wild-type or spt2Δ strain and performed Northern blot analyses. We observed a high level of SER3 transcript in the spt2Δ strain, while no detectable amount of such transcript was observed in the wild-type cells (Fig. 1B). This is consistent with our previous observations (24). Interestingly, although the SER3 mRNA level is dramatically increased in spt2Δ cells, the amount of ncRNA SRG1 appears only slightly reduced in this mutant (Fig. 1B). We decided to quantify precisely the level of both SER3 and SRG1 transcripts by RT-qPCR. As indicated in Fig. 1C and D, the deletion of SPT2 resulted in a dramatic increase of SER3 mRNA (30- to 40-fold induction) but only a 20% reduction of the noncoding SRG1 RNA steady-state level.
Recent observations showed that Spt2 plays a significant role in the posttranscriptional events and that it is necessary for the polyadenylation of RNAP II transcripts (11). Therefore, it is possible that the slight reduction of ncRNA SRG1 is the consequence of a posttranscriptional defect linked to the SPT2 deletion. To explore the possible role of Spt2 in the stability of the noncoding SRG1 RNA transcripts, we conducted an experiment to measure the half-life of the ncRNA SRG1 in wild-type or spt2Δ strains (see Fig. S1 at http://www.crc.ulaval.ca/nourani/supplemental_data). Our data indicate that SRG1 ncRNA stability is not affected by the loss of Spt2.
Our results show that SRG1 ncRNA levels are slightly affected by the loss of Spt2. Importantly, this effect is not associated with the posttranscriptional function of Spt2, since no defect in the stability of this transcript was observed (see Fig. S1 at the URL above). This finding suggests that a SRG1 ncRNA drop in the spt2Δ strain could be the consequence of a decrease in transcription activity. We wanted to analyze, using different methods, the transcription activity at the SRG1 region in wild-type and spt2Δ strains. For that, we first measured the transcription activity in different regions of the SRG1-SER3 locus using run-on assays. Radiolabeled RNA was extracted from the wild-type strain or the spt2Δ mutant and blotted on membranes containing four probes covering the SRG1-SER3 locus and a control probe located in the ACT1 gene (Fig. 1F). Not surprisingly, as a consequence of the SER3 derepression in the spt2Δ strain, we detected a higher signal at the SER3 open reading frame (probe 4, Fig. 1F) in this mutant. Importantly, deletion of the SPT2 gene is associated with a reduction of probe 1, 2, and 3 run-on signals, indicating a lower transcription activity at the intergenic region (SRG1) in the absence of Spt2 (Fig. 1F).
In order to confirm our data, we extended our analysis and asked whether the reduction of transcription activity in the absence of Spt2 is associated with the reduction of the RNAP II level at SRG1. For that we measured the RNAP II occupancy in the wild-type or spt2Δ strain by Rpb1 chromatin immunoprecipitation assays (Fig. 1E). The deletion of SPT2 led to a decrease of RNAP II association at the 5′ region of SRG1, indicating a correlation between the transcriptional activity and the level of RNAP II at this location. Therefore, we concluded that Spt2 is necessary for normal association of the transcription machinery at this location.
A different hypothesis could explain the reduction in transcription activity at this location. Spt2 could be required for the first steps of initiation or during early elongation of transcription at the intergenic region SRG1. One of the most important events during initiation is the recruitment of TBP to the TATA box (32). We analyzed the recruitment of TBP-HA to both SRG1 TATA and SER3 TATA in the wild-type and spt2Δ strains (see Fig. S2 at http://www.crc.ulaval.ca/nourani/supplemental_data). Our experiment shows clearly that TBP-HA association at SRG1 TATA is similar in both strains, indicating that Spt2 loss affects transcription of ncDNA SRG1 at a post-TBP recruitment step.
Spt2 role on SRG1 transcription cannot explain the loss of SER3 repression.
The SER3 gene is repressed presumably by transcription interference mechanism, whereby passages of RNAP II transcribing the SRG1 intergenic region obstruct the SER3 promoter, inhibiting the interaction of activators and initiation factors with their respective binding sites (21). If the transcription interference, in this case, was achieved merely by maintaining a certain transcription rate, then it is possible that the small reduction in transcriptional activity observed at the intergenic region in the spt2Δ mutant can disturb the transcription interference and consequently compromise SER3 repression. We reasoned that, if this hypothesis was true, increasing the SRG1 ncRNA transcription in the spt2Δ mutant to wild-type level would suppress the phenotype associated with this mutation and restore SER3 repression. We therefore asked whether a higher level of transcriptional activity at the SRG1 intergenic region is capable of restoring SER3 repression in spt2Δ. To address this, we used an experimental system (Fig. 2 A) in which we replaced the SRG1 promoter by the strong inducible GAL1 promoter in the wild type and the spt2Δ mutant (pGAL1-SRG1) and induced transcription of SRG1. Our goal was to increase the transcription activity at the intergenic region in the spt2Δ mutant in order to reach levels equal to or higher than those found in wild-type cells. We first compared the level of transcription activity under inducing conditions in wild-type, spt2Δ, and spt2Δ pGAL1-SRG1 strains by analyzing RNAP II occupancy at the SRG1 intergenic region via Rpb1 ChIP assays (Fig. 2B). RNAP II occupancy at the SRG1 intergenic region in the spt2Δ pGAL1-SRG1 strain was significantly higher than that in wild-type or spt2Δ cells, where SRG1 is transcribed from its own promoter. This indicates that replacing the SRG1 promoter by that of GAL1, in spt2Δ cells, results in higher transcription activity at the SRG1 intergenic region. We next wanted to know whether this higher transcription activity correlated with higher ncRNA levels. Total RNA was extracted from wild-type, spt2Δ, and spt2Δ pGAL1-SRG1 cells grown under galactose-inducing conditions, and the ncRNA SRG1 levels were analyzed by Northern blotting (Fig. 2C). As expected, in the spt2Δ mutant, the GAL1 promoter produced a higher level of the ncRNA SRG1 transcript than the SRG1 promoter in both wild-type and spt2Δ mutant strains (compare lanes 1 and 2 to 4 in Fig. 2C). This observation was confirmed by RT-qPCR (see Fig. S4 at http://www.crc.ulaval.ca/nourani/supplemental_data). We conclude that the GAL1 promoter induces a higher transcription level at SRG1 in spt2Δ cells. Surprisingly, analyses of the SER3 transcript level by Northern blotting or RT-qPCR (Fig. 2C and D; also see Fig. S3 at the URL above) indicated that this higher level of transcription activity observed in spt2Δ pGAL1-SRG1 was not associated with repression of the SER3 gene. We observed instead that higher transcription activity at ncDNA SRG1 resulted in further derepression of SER3. Therefore, suppression of the SRG1 transcription defect associated with the spt2Δ mutation does not restore SER3 repression. We conclude that the reduction of transcription activity observed in spt2Δ cells cannot explain the failure of SER3 repression by the transcription interference mechanism. Interestingly, higher transcription activity in wild-type cells was also associated with SER3 activation (Fig. 2C, compare lanes 1 to 3). This indicates that high SRG1 transcription induced by the GAL1 promoter is not sufficient by itself for the repression of SER3.
FIG. 2.
Increasing the intergenic transcription of ncDNA SRG1 does not restore SER3 repression in the spt2Δ mutant. (A) Diagram showing the pGAL1-SRG1 construct. WT, wild type. (B) GAL1 promoter drives a higher level of transcription at ncDNA SRG1. Yeast cells from the wild-type strain (YAN1034), the spt2Δ mutant (YAN1035), wild-type pGAL1-SRG1 (YAN 1040), or the spt2Δ strain containing the pGAL1-SRG1 construct (YAN1039) were grown in YP galactose medium to mid-log phase and then cross-linked with 1% formaldehyde. Chromatin immunoprecipitations were performed using the 8WG16 antibody against Rpb1. The fold enrichment is the ratio of the percent immunoprecipitation (%IP) of ncDNA SRG1 region to the %IP of the nontranscribed control region (NoORF). The values shown represent the average results with standard errors from three independent experiments. (C) Overproduction of the ncRNA SRG1 in the spt2Δ strain containing the pGAL1-SRG1 construct is not associated with repression of SER3. Total RNA was extracted from cells of wild-type (YAN1034), spt2Δ (YAN1035), wild-type pGAL1-SRG1 (YAN 1040), and spt2Δ pGAL1-SRG1 (YAN1039) strains grown as indicated above. The RNA was analyzed by Northern blotting with probes against SER3 and SRG1. SCR1 served as a loading control. (D) Quantification of the SER3 mRNA by RT-qPCR. Total RNA analyzed by Northern blotting as shown in panel C was used to produce cDNA that was quantified by qPCR. The relative level is the ratio of the indicated SER3 mRNA to the SCR1 transcript level. The values shown represent the average results with standard errors from three independent experiments. (E) High ncRNA SRG1 levels in serine-rich media are not associated with SER3 repression in the spt2Δ mutant. Yeast cells from the wild-type (YAN1034) or spt2Δ (YAN1035) strains were grown, respectively, in YPD or SD medium supplemented with serine. Total RNA was extracted and analyzed by Northern blotting with probes against SER3 and SRG1. SCR1 served as a loading control. Lanes 2 and 3 are duplicates that represent results of two independent experiments.
Remarkable changes in intergenic SRG1 transcription are observed when different serine levels are present in the growth media (23). Therefore, we analyzed the SRG1 and SER3 transcript levels during a time course with different serine concentrations (see Fig. S5 at http://www.crc.ulaval.ca/nourani/supplemental_data). Our data clearly show that the absence of serine results in a decreased level of SRG1 and induction of SER3 transcription in the wild-type strain. When serine was added back, we observed a dramatic increase of SRG1 level that correlated with SER3 repression (see Fig. S5). Interestingly, in the spt2Δ mutant, the serine regulation of ncRNA SRG1 is similar to that in the wild-type strain. However, in this mutant SER3 is never completely repressed, even after a dramatic increase of the ncRNA level in the presence of serine (see Fig. S5). This observation is highlighted in Fig. 2E, where we compare conditions in which the level of ncRNA SRG1 is significantly higher in the spt2Δ strain than in the wild-type strain. Our Northern blot (Fig. 2E) and RT-qPCR (data not shown) analyses demonstrate that the levels of the SRG1 ncRNA are lower in wild-type cells in the presence of low levels of serine (YPD medium) than those in the spt2Δ mutant in the presence of high levels of serine (SD medium with added serine). However, in contrast to the level in the wild type, the higher level of the SRG1 ncRNA in spt2Δ cells is not associated with better repression of SER3. This indicates a disconnection between intergenic SRG1 transcription levels and SER3 regulation in the spt2Δ mutant and confirms that the small reduction of ncRNA transcription observed in these cells cannot explain by itself the loss of SER3 repression. The transcription of the SRG1 ncDNA alone is not able to prevent expression of SER3 when SPT2 is mutated. Conversely, the presence of Spt2 in wild-type cells is not by itself sufficient for repression of SER3 when the intergenic SRG1 region is not transcribed (21). Together, these observations indicate that both intergenic transcription and Spt2 are necessary for repression of SER3. Importantly, it also follows that the mere passage of RNAP II and production of ncRNA are not sufficient for silencing of SER3. Other transcription-associated events involving Spt2 must be required to achieve full repression of the downstream gene.
Mutation of the histone chaperone SPT6 has a similar defect in the repression of SER3.
Our present data show that SER3 regulation in spt2Δ cells is not linked to the modulation of ncRNA SRG1 transcription. Therefore, the mechanism used by the elongation factor Spt2 to repress SER3 remains unclear. Interestingly, we previously showed a functional link between the histone chaperone Spt6 and Spt2 (24). Moreover, we found that Spt6 is required for the recruitment of Spt2 to the transcribed regions of some active genes (24). To further understand how Spt2 regulates the SER3 gene, we asked if the recruitment of this factor to the SRG1 intergenic region is under the control of Spt6. To address this question, we analyzed the association of Spt2-13Myc with SRG1 by ChIP assays in the wild-type strain, the spt6-1004 strain, and a control untagged strain (Fig. 3 A). In the wild-type strain, the Spt2-13Myc signal at the SRG1 intergenic region was 10-fold higher than that of the untagged control, confirming a strong specific recruitment to SRG1. In contrast, the association of Spt2-13Myc with SRG1 in spt6-1004 was very weak, indicating that Spt6 plays an important role in the recruitment of Spt2 to the SRG1 intergenic region. We next asked if Spt6 recruitment to SRG1 is dependent on the Spt2 protein. As shown in Fig. 3B, deletion of SPT2 is associated with partial loss of Spt6-Flag occupancy at SRG1. We therefore conclude that Spt2 and Spt6 are dependent on each other for their recruitment to SRG1.
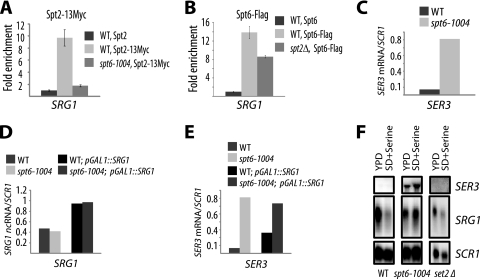
FIG. 3.
Spt6 regulates SER3 independently of SRG1 transcription level. (A) Recruitment of Spt2 to the ncDNA SRG1 is dependent on Spt6. ChIP assays of Spt2-13Myc were performed with chromatin extracted from wild-type (WT) (YAN1040), spt6-1004 (YAN1041), and untagged (YAN1042) strains. The fold enrichment is the ratio of the percent immunoprecipitation (%IP) of the ncDNA SRG1 region to the %IP of the nontranscribed control region (NoORF). The values shown represent the average results with standard errors from three independent experiments. (B) Recruitment of Spt6 to the ncDNA SRG1 is partially dependent on Spt2. ChIP assays of Spt6-Flag were performed with chromatin extracted from wild-type (YAN1057), spt2Δ (YAN1058), and untagged (YAN1042) strains. The fold enrichment is the ratio of %IP of the ncDNA SRG1 region to the %IP of the nontranscribed control region (NoORF). The values shown represent the average results with standard errors from three independent experiments. (C) SER3 is derepressed in the spt6-1004 mutant. Total RNA extracted from the wild-type (YAN1034) or the spt6-1004 (YAN1043) strain was used to produce cDNA that was subsequently quantified by qPCR. The SER3 mRNA relative level is the ratio of the SER3 mRNA to the SCR1 transcript level. The values shown represent the average results of two independent experiments. (D) GAL1 promoter increases the level of the ncDNA SRG1 transcripts in the spt6-1004 mutant. Total RNAs extracted from the wild-type (YAN1034), spt6-1004 (YAN1043), wild-type pGAL1-SRG1 (YAN 1040), and spt6-1004 pGAL1-SRG1 (YAN1044) strains were used in reverse transcription reactions to produce cDNA that was quantified by qPCR. The relative level is the ratio of SRG1 ncRNA to the SCR1 transcript level. The values represent the average results of two independent experiments. (E) Overproduction of the ncRNA SRG1 in the spt6-1004 strain does not lead to repression of SER3. The SER3 mRNA was quantified using the cDNA obtained in the experiment shown in panel D. The SER3 relative level is the ratio of SER3 mRNA to the SCR1 transcript level. (F) Deletion of the SET2 gene has no effect on SER3 repression. Yeast cells from the wild-type (YAN1034), spt2Δ (YAN1035), and set2Δ (YAN1051) strains were grown, respectively, in YPD or SD medium supplemented with serine. Total RNA was extracted and analyzed by Northern blotting with probes against SER3 and SRG1. SCR1 served as a loading control. All experiments using spt6-1004 strains and described in this figure were performed with cells grown at 30°C.
Given the role of Spt2 in the regulation of SER3 and the fact that its function depends on Spt6 integrity, it became tempting to speculate that similarly to Spt2, Spt6 could be involved in the regulation of SER3 by transcription of ncDNA SRG1. To test this, we first analyzed the SER3 transcript levels in wild-type and spt6-1004 strains by RT-qPCR and found a dramatic increase of the SER3 transcript in spt6-1004 cells (Fig. 3C). Under these conditions, the derepression of SER3 was also associated with a small reduction in intergenic SRG1 transcription, as evidenced by a decreased ncRNA level and a reduced RNAP II occupancy (data not shown). Interestingly, as observed for Spt2, artificially increasing SRG1 intergenic transcription in spt6-1004 cells by replacing the SRG1 promoter with that of GAL1 resulted in a higher transcription activity without restoring the expected repression of SER3 (Fig. 3D and E). Thus, similarly to Spt2, Spt6 plays a major role in the regulation of SER3 by ncRNA SRG1. The transcription of SRG1 alone is not able to prevent expression of SER3 when SPT2 or SPT6 is mutated. In addition, the presence of Spt2 or Spt6 in wild-type cells is not by itself sufficient for repression of SER3 when intergenic SRG1 region is not actively transcribed (21). Collectively, these observations indicate that in addition to RNAP II passages and production of ncRNA, other transcription-associated events involving Spt2 and the histone chaperone Spt6 must be required to achieve full repression of the downstream gene.
Histone H3 methylation is not involved in the regulation of SER3 by transcription of ncDNA SRG1.
Spt6 is a transcription elongation factor, mainly known to be involved in chromatin modulation associated with elongation (15). In addition to functioning together with Spt2, Spt6 controls the histone H3-lysine 36 methylation by the Set2 enzyme (4, 5, 45). Interestingly, a number of recent studies have reported a link between regulation by transcription of ncRNA and chromatin modifications, specifically histone H3 methylation (reviewed in reference 10). Indeed, regulation of GAL1 and GAL10 by the ucut Gal1-10 ncRNA requires methylation of histone H3-K36 and H3-K4 (13). Therefore, the phenotype observed in spt6-1004 could be associated with the known defects in histone H3-K36 methylation in this mutant (4, 5, 45). To address this possibility directly, we analyzed by Northern blotting the level of SER3 and SRG1 in wild-type, spt6-1004, and set2Δ strains in media containing low or high concentrations of serine (Fig. 3F). In both conditions, the SER3 transcript was not detected in wild-type strains, whereas appreciable amounts were visible in spt6-1004 cells. Importantly, no SER3 transcripts were observed in cells bearing the deletion of the SET2 gene. Moreover, we observed a similar regulation of SER3 and SRG1 in wild-type and set2Δ strains, while the SPT6 mutation resulted in constitutive transcription of SER3 (see Fig. S6 at http://www.crc.ulaval.ca/nourani/supplemental_data). Thus, the Spt6 function in regulating SER3 cannot be attributed to its role in methylation of H3-K36. In addition, we also analyzed the role of histone H3-lysine 4 methylation in SER3 regulation by measuring the SER3 mRNA levels by RT-qPCR in wild-type cells and spt2Δ and set1Δ mutants (see Fig. S7 at the URL above). This experiment showed that loss of H3-K4 methylation had no significant impact on SER3 repression. Our observations clearly indicate that in contrast to regulation of GAL1 and GAL10 by ncRNA, histone H3-K4 and H3-K36 methylation by Set1 and Set2, respectively, are not required for normal SER3 repression by ncRNA SRG1.
Spt2 is required for maintaining proper nucleosomal structure at the SRG1 intergenic region.
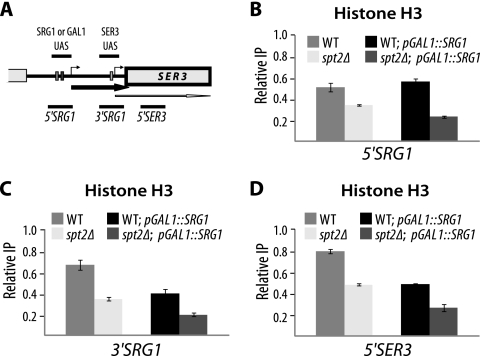
Mutations in SPT2 or SPT6 result in spurious transcription from cryptic promoters located within coding regions (15, 24). Presumably, in the absence of normal Spt2 activity, the chromatin structure modified by the passage of transcription machinery is not properly restored, leading to the derepression of cryptic promoters. Importantly, Spt2 was shown to control the level of histone H3 at transcribed regions of GAL1 and PMA1 genes (24). This led us to imagine that the role of Spt2 in regulation of SER3 could have a link with its function in modulating chromatin structure at transcribed regions. To address this question, we first determined whether the deletion of SPT2 gene affects the histone H3 levels at the SRG1-SER3 locus (Fig. 4 A). To this end, we conducted histone H3 ChIP assays at three different locations around SRG1-SER3 in wild-type and spt2Δ strains. As shown in Fig. 4B, C, and D, H3 occupancy is significantly reduced at this locus in Spt2-deficient cells. This result indicates that chromatin structure within the SRG1 transcribed region corresponding to the SER3 promoter could be significantly affected by the loss of Spt2. Interestingly, increasing transcription activity at SRG1 by inserting the strong GAL1 promoter does not suppress the loss of histone H3 associated with mutation of SPT2 at the SRG1 transcribed region (see Fig. 4B, C, and D). Therefore, the significant histone H3 occupancy change observed in spt2Δ cells is independent of the transcription rate. This important observation indicates clearly that spt2Δ mutation effect on SRG1-SER3 chromatin structure is not mediated through the modulation of the SRG1 transcription activity. It should be noted that inserting the GAL1 promoter at SRG1 in wild-type cells resulted in the reduction of histone H3 level in the 3′SRG1 and 5′SER3 regions (see Fig. 4C and D). This effect on nucleosome occupancy at SRG1 could explain the SER3 derepression observed in wild-type cells and reported in Fig. 2C.
FIG. 4.
Spt2 controls the histone H3 occupancy at the SRG1-SER3 locus. (A) Diagram showing the SRG1-SER3 regions analyzed by histone H3 ChIP. (B, C, and D) Spt2 is required to maintain normal histone H3 level at the SRG1 3′-end region corresponding to the SER3 promoter. Histone H3 ChIP assays were conducted using chromatin extracted from wild-type (WT) (YAN1034) and spt2Δ (YAN1035) strains with or without the pGAL1-SRG1 construct. For each strain, the value shown represents the ratio of the percent immunoprecipitation (%IP) at the indicated region to the %IP at NoORF. The values shown represent the average results with standard errors from three to six independent experiments.
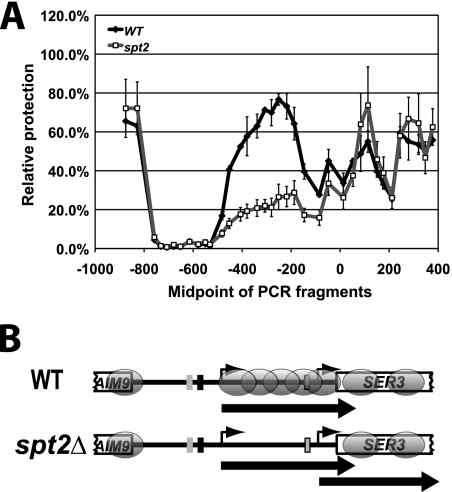
To further study the role of Spt2 in maintaining the nucleosomal structure of SRG1-SER3, we determined precise nucleosome positions at this locus by a nucleosome scanning assay similar to those previously described (3, 17, 42). Using this approach first in wild-type cells, we identified two sharp peaks of MNAse protection within the 5′ end of the SER3 open reading frame, indicating the presence of two well-positioned nucleosomes (Fig. 5 A, black line, and Fig. 5B). In addition, we observed a broad region of MNase protection that overlaps the SRG1 transcribed region, which is consistent with the presence of nucleosomes that are randomly positioned. Importantly, this broad region of MNase protection is greatly reduced in spt2Δ cells (Fig. 5A, gray line), indicating a loss of nucleosomes over the SRG1 transcribed region. Taken together with our histone H3 ChIP results, our data suggest that there is a significant disruption of nucleosomal structure at SRG1 in spt2Δ cells. Moreover, our data also show that this effect is independent of the level of SRG1 transcription.
FIG. 5.
Spt2 controls the nucleosomal structure at the ncDNA SRG1. (A) spt2Δ mutation is associated with loss of specific nuclesomes at the ncDNA SRG1 region corresponding to the SER3 promoter. A nucleosome scanning assay was performed on wild-type (WT) (OY8 or KY766) and spt2Δ (YAN16 or OY8spt2) cells that were grown in YPD (SER3 repressed) at 30°C. Using qPCR, the relative MNase protection of each SER3 template was calculated as a ratio to that of a GAL1 promoter template (GAL1 NB) found within a well-positioned nucleosome in the GAL1-10 promoter. Each point on the graph shows the mean ± standard error of the mean result from three independent experiments that are plotted at the midpoint of each PCR product. (B) Diagram of the SER3 locus showing nucleosome positions (ovals) extrapolated from the nucleosome scanning experiments.
Mutations in SPT2 and SPT6 compromise SRG1-mediated inhibition of activator binding.
Spt2 is required for normal positioning of nucleosomes in the SRG1 transcribed region that corresponds to the SER3 promoter (here called SRG1 3′). In the absence of these nucleosomes, it is possible that transcription activators governing the induction of SER3 can freely bind their target sites. Because the SER3 regulators are not known, we decided to use a strain (see Fig. 6 A) in which the SRG1 promoter was inserted in front of the GAL7 promoter region (21). It was previously shown using this construction that SRG1 transcription reduces Gal4 binding to the GAL7 upstream activation sequence (UAS) (21). We asked if SPT2 or SPT6 mutations impaired the inhibition of Gal4 binding by SRG1 transcription. Therefore, we performed Gal4 ChIP assays in wild-type, spt6-1004, and spt2Δ strains containing the SRG1 promoter in front of the GAL7 UAS region. As shown in Fig. 6B, Gal4 binding is significantly increased in the spt2Δ and spt6-1004 mutants. Thus, SPT2 or SPT6 mutations compromise the Gal4 inhibition achieved by SRG1 transcription at the GAL7 UAS and enhance significantly the accessibility of this region to activators. Importantly, in both spt2Δ and spt6-1004 mutants, Gal4 binding does not increase at the normal GAL7 promoter (see Fig. S8 at http://www.crc.ulaval.ca/nourani/supplemental_data). We next asked whether these mutations alter the chromatin structure at the SRG1::GAL7 promoter. As shown in Fig. S9 at the URL above, both mutations are associated with a significant loss of histone H3 occupancy at SRG1::GAL7. Collectively, these observations suggest that a loss of nucleosomes at the SRG1 region observed in Spt2-deficient cells compromise the SER3 promoter occlusion to activators.
FIG. 6.
Mutations of SPT2 or SPT6 enhance the accessibility of pSRG1-GAL7 UAS to the Gal4 activator. (A) Diagram showing the pSRG1-GAL7 construct used to analyze Gal4 binding in the presence of SRG1 transcription. (B) Gal4 binding to the pSRG1-GAL7 UAS is increased in the spt2Δ and spt6-1004 mutants. ChIP assays using anti-Gal4 antibody were conducted on chromatin extracted from wild-type (WT) (FY2257), spt2Δ (YAN1045), and spt6-1004 (YAN1046) strains. For each strain, the fold enrichment shown is relative to the fold enrichment calculated for the wild-type strain and arbitrarily set at 1.0.
Nucleosome deposition in the wake of RNAP II passage at the SRG1 intergenic region is significantly affected in spt2Δ cells.
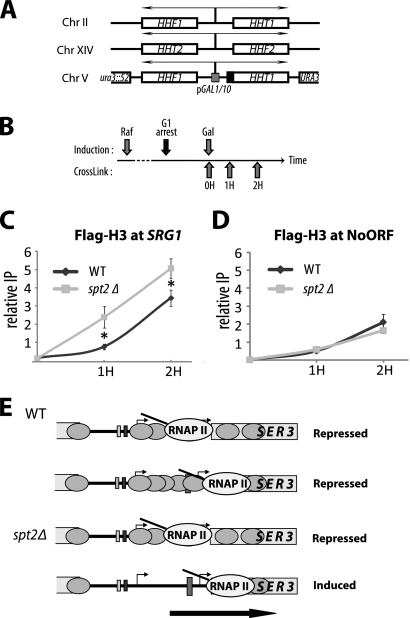
We established that Spt2 has an important impact on the chromatin structure of the SRG1 intergenic region. However, how this elongation factor participates in the maintenance of this chromatin structure remained an open question. Interestingly, this genomic region is robustly transcribed and thus RNAP II activity could lead to displacement of key nucleosomes in this region that must be subsequently reassembled. One could imagine that Spt2 plays a role in nucleosome redeposition in the wake of transcription machinery and that the absence of this factor could lead to a deficient restoration of nucleosomes. To directly test this hypothesis we first used an experimental system described in a number of studies (7, 36). Briefly, through histone H3 ChIP assays, we measured the kinetics of histone H3 redeposition in the transcribed region of the large (8-kb) model gene FMP27 that is driven by the GAL1 promoter (36). The rapid repression of GAL1-FMP27 obtained by adding glucose to the growth medium allowed us to study the rate with which histone H3 is deposited following the last wave of transcription (Fig. 7 A). In wild-type cells, upon the addition of glucose, histone H3 levels increased rapidly and reached their maximum at the middle region of GAL1-FMP27 within 4 min (Fig. 7B). This is consistent with previous studies (36). Deletion of SPT2 was associated with a significant defect in the H3 redeposition at the GAL1-FMP27 region after glucose repression. As shown in Fig. 7B, although glucose repression was associated with a small increase of histone H3 occupancy in spt2Δ cells, the recovery of nucleosomes in this strain is slower and did not reach the wild-type level even after 20 min of repression. We therefore conclude that the loss of Spt2 has a substantial effect on the nucleosome deposition associated with transcription elongation at GAL1-FMP27.
FIG. 7.
Spt2 is required for nucleosome reassembly in the wake of RNAP II at the SRG1 intergenic region. (A) Diagram explaining the experimental procedure designed to analyze histone H3 recovery upon repression of GAL1-FMP27. WT, wild type. (B) spt2Δ mutation affects significantly histone H3 redeposition associated with the last wave of transcription at the GAL1-FMP27 transcribed region. Yeast cells from wild-type (YAN1047) or spt2Δ (YAN1048) strains were grown in galactose medium to mid-log phase. Glucose was then added to the medium, and the cells were cross-linked and harvested at the indicated time points. Histone H3 level was analyzed by ChIP assays using chromatin extracted from wild-type or spt2Δ strains. For each strain, the value of the ratio IP/input calculated for the time zero min was arbitrarily set at 1. (C) Diagram explaining the experimental procedure designed to analyze histone H3 recovery at the SRG1 3′ end upon repression of pGAL1-SRG1. (D) spt2Δ mutation affects significantly histone H3 redeposition associated with the last wave of transcription at the SRG1 region corresponding to the SER3 promoter. The experiment was conducted as described for panel B with chromatin extracted from wild-type (YAN1040) or spt2Δ cells (YAN1039). The ratio IP/input calculated for time zero in the wild type was arbitrarily set at 1. (E) Spt2 delays maximal SER3 induction. The SER3 mRNA and SCR1 levels were analyzed by RT-qPCR using total RNA extracted from cells treated as described for panel B. For each strain, the value of the ratio SER3 mRNA/SCR1 calculated for time zero was arbitrarily set at 1. The values represent the average of two to three independent experiments. Note that at time zero, as expected, the absolute levels in the spt2Δ strain are higher than those observed in wild-type cells.
Altogether, our findings suggest the possibility that Spt2 is required for nucleosome redeposition in the wake of transcription at the SRG1 ncDNA. To address this possibility, we used the same approach described in the legend to Fig. 7A by replacing the SRG1 promoter with that of GAL1 and following the histone H3 deposition after the last wave of transcription upon glucose addition (Fig. 7C). We analyzed these kinetics at the SRG1 transcribed region corresponding to the SER3 promoter, and the results are reported in Fig. 7D. In wild-type cells, we observed a very rapid increase in histone H3 occupancy that reached its maximum level after 4 to 6 min of repression. Surprisingly, in contrast to the results for GAL1-FMP27, the level of histone H3 started to drop and returned to a low level quickly, indicating that nucleosomes that reassembled following RNAP II passage at the ncDNA SRG1 are very unstable compared to those at FMP27. Interestingly, the histone H3 deposition associated with the last wave of transcription was severely impaired in spt2Δ cells, indicating an important role of Spt2 in transcription-dependent nucleosome deposition at the region corresponding to the SER3 promoter. Importantly, the histone H3 steady-state level in the pGAL1-SRG1 spt2Δ strain grown in glucose almost reaches the wild-type level (see Fig. S10 at http://www.crc.ulaval.ca/nourani/supplemental_data).
In addition to monitoring histone H3, we analyzed SER3 transcript levels upon glucose repression of GAL1-SRG1 (Fig. 7E). In this experiment, we focused on the degree of induction in each strain and not on the absolute levels, which are higher in spt2Δ cells. Upon glucose addition, we observed a rapid induction of SER3 transcripts in wild-type and spt2Δ cells. This induction quickly reached a plateau in the spt2Δ mutant, while it continued to increase in wild-type cells. The graph in Fig. 7E shows that maximum induction of SER3 was reached in cells lacking Spt2 after only 10 min. In contrast to this, SER3 induction continued for at least 10 more minutes in wild-type cells and did not reach a plateau even after 20 min. This delay could be due to the deposition of nucleosomes in the wild-type cells during the first 4 to 6 minutes after GAL1 promoter shutdown. Later, these nucleosomes are displaced, allowing a further induction of SER3. Therefore, our observation suggests that nucleosome deposition associated with the last wave of transcription delays SER3 induction in wild-type cells. Since the spt2Δ mutant is deficient in nucleosome deposition, we did not observe any delay in maximal induction of SER3 and a plateau was reached after 10 min of glucose repression. Altogether, our data strongly suggest that Spt2 is required for the assembly of nucleosomes associated with transcription of ncDNA SRG1 and that these nucleosomes play a crucial role in the normal repression of SER3.
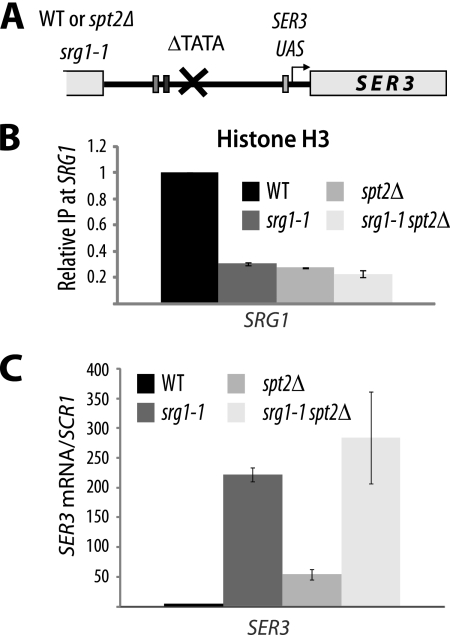
Our data strongly suggest that Spt2, together with SRG1 transcription, maintains higher nucleosome occupancy over the SER3 promoter and therefore represses its transcription. To confirm this, we analyzed histone H3 occupancy in the srg1-1 mutant. In this strain (Fig. 8 A), mutation of the SRG1 TATA box abolishes its transcription (21). As shown in Fig. 8B, in the absence of SRG1 transcription, the levels of histone H3 at the SRG1 region corresponding to the SER3 promoter dropped significantly, indicating that chromatin structure in this region is dependent on transcription. Interestingly, deletion of the SPT2 gene did not result in a further decrease in histone H3 occupancy (Fig. 8B). This observation shows that the role of Spt2 in maintaining chromatin structure at the SER3 promoter is dependent on SRG1 transcription. Moreover, we found that deletion of SPT2 did not affect significantly the SER3 transcript level in the srg1-1 mutant (Fig. 8C), suggesting therefore that the role of Spt2 in the regulation of SER3 is associated with SRG1 transcription.
FIG. 8.
Chromatin structure at SRG1 depends on active transcription. (A) Diagram showing the srg1-1 construct. WT, wild type. (B) Mutation of SRG1 TATA is associated with a significant loss of histone H3 at the SRG1 3′-end region corresponding to the SER3 promoter. Histone H3 ChIP assays were conducted using chromatin extracted from wild-type (YAN1053), srg1-1 (YAN1054), spt2Δ (YAN1055), and srg1-1 spt2Δ (YAN1056) strains. For each strain, the value shown represents the ratio IP/input relative to the same ratio calculated for the corresponding wild-type strain and arbitrarily set at 1.0. The values shown are the averages and standard errors of results from three independent experiments. (C) The spt2Δ mutation in the srg1-1 strain does not increase significantly SER3 derepression. Total RNA extracted from wild-type (YAN1053), srg1-1 (YAN1054), spt2Δ (YAN1055), and srg1-1 spt2Δ (YAN1056) strains was used to produce cDNA that was subsequently quantified by qPCR. The SER3 mRNA relative level is the ratio of the SER3 mRNA to the SCR1 transcript level. The values shown are the averages and standard errors of results from three independent experiments.
Spt2 contributes to redeposition of nucleosomes displaced by SRG1 ncRNA transcription.
As shown earlier, there are nucleosomes randomly positioned at the SRG1 ncDNA region corresponding to the SER3 promoter. These nucleosomes are displaced and reassembled by continuous passages of RNAP II. We first wanted to know whether this chromatin modulation involves a mechanism that recycles the histones of these nucleosomes or uses new histones. As our data show that Spt2 participates in the deposition of these nucleosomes, we asked specifically whether Spt2 favors the recycling or the use of new histones. To answer these questions, we used an experimental system previously described by us and others (6, 33, 34). In this system, there are two different sources of histone H3 in the cell: the endogenous untagged histone H3 and a galactose-inducible form fused to the Flag tag that is coexpressed with histone H4 (Fig. 9 A). In order to eliminate the contribution of DNA replication-dependent histone deposition, exponentially growing cells containing the described construction (Fig. 9B) are blocked in G1 with α-factor. After incubation with α-factor, cells are either left untreated or induced to express Flag-H3 prior to formaldehyde treatment to cross-link chromatin. Next, the levels of Flag-H3 are assayed by standard ChIP-qPCR at the SRG1 ncDNA region corresponding to the SER3 promoter and at a control nontranscribed intergenic region of chromosome V (NoORF) (Fig. 9C and D). As shown in Fig. 9C and D, after induction of the new histone H3 in wild-type cells, we detected higher levels of its incorporation at SRG1 than in the control region (NoORF). Thus, SRG1 ncDNA is associated with a significant replication-independent histone H3 turnover, indicating the existence of a transcription-dependent chromatin assembly mechanism that deposits new nucleosomes. Surprisingly, deletion of SPT2 resulted in higher deposition of new histone H3 at ncDNA SRG1. Moreover, the low level of total H3 in Spt2-deficient cells suggests that, in the spt2Δ mutant, the vast majority of nucleosomes at SRG1 are refolded using newly synthesized histones. This indicates that Spt2 inhibits nucleosome assembly that uses newly synthesized histones. Because the overall nucleosome deposition is reduced in spt2Δ cells (Fig. 7), our data suggest that, similarly to yFACT at the transcribed regions of some genes (14), Spt2 favors a dominant nucleosome assembly pathway that uses old histones displaced by elongating RNAP II. In Spt2-deficient cells, the induction of another chromatin assembly pathway that uses new histones is not able to overcome this loss. Thus, Spt2 participates in the major mechanism that refolds chromatin structure in the wake of RNAP II at SRG1. This mechanism recycles histones of nucleosomes previously displaced by the transcription machinery and shapes specific chromatin structure that is crucial for SER3 repression.
FIG. 9.
Spt2 is involved in a nucleosome reassembly pathway that uses old histones at SRG1. (A) Diagram representing the different sources of histone H3 and H4 in the yeast strains used in the experiment. (B) Diagram representing the experimental procedure. (C and D) Spt2 inhibits the incorporation of new histone H3 at the SRG1 region corresponding to the SER3 promoter. Yeast cells from the wild-type (WT) (YAN1049) or spt2Δ strains (YAN1050) containing the construction encoding histone Flag-H3 under the control of the GAL1 promoter were grown in raffinose-containing medium to mid-log phase. After G1 arrest by α-factor, the cells were formaldehyde fixed or shifted to galactose medium for 60 or 120 min prior to formaldehyde treatment. ChIP assays were then performed using anti-Flag antibody. The values shown (relative IP/input) represent the averages and standard errors of results from three independent experiments. *, P < 0.05. (E) Model of SER3 regulation by ncRNA SRG1 transcription.
DISCUSSION
The modulation of gene expression by ncRNA is an emerging field that has focused mostly on transregulatory mechanisms, including repression by RNAi pathways. However, a growing amount of evidence points toward the existence of various cis-directed mechanisms where the process of ncRNA production itself, and not the product, contributes to the regulation of a gene. Interestingly, several observations introduced the notion that the chromatin environment created by the transcription of ncRNA may have a regulatory role (10). These studies involved different mechanisms where ncRNA transcription displaces nucleosomes or modifies histones to regulate a nearby gene (12, 13, 30). Here we show that SRG1 ncRNA transcription deposits nucleosomes that participate in the repression of the adjacent SER3 gene. Our study also brings new insights into the molecular mechanism of transcription interference. Different models of transcription interference have been proposed, including promoter competition, occlusion by elongating RNAP II, or collision between transcription machineries (reviewed in reference 37). We provide evidence that transcription interference in the SRG1-SER3 system involves deposition of specific nucleosomes in the wake of an elongating RNAP II. Importantly, and in contrast to previous observations, our work shows that histone H3-K4 methylation and H3-K36 methylation are not required for the normal SER3 regulation by ncRNA transcription. This suggests that the most important factor in this repression is the deposition of nucleosomes and not their posttranslational modifications that could alter their dynamic properties. Finally, we show that the elongation factor Spt2 and the histone chaperone Spt6 play central roles in this regulatory system. Our data indicate that Spt2 participates in the shaping of the transcription-dependent chromatin structure over the SRG1 transcribed region that controls the activity of SER3 promoter.
Before this work was completed, the working model of SER3 regulation had been based on the canonical mechanism of transcription interference, whereby successive passages of RNAP II transcribing the SRG1 intergenic region obstruct the SER3 promoter, inhibiting the interaction of activators and initiation factors with their respective binding sites. In the spt2Δ or spt6-1004 mutant strains grown in standard rich medium, we observed a small but consistent reduction of transcription activity at the SRG1 intergenic region that produces the ncRNA. It was easy to imagine that if regulation by transcription interference was achieved merely by maintaining a certain transcription rate involving the passage of a specific number of RNAP II complexes, a small reduction in the number of these complexes traversing the SER3 promoter could compromise the interference. However, direct and indirect experimental evidence quickly challenged this simple view. First, our work showed that under conditions where SRG1 transcription in the spt2Δ mutant is higher than that in wild-type cells, the SER3 gene remained active (Fig. 2E; also see Fig. S5 at http://www.crc.ulaval.ca/nourani/supplemental_data). Second, increasing artificially the transcription rate of the SRG1 ncRNA in the spt2Δ mutant did not restore normal repression (Fig. 2C and D). Third, in spt6-1004 cells, we observed similar defects in SER3 repression. This phenotype cannot be explained by variations in SRG1 transcription (Fig. 3). Fourth, changing the transcription in the wild type from a high level (SRG1 promoter) to a very high level (GAL1 promoter) did not further repress SER3 transcription. In fact, we observed the opposite result, since insertion of the GAL1 promoter was associated with SER3 derepression (Fig. 2C, 2D, and 3E; also see Fig. S3 at the URL above). These data suggest a more complicated mechanism of transcriptional interference in which the passages of RNA polymerase II alone are not sufficient to repress SER3 transcription. Thus, the presence or absence of normal Spt2 and Spt6 function could slightly affect the production of the ncRNA under some conditions, but this marginal role could not explain the loss of SER3 repression. Finally, taken together, these observations indicate that SER3 repression is not only mediated by the production of the SRG1 ncRNA or the frequency of transcription machinery passages over the SER3 promoter. Instead, it is the entire process of transcription and its associated events such as chromatin modulation that are involved in the interference with the SER3 promoter. Our observations support an important role of Spt2 in the chromatin modulations associated with SRG1 transcription that contributes directly to the repression of SER3.
Perhaps the most significant observation in this work is the one related to the chromatin structure in the SRG1 intergenic region (Fig. 4 and 5). Nucleosome scanning assays conducted in wild-type cells indicate that this region contains a large peak representing the presence of nucleosomes randomly positioned within this short DNA segment (Fig. 5). Importantly, our data indicate clearly that nucleosomes are assembled in the wake of transcription at this region (Fig. 7). However, shortly after their deposition, these nucleosomes appear to be displaced. Therefore, our findings suggest that continuous transcription of intergenic SRG1 is required for the maintenance of nucleosomes at this region. Interestingly, mutation of the SRG1 TATA box abolishes its transcription and results in a clear depletion of histone H3 at this region (Fig. 8). This is consistent with a recent study showing that nucleosomes over the SER3 UAS (upstream activation sequence) are present when SRG1 is transcribed, while inhibition of the intergenic transcription in the absence of serine is associated with a loss of nucleosomes at this position (9a).
The precise mechanism by which Spt2 contributes to the shaping of the nucleosomal structure at the SRG1 ncDNA remains an interesting question. Several hypotheses could be proposed. It is possible that Spt2 acts after chromatin synthesis and stabilizes the nucleosomes deposited in the wake of RNAP II. Alternatively, Spt2 could be directly involved in transcription-dependent chromatin assembly associated with SRG1 transcription. The latter possibility is likely to be the case for several reasons. First, Spt2 has two HMG-like domains and could, similarly to other HMG-box proteins, assist histone chaperones in nucleosomal assembly (9, 26). Second, it has functional genetic and physical interactions with the histone chaperone Spt6 which are consistent with such a role (reference 24 and unpublished data). Third, it binds four-way junction DNA, a structure similar to that found at the entrance/exit point of DNA from a nucleosome (25, 46). Consistently, we found that it interacts with mononucleosomes in vitro (unpublished data). Future studies focused on Spt2 should help to determine its precise role in chromatin remodeling in the wake of RNAP II.
Our analyses of the replication-independent histone H3 incorporation indicate the existence of at least two different mechanisms involved in the control of transcription-dependent nucleosome deposition at the ncDNA SRG1. First, we showed that SRG1 ncDNA incorporates a high level of new histones in G1-arrested cells, indicating that transcription at this region is coupled to a replication-independent chromatin assembly pathway that uses new histones. Second, our analyses of histone H3 deposition outside S-phase show that the absence of Spt2 is associated with both a preferential deposition of newly synthesized H3 and an overall loss of histone H3 (Fig. 4 and 9) at the SRG1 ncDNA. This indicates that the loss of Spt2 affects an additional mechanism(s) that recycles old histone H3 to reassemble nucleosomes displaced by RNAP II at SRG1. This observation is similar to the data recently reported on the yFACT histone chaperone (14). Indeed, the Strubin group clearly showed that newly synthesized histone H3 deposition is induced on a number of genes in the spt16-197 mutant, while nucleosomal occupancy decreases overall (14). Without excluding other possibilities, one simple explanation of such observations is that two different chromatin assembly pathways coexist at SRG1. One uses old histones and recycles them to redeposit nucleosomes displaced by RNAP II and requires Spt2. The other pathway uses newly synthesized histones and is not dependent on Spt2. At SRG1, one possibility is that the recycling pathway is dominant, and the induction of another pathway using newly synthesized histone H3 cannot overcome the loss of Spt2. Whether this effect is more general and could be applied to other genomic loci would be interesting to assess. It is likely that this mechanism also functions outside the SRG1-SER3 locus. Indeed, analyses at other transcribed regions showed that spt2Δ mutation is also associated with the induction of newly synthesized H3 deposition (P. Thebault, unpublished data). Finally, given the fact that Spt2 and Spt16 defects have similar phenotypes on histone H3 dynamics at different transcribed regions, it will be interesting to test whether yFACT depletion has any role on SRG1 histone H3 turnover or chromatin structure and therefore on SER3 regulation.
Finally, our data suggest a new model of regulation by ncRNA transcription that involves nucleosomes deposited in the wake of the transcription machinery (Fig. 9E). We propose that ncRNA SRG1 transcription achieves SER3 repression mainly by creating a repressive chromatin structure. Therefore, in contrast to what has been imagined, transcription interference within the SRG1-SER3 system is not only achieved by RNAP II passing through the SER3 promoter but is also mediated by the nucleosomes inserted behind RNAP II complexes as they leave this regulatory region. These nucleosomes are important for the repression of SER3 transcription that could possibly be initiated in the time frame between two successive rounds of ncRNA SRG1 transcription. In the spt2Δ mutant, these nucleosomes are not assembled correctly, creating an open chromatin context for the SER3 activator(s). Importantly, and in contrast to previous observations, our work shows that histone H3-K4 and H3-K36 methylation is not required for the normal SER3 regulation by ncRNA transcription, suggesting that the most important factor is the deposition of nucleosomes and not modifications that play a role in the dynamic properties of these nucleosomes. Overall, we provide strong evidence that Spt2 participates in a regulation by transcription interference via a novel mechanism involving a complex interplay between RNAP II and chromatin dynamics associated with its passage.
Acknowledgments
We thank Nikita Avvakumov and Jacques Côté for helpful comments and advice on the manuscript.
This work was supported by a CIHR grant to A.N. A.N. holds a Canadian Research Chair, and G.B. holds an NSERC Ph.D. fellowship.
Footnotes
Published ahead of print on 10 January 2011.
REFERENCES
- 1.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baxter, B. K., and E. A. Craig. 1998. Suppression of an Hsp70 mutant phenotype in Saccharomyces cerevisiae through loss of function of the chromatin component Sin1p/Spt2p. J. Bacteriol. 180:6484-6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brickner, D. G., et al. 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrozza, M. J., et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581-592. [DOI] [PubMed] [Google Scholar]
- 5.Cheung, V., et al. 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6:e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dion, M. F., et al. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315:1405-1408. [DOI] [PubMed] [Google Scholar]
- 7.Fleming, A. B., C. F. Kao, C. Hillyer, M. Pikaart, and M. A. Osley. 2008. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell 31:57-66. [DOI] [PubMed] [Google Scholar]
- 8.Floer, M., et al. 2010. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell 141:407-418. [DOI] [PMC free article] [PubMed]
- 9.Formosa, T., et al. 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20:3506-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Hainer, S. J., J. A. Pruneski, R. M. Monteverde, and J. A. Martens. 2011. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 25:29-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartzog, G. A., and J. A. Martens. 2009. ncRNA transcription makes its mark. EMBO J. 28:1679-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hershkovits, G., H. Bangio, R. Cohen, and D. J. Katcoff. 2006. Recruitment of mRNA cleavage/polyadenylation machinery by the yeast chromatin protein Sin1p/Spt2p. Proc. Natl. Acad. Sci. U. S. A. 103:9808-9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirota, K., et al. 2008. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature 456:130-134. [DOI] [PubMed] [Google Scholar]
- 13.Houseley, J., L. Rubbi, M. Grunstein, D. Tollervey, and M. Vogelauer. 2008. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell 32:685-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamai, A., A. Puglisi, and M. Strubin. 2009. Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Mol. Cell 35:377-383. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan, C. D., L. Laprade, and F. Winston. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301:1096-1099. [DOI] [PubMed] [Google Scholar]
- 16.Kruger, W., and I. Herskowitz. 1991. A negative regulator of HO transcription, SIN1 (SPT2), is a nonspecific DNA-binding protein related to HMG1. Mol. Cell. Biol. 11:4135-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, W., et al. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 39:1235-1244. [DOI] [PubMed] [Google Scholar]
- 18.Lohr, D. 1984. Organization of the GAL1-GAL10 intergenic control region chromatin. Nucleic Acids Res. 12:8457-8474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Longtine, M. S., et al. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 20.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 21.Martens, J. A., L. Laprade, and F. Winston. 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429:571-574. [DOI] [PubMed] [Google Scholar]
- 22.Martens, J. A., and F. Winston. 2002. Evidence that Swi/Snf directly represses transcription in S. cerevisiae. Genes Dev. 16:2231-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martens, J. A., P.-Y. J. Wu, and F. Winston. 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 19(22):2695-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nourani, A., F. Robert, and F. Winston. 2006. Evidence that Spt2/Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:1496-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novoseler, M., G. Hershkovits, and D. J. Katcoff. 2005. Functional domains of the yeast chromatin protein Sin1p/Spt2p can bind four-way junction and crossing DNA structures. J. Biol. Chem. 280:5169-5177. [DOI] [PubMed] [Google Scholar]
- 26.Orphanides, G., W. H. Wu, W. S. Lane, M. Hampsey, and D. Reinberg. 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400:284-288. [DOI] [PubMed] [Google Scholar]
- 27.Perez-Martin, J., and A. D. Johnson. 1998. Mutations in chromatin components suppress a defect of Gcn5 protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1049-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson, C. L., W. Kruger, and I. Herskowitz. 1991. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell 64:1135-1143. [DOI] [PubMed] [Google Scholar]
- 29.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinskaya, M., S. Gourvennec, and A. Morillon. 2009. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 28:1697-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollard, K. J., and C. L. Peterson. 1997. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol. Cell. Biol. 17:6212-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pugh, B. F. 2000. Control of gene expression through regulation of the TATA-binding protein. Gene 255:1-14. [DOI] [PubMed] [Google Scholar]
- 33.Rufiange, A., P. E. Jacques, W. Bhat, F. Robert, and A. Nourani. 2007. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell 27:393-405. [DOI] [PubMed] [Google Scholar]
- 34.Schermer, U. J., P. Korber, and W. Horz. 2005. Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol. Cell 19:279-285. [DOI] [PubMed] [Google Scholar]
- 35.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwabish, M. A., and K. Struhl. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24:10111-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shearwin, K. E., B. P. Callen, and J. B. Egan. 2005. Transcriptional interference—a crash course. Trends Genet. 21:339-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sikdar, N., S. Banerjee, H. Zhang, S. Smith, and K. Myung. 2008. Spt2p defines a new transcription-dependent gross chromosomal rearrangement pathway. PLoS Genet. 4:e1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sternberg, P. W., M. J. Stern, I. Clark, and I. Herskowitz. 1987. Activation of the yeast HO gene by release from multiple negative controls. Cell 48:567-577. [DOI] [PubMed] [Google Scholar]
- 40.Uhler, J. P., C. Hertel, and J. Q. Svejstrup. 2007. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc. Natl. Acad. Sci. U. S. A. 104:8011-8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.West, K. L. 2004. HMGN proteins play roles in DNA repair and gene expression in mammalian cells. Biochem. Soc. Trans. 32:918-919. [DOI] [PubMed] [Google Scholar]
- 42.Whitehouse, I., and T. Tsukiyama. 2006. Antagonistic forces that position nucleosomes in vivo. Nat. Struct. Mol. Biol. 13:633-640. [DOI] [PubMed] [Google Scholar]
- 43.Winston, F., D. T. Chaleff, B. Valent, and G. R. Fink. 1984. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics 107:179-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winston, F., C. Dollard, and S. L. Ricupero-Hovasse. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53-55. [DOI] [PubMed] [Google Scholar]
- 45.Youdell, M. L., et al. 2008. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol. Cell. Biol. 28:4915-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zlatanova, J., and K. van Holde. 1998. Binding to four-way junction DNA: a common property of architectural proteins? FASEB J. 12:421-431. [PubMed] [Google Scholar]