Abstract
The phosphorylation of heterochromatin protein 1 (HP1) has been previously described in studies of mammals, but the biological implications of this modification remain largely elusive. Here, we show that the N-terminal phosphorylation of HP1α plays a central role in its targeting to chromatin. Recombinant HP1α prepared from mammalian cultured cells exhibited a stronger binding affinity for K9-methylated histone H3 (H3K9me) than that produced in Escherichia coli. Biochemical analyses revealed that HP1α was multiply phosphorylated at N-terminal serine residues (S11-14) in human and mouse cells and that this phosphorylation enhanced HP1α's affinity for H3K9me. Importantly, the N-terminal phosphorylation appeared to facilitate the initial binding of HP1α to H3K9me by mediating the interaction between HP1α and a part of the H3 tail that was distinct from the methylated K9. Unphosphorylatable mutant HP1α exhibited severe heterochromatin localization defects in vivo, and its prolonged expression led to increased chromosomal instability. Our results suggest that HP1α's N-terminal phosphorylation is essential for its proper targeting to heterochromatin and that its binding to the methylated histone tail is achieved by the cooperative action of the chromodomain and neighboring posttranslational modifications.
The formation of the higher-order chromatin structure, the so-called heterochromatin, is critical for genomic stability and transcriptional silencing (19, 21, 30). Heterochromatin protein 1 (HP1) is a nonhistone chromosomal protein that was initially discovered in Drosophila melanogaster as a factor that is enriched at highly condensed, transcriptionally inert regions of polytene chromosomes (24, 25). HP1 is an evolutionarily conserved molecule whose homologues and isoforms have been previously identified in a wide range of eukaryotic organisms (9). As well as being an architectural component of heterochromatin, HP1 is known to play crucial roles in heterochromatin-mediated gene silencing. Profound effects of HP1 deletion, mutation, or overexpression on transcriptional silencing have been previously demonstrated in Drosophila (11, 12), Schizosaccharomyces pombe (13), and mammals (17). Moreover, an ever-expanding list of HP1-interacting proteins indicates that HP1 may also be involved in the regulation of several biological processes, including transcriptional activation, faithful chromosome segregation, and DNA damage repair (8, 19, 21, 30).
Structurally, all HP1 homologues possess two functionally distinct globular domains, the N-terminal chromodomain (CD) (39) and the C-terminal chromoshadow domain (CSD) (1), which are linked by an unstructured hinge region. The CD recognizes and interacts with methylated lysine 9 of the histone H3 N-terminal tail, which is a characteristic modification in heterochromatic regions (4, 27, 35). The CSD, on the other hand, is responsible for dimer formation and is involved in HP1's interactions with a large variety of proteins (5, 44, 46). Because of its abilities to bind directly to chromatin and to form dimers and oligomers, HP1 is thought to contribute to the formation and/or stabilization of higher-order chromatin structure by cross-linking methyl H3K9-marked nucleosomes (4).
In contrast to the cytological appearance of heterochromatin, the interaction between HP1 and chromatin is not static but highly dynamic (6, 16). The kinetics of the interaction can change according to cellular circumstances; for example, alterations in the kinetics of the HP1-chromatin interaction were previously demonstrated during T-lymphocyte activation (16) and embryonic stem (ES) cell differentiation (33). Several mechanisms for these changes in HP1-chromatin interaction kinetics have been proposed. For example, alterations in the histone modifications or in the levels of other HP1-interacting partners appear to change the HP1-chromatin interaction (18, 22, 33). Alternatively, posttranslational modification of the HP1 protein itself may modulate HP1's dynamics and functions. Drosophila studies have revealed that HP1 is highly phosphorylated in vivo (10) and that phosphorylation of HP1 influences its heterochromatin binding and silencing activity (47, 48), although elucidation of the underlying molecular mechanisms remains elusive.
There is accumulating evidence that mammalian HP1 homologues are also highly phosphorylated (28, 34, 38), suggesting that the mammalian HP1 function is also regulated by phosphorylation. For instance, HP1γ Ser-83 phosphorylation impairs HP1γ's silencing activity and serves as a marker for transcriptional elongation (29). In addition, HP1β Ser-51 phosphorylation has previously been linked to HP1β mobilization during the initiation of the DNA damage response (3). Nevertheless, the biological functions of the HP1 phosphorylation in mammals remain largely unclear.
In this study, we show that mammalian HP1α is phosphorylated at its N-terminal serine residues and that this phosphorylation plays a critical role in modulating HP1α's activity to efficiently bind to the methylated histone H3 tail. Interestingly, the N-terminal phosphorylation appears to persist during the cell cycle and form an essential structural unit of HP1α to ensure its chromatin binding.
MATERIALS AND METHODS
Plasmid construction.
Mouse HP1α, HP1β, and HP1γ cDNAs were amplified from an NIH 3T3 cDNA library by PCR, using the Expand High Fidelity PCR system (Roche). The amplified HP1 cDNAs were first cloned into the TOPO pCRII vector (Clontech) and then subcloned into each of mammalian expression vectors pEGFP-C1 (Clontech), pFLAG-C1 (the enhanced green fluorescent protein [EGFP] coding region of pEGFP-C1 was replaced with the FLAG epitope), EGFP-pCAGI, and FLAG-pCAGI or subcloned into a bacterial expression vector, pCold II (TaKaRa). To express 6×His-tagged HP1α, HP1β, and HP1γ in NIH 3T3 cells, the HP1α, HP1β, and HP1γ cDNA with a 6×His coding region was amplified from HP1α/β/γ-pCold II by PCR and cloned into pCAG1. To produce a recombinant CD of HP1α, HP1β, and HP1γ, the corresponding region was amplified by high-fidelity PCR from full-length HP1 cDNA and subcloned into pCold I vector (TaKaRa). Mutagenesis of the HP1α cDNA sequence was performed according to the manufacturer's instructions (Stratagene). Unless otherwise mentioned, all of the PCRs were carried out using Pfu Turbo DNA polymerase (Stratagene). The mouse CK2α and CK2β cDNAs were synthesized as described above, and each open reading frame (ORF) was cloned into a pRSFDuet-1 vector (Merck).
Recombinant protein production.
Recombinant 6×His-tagged proteins (full-length HP1 and its CD) were expressed in Escherichia coli BL21(DE3) and purified by nickel-nitrilotriacetic acid (Ni-NTA) chromatography according to the instructions of the manufacturers (TaKaRa and Clontech). For fluorescence anisotropy, the HP1-CD was further purified by gel filtration chromatography (Superdex 200; GE Healthcare) equilibrated with GF buffer (50 mM potassium phosphate buffer [pH 8.0], 25 mM NaCl, 1 mM dithiothreitol [DTT]). For production of CK2-phosphorylated HP1αCD, E. coli BL21(DE3) cells were simultaneously transformed with HP1αCD-pCold I and pRSFDuet-FLAG-CK2α/β plasmids and selected with ampicillin (for pCold I) and kanamycin (for pRSFDuet). Phosphorylated 6×His-tagged HP1αCD was purified as described above. For the in vitro kinase reaction, 5 μg of the recombinant HP1 proteins was treated with 100 U of recombinant CK2 (New England BioLabs) in CK2 buffer (20 mM Tris-HCl [pH 7.5], 50 mM KCl, 10 mM MgCl2, 200 mM ATP) for 3 h at 30°C.
Cell culture and transfection.
NIH 3T3, HeLa, U2OS, and MEF cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FBS). For serum starvation, cells at 90% confluence were grown for another 24 to 48 h in DMEM supplemented with 0.5% FBS. G2/M-phase arrest was achieved by treating the cells with 200 ng/ml of nocodazole (Sigma) for 4 to 6 h. Transfection of plasmid DNA into NIH 3T3 cells was carried out using Lipofectamine LTX and Plus reagent (Invitrogen). After 24 h, the cells were harvested and used for further experiments.
Prediction of phosphorylation sites.
The mouse HP1α phosphorylation sites were predicted using NetPhos version 2.0 software (http://www.cbs.dtu.dk/services/NetPhos/) and the ELM functional site prediction program (http://elm.eu.org/). The predicted sites were compared to known mammalian HP1α phosphorylation sites (http://www.phosphosite.org/), and those serine residues that were likely to be phosphorylated in vivo were selected for mutagenesis.
Antibodies.
The antibodies used in this study were as follows: anti-HP1α (2HP-2G9 [Euromedex] and MAB3446 [Chemicon]), anti-HP1β (1MOD-1A9; Euromedex), anti-HP1γ (2MOD-1G6; Euromedex), anti-phospho-histone H3 (catalog no. 16-657 [Millipore] and 06-570 [Upstate]), peroxidase-conjugated anti-histone H3 (ab21054; Abcam), anti-trimethyl H3K4 (ab8580; Abcam), anti-trimethyl H3K9 (catalog no. 07-442; Millipore), anti-CK2α (1AD9; Calbiochem), anti-α-tubulin (T5168; Sigma), anti-TIF1β (MAB3662; Millipore/Chemicon), peroxidase-conjugated anti-FLAG M2 (A8592; Sigma), anti-6×His (catalog no. 34660 [Qiagen] or D291-07 [MBL]), peroxidase-conjugated anti-rabbit IgG (A6667; Sigma), peroxidase-conjugated anti-mouse IgG (catalog no. 112-035-072; Jackson ImmunoResearch), Alexa 546-conjugated goat anti-rabbit IgG (Molecular Probes), and Cy3-conjugated anti-rabbit IgG (Invitrogen).
Immunoblotting.
Mouse and human cell lines were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) supplemented with protease inhibitor cocktail (Complete [EDTA-free]; Roche) and 10 mM sodium fluoride (NaF). For dephosphorylation, an aliquot of whole-cell lysate (WCL) was incubated with 0.08 U/ml shrimp alkaline phosphatase (SAP) (TaKaRa) for 3 to 6 h at 37°C. Western blotting was performed according to a standard protocol. Alternatively, the blocking and antibody treatments were carried out using the SNAP identification system (Millipore). The intensity of each protein band was quantified using NIH ImageJ software, and the relative levels of Western blotting signals are presented in the figures. For polyacrylamide gel electrophoresis using a chemical reagent called Phos-tag (26) (here, we refer to SDS-PAGE analysis using this reagent as “phos-tag-PAGE”), 50 or 80 μM phos-tag-acrylamide (Wako) and an equal volume of 10 mM MnCl2 were added to a 10% (for full-length domains) or 12% (for CD) SDS-PAGE gel. Prior to the transfer of proteins, the phos-tag gels were incubated with transfer buffer containing 5 mM EDTA for 20 min and subsequently washed with fresh transfer buffer for 15 min.
Peptide pulldown assays.
Peptides were synthesized by Sigma Genosys. The peptides used in this study were unmodified H3 (H3unmod [ARTKQTARKSTGGKAPRKQL-C]), K9-trimethylated H3 (H3K9me3 [ARTKQTARKme3STGGKAPRKQL-C]), unmodified mutant H3 (ARTKQTARKme3STGGQAPRQQL-C), and K9-trimethylated mutant H3 [ARTKQTARKme3STGG(A/Q)APR(A/Q)QL-C]. Peptides (50 nmol) were covalently linked to 500 μl (bed volume) of Sulfo-link Sepharose resin (Pierce) via an artificially added C-terminal cysteine residue (peptide beads). Then, 1 pmol of recombinant HP1 was incubated with 10 μl (bed volume) of the peptide beads (protein:peptide ∼ 1:1,000) in 150 μl of binding buffer (20 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 0.1% Triton X) for 1 h at room temperature (RT). The beads were then washed twice with 500 μl of binding buffer. The bound proteins were eluted by mixing the beads with 45 μl of SDS sample buffer and boiling. Equal volumes of input, unbound, and bound fractions were analyzed by immunoblotting using antibodies against each HP1 isoform. Since input and unbound fractions (150 μl) were mixed with 30 μl of 6× SDS sample buffer, the isovolume-bound fraction corresponded to 4× the input. For quantitative pulldown assays, 1 pmol of protein was incubated with a 500-, 1,000-, 5,000-, or 10,000-fold molar excess of peptide beads. For the elution analysis, four aliquots of 1 pmol of recombinant protein were first bound to 10 μl of H3K9me3 beads as described above. Following the washes, the beads were then incubated in 150 μl of binding buffer containing a 0-, 100-, 1,000-, or 10,000-fold molar excess of free H3K9me3 peptides for 1 h. The beads were washed twice with 500 μl of binding buffer, and the bound proteins were eluted and analyzed.
Immunopurification of 6×His- and FLAG-tagged HP1 from NIH 3T3 cells.
NIH 3T3-expressed 6×His-HP1α, -HP1β, and -HP1γ were purified using a different procedure from that used for the bacterially expressed counterpart. NIH 3T3 cells expressing 6×His-HP1 were lysed in NP-40 lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40) supplemented with protease inhibitor cocktail (Complete [EDTA-free]; Roche) and 50 mM NaF and were subjected to sonication (5 pulses at 30% amplitude) on ice. 6×His-HP1 was purified using a His-tagged protein purification kit (MBL) according to the manufacturer's instructions. The protein concentration was estimated by comparing the intensity of silver- or CBB (Coomassie)-stained protein bands with that of bands corresponding to bacterially produced HP1 of a known concentration. For FLAG-tagged HP1, the cells were resuspended in IP lysis buffer (50 mM HEPES [pH 7.9], 10% glycerol, 250 mM NaCl, 0.1% Triton X) supplemented with protease inhibitor cocktail and 50 mM NaF and were further lysed by three freeze-thaw cycles. FLAG-tagged HP1 was purified with anti-FLAG M2 affinity gel and eluted by adding 1× FLAG peptide (Sigma).
MS.
Immunoaffinity-purified 6×His- or FLAG-tagged HP1α was concentrated using a Microcon system (Millipore), dialyzed against phosphate-buffered saline (PBS), and resolved by 15% SDS-PAGE. After Coomassie staining, the corresponding band was excised from the gel and subjected to in-gel reduction with 10 mM DTT, alkylation with 55 mM iodoacetamide, and digestion with 10 ng/μl modified trypsin (Promega) at 37°C for 16 h. After in-gel digestion, the phosphorylated peptides were concentrated using a Titansphere Phos-Tio kit (GL Science Inc.). The collected phosphorylated peptides were subjected to mass spectrometry (MS) analysis as described previously (41). For phosphorylated peptides, the ion trap was programmed to carry out four successive scans consisting of, first, a full MS scan over a range of 450 to ∼2,000 m/z, second and third, data-dependent scans of the top two most abundant ions obtained in the first scan, and fourth, MS/MS/MS analysis for determination of MS/MS spectra, with neutral loss corresponding to the phosphate groups obtained in the second and third scans.
Fluorescence anisotropy.
Fluorescein-conjugated peptides for unmodified H3 (fluorescein-C-[GARTKQTARKSTGGKAY]) and K9-trimethylated H3 (fluorescein-C-[GARTKQTARKme3STGGKAY]) were synthesized, and the yields were determined by absorption spectroscopy at A280, using the absorbance of an artificially added C-terminal tyrosine residue. The fluorescence intensity of fluorescein in FP buffer (50 mM potassium phosphate [pH 7.0], 25 mM NaCl, 1 mM DTT) at 30°C was observed using an FP-6500 fluorescence spectrometer (JASCO) or an F2700 fluorescence spectrophotometer (Hitachi High-tech). The temperature was controlled by circulating water around a water-jacketed cell holder. Linear polarization filters (HN32 from Polaroid Co.) were inserted in both the excitation and emission light paths, and their orientations were manually controlled in each set of measurements to calculate the anisotropy values. The excitation and emission wavelengths were 420 and 520 nm, respectively. The fluorescein-labeled peptide (10 nM) was incubated with increasing concentrations of recombinant HP1CD. The fluorescence anisotropy values (R) were plotted against the protein concentrations. Curve-fitting analyses and Kd (dissociation constant) estimations were carried out using IGOR Pro software (version 6.05A; WaveMatrics). The observed data fit very well into the equations (assuming a 1:1 binding stoichiometry ratio) as shown below:
 |
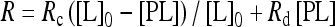 |
where PL, L0, and P0 represent the equilibrium concentration of the protein-ligand complex, the total ligand-peptide concentration (constant), and the total protein concentration, respectively. R represents the fluorescence anisotropy at a certain protein concentration. Rc and Rd represent the reference fluorescence anisotropy values seen when 100% of the fluorescent peptide molecules are complexed with protein molecules and when no protein is present (i.e., there is no peptide-protein complex) in the reaction, respectively.
Immunofluorescence microscopy.
Cells grown on coverslip-bottomed culture dishes (MatTek) were washed briefly with phosphate-buffered saline (PBS) and fixed with 3.7% formaldehyde for 10 min at RT. The fixed cells were permeabilized with 0.5% Triton X-PBS for 5 min at RT and blocked with 2.5% bovine serum albumin (BSA) in PBS for 30 min at RT. The cells were then incubated with an anti-phosphohistone H3 antibody (1/500 in blocking buffer with 0.05% Tween 20) for 2 h and subsequently with Cy3-conjugated anti-rabbit IgG (1/500 in blocking buffer with 0.05% Tween 20) for 1 h at RT. After each staining step, the cells were washed three times with PBST (PBS containing 0.05% Tween 20). The cells were counterstained with DAPI (4′,6-diamidino-2-phenylindole) (1 mg/ml)-PBST and mounted in antifade reagent (Invitrogen). Pictures were acquired with an Olympus microscope and analyzed using MetaMorph software (Universal Imaging), NIH ImageJ software, or DeltaVision softWoRx software (Applied Precisions). For categorization of EGFP-HP1α localization, the brightest EGFP spot in the nucleus was chosen, and the fluorescent intensity (FI) of the EGFP at that spot and the FI of background (euchromatic) area were measured with a line-plotting tool. In cases in which there was no clear EGFP spot, the most DAPI-dense area was analyzed. The FI ratio of the brightest EGFP-HP1α spot and background region was calculated, and the EGFP-HP1α localization patterns were categorized as follows: strong (FI ratio ≥ 1.8), weak (1.8 > FI ratio > 1), or diffused (1 ≥ FI ratio). The cutoff ratio was set at 1.8 because over 80% of the cells expressing wild-type EGFP-HP1α (EGFP-HP1αWT) consistently gave a ratio equal to or higher than 1.8.
Chromatin fractionation assay.
The chromatin fractionation was carried out essentially as described previously (46). NIH 3T3 cells stably expressing FLAG-tagged HP1αWT or mutant HP1αS14A were transfected with 100 pmol of control siRNA (small interfering RNA) (siTrio negative control; B-Bridge International) or siRNA against the HP1α 3′ untranslated region (UTR) (siHP1α [sense, 5′-ggaucagucugagaguuaTT-3′; antisense, 5′-uaacucucagacuguauccTT-3′; uppercase represents artificially added 3′ overhangs]) (B-Bridge International) by the use of a 12-well plate and Lipofectamine RNAiMAX (Invitrogen). After 48 h, the siRNA-treated cells were washed twice with PBS and incubated in PBS with 0.1% Triton X for 10 min at 4°C. The soluble fraction was collected by centrifugation at 2,000 rpm (390 × g) for 2 min. The remaining pellet was washed once with PBS and lysed in SDS-sample buffer.
Establishment of stable NIH 3T3 cell clones.
FLAG-HP1α (WT or S11-14A)-expressing plasmid (pCAGI/puromycin) was linearized by FspI digestion and introduced into NIH 3T3 cells by the use of Lipofectamine LTX and Plus reagent (Invitrogen). After 48 h, the cells were diluted and subcultured, and stably transfected cells were selected by treatment with 3 μg/ml puromycin for 72 h. Puromycin-resistant clones were isolated and subcultured in a culture medium without antibiotics. Expression of FLAG-HP1α was checked by Western blotting and immunofluorescence analysis using anti-HP1α and anti-FLAG antibodies, respectively.
Coimmunoprecipitation of native nucleosomes with FLAG-HP1α.
ΝΙΗ 3T3 cells stably expressing FLAG-HP1αWT or S11-14A were lysed in 600 μl of IP lysis buffer containing 5% glycerol with brief sonication. To solubilize nucleosomes and chromatin-bound proteins, lysates were supplemented with 0.6 μl of 1 M CaCl2 and treated with 120 U of micrococcal nuclease (TaKaRa) for 30 min at room temperature. Nuclease digestion was quenched by the addition of 6 μl of 0.5 M EDTA. The lysates were precleared with Sepharose 4B (GE Healthcare) and then incubated with anti-FLAG M2 affinity gel. The beads were extensively washed with IP lysis buffer, and FLAG-HP1α complexes were eluted by adding 1× FLAG peptide (Sigma). The eluates were analyzed by Western blotting using anti-histone H3 antibodies.
Preparation of chromosome spreads.
Cells were treated with 50 ng/ml demecolcine (Sigma) for 1 h, and single-cell suspensions were prepared by trypsin treatment. The cells were swelled by treatment with 7 ml of 75 mM KCl for 10 min at 37°C, mildly prefixed by adding 0.8 ml of fixative (methanol:acetic acid = 3:1), and collected by centrifugation at 800 rpm (120 × g) for 3 min. The cells were then fixed by the addition of ice-cold fixative. The fixed cells were spotted onto a glass slide, dried in air, and rinsed with 70% acetic acid. The cells were then stained with DAPI, mounted in Slowfade reagent (Invitrogen), and observed under a fluorescence microscope using a UPlanSApo lens (Olympus) (100× objective).
RESULTS
Bacterially produced HP1α shows poor binding to the K9-methylated histone H3 tail.
Mammalian cells express three HP1 isoforms, HP1α (Cbx5), HP1β (Cbx1), and HP1γ (Cbx3). These proteins are thought to target chromatin via an interaction between the chromodomain (CD) and K9-trimethylated histone H3 (H3K9me3), but the molecular basis of their distinct localization and cellular functions remains largely unknown. To investigate the chromatin-targeting mechanisms of these isoforms, we produced recombinant mouse HP1α, -β, and -γ by the use of E. coli cells (Fig. 1A) and examined their ability to interact with H3K9me3 in a peptide pulldown assay using peptide-cross-linked beads (Fig. 1B).
FIG. 1.
Recombinant HP1α produced in E. coli displays lower binding affinity for H3K9me3 than NIH 3T3 cell-produced HP1α. (A) Recombinant mouse HP1α, -β, and -γ prepared from E. coli cells. 6×His-tagged mouse HP1α, -β, and -γ were produced in E. coli cells and purified using metal affinity chromatography. The purified proteins were resolved by 8 to 15% SDS-PAGE and visualized by Coomassie staining. (B) In vitro pulldown assays of mouse HP1 isoforms. Bacterially produced mouse HP1 isoforms (1 pmol) were incubated with unmodified H3 (H3unmod.) peptide or K9-trimethylated H3 (H3K9me3) peptide cross-linked beads (approximately 1 nmol of peptide). Input (I), unbound (U [1× input]), and bound (B [4× input]) fractions were resolved by SDS-PAGE and analyzed by Western blotting (WB) using cognate antibodies. Unsaturated protein bands on X-ray films were quantified using NIH ImageJ software. The ratios of unbound and bound fractions to input are shown beneath each lane. (C) Recombinant mouse HP1α prepared from E. coli and NIH 3T3 cells. 6×His-tagged HP1α was transiently expressed in NIH 3T3 cells and purified using anti-His-tag agarose. The proteins, accompanied by the bacterially expressed HP1α (see panel A), were resolved by 8 to 15% SDS-PAGE and visualized by Coomassie staining. (D) In vitro pulldown assays of recombinant mouse HP1α prepared from E. coli or NIH 3T3 cells. Each recombinant HP1α was subjected to a pulldown assay with the H3unmod. or H3K9me3 peptide as described for panel B and analyzed by Western blotting using anti-HP1α antibodies.
Both recombinant HP1β and HP1γ strongly bound to the H3K9me3 peptide and showed only negligible binding to unmodified H3 peptide. HP1α also bound to H3K9me3, but its binding ability was clearly weaker than that of HP1β or HP1γ, as evidenced by the larger unbound (U) fraction (Fig. 1B). While a similar result indicating that HP1αCD shows weaker binding to H3K9me3 than HP1βCD or HP1γCD was previously reported (18), these results seemingly contradict the observation that mammalian HP1α shows a clear heterochromatic localization (34). To further examine the interaction between HP1α and H3K9me3, we produced HP1α in NIH 3T3 cells and subjected it to affinity purification for pulldown assays (Fig. 1C). Interestingly, the HP1α prepared from NIH 3T3 cells bound to the H3K9me3 more strongly than that produced from E. coli (Fig. 1D), with a binding affinity comparable to that of recombinant HP1β and HP1γ (compare Fig. 1B and 1D). This phenomenon was specific to HP1α; no clear difference in levels of affinity was observed between E. coli- and NIH 3T3-expressed proteins for HP1β and HP1γ (data not shown). In addition, the HP1α prepared from NIH 3T3 cells migrated more slowly in an SDS-PAGE gel than the E. coli-expressed HP1α (Fig. 1C), although both contained the same peptide sequence. Together, these results suggested that some kind of posttranslational modification(s) of HP1α modulates its binding to H3K9me3.
Mouse HP1α exhibits two distinct types of phosphorylation.
The difference in the apparent molecular sizes of E. coli- versus NIH 3T3-produced HP1α was only several kilodaltons (Fig. 1C), and phosphorylation is a candidate for such posttranslational modifications. When endogenous HP1α was treated with shrimp alkaline phosphatase (SAP), a similar band shift in the SDS-PAGE gel was observed (Fig. 2A, mock), suggesting the involvement of phosphorylation in the recombinant HP1α migration change. Recent proteomics studies identified phosphorylation sites of human HP1α (28, 38), but the relative abundances, dynamics, and physiological roles of these phosphorylations remained unclear. Therefore, we investigated the HP1α phosphorylation patterns by the use of mouse NIH 3T3 cells.
FIG. 2.
Identification of phosphorylation sites of mouse HP1α. (A) The phosphorylation patterns of mouse HP1α analyzed by phos-tag Western blotting (phos-tag WB). Whole-cell lysates (WCLs) of NIH 3T3 cells were resolved on an SDS-PAGE gel containing phos-tag acrylamide (phos-tag) or on a standard SDS-PAGE gel (mock). As an unphosphorylated control, SAP-treated WCL was loaded on the same gel. HP1α was detected using an anti-HP1α antibody. (B) Phosphorylation patterns of mouse HP1α in cells arrested at different cell cycle stages. WCLs were prepared from NIH 3T3 cells grown under the indicated conditions and analyzed by phos-tag WB. The arrowhead indicates an HP1α with basal band shift. The asterisk represents an HP1α species with metaphase-associated phosphorylation. The same membrane was immunoblotted with anti-α-tubulin and anti-TIF1β antibodies for loading controls. (C) Amino acid sequence of mouse HP1α. Light- and dark-shaded boxes indicate the CD and CSD, respectively. The sites of serine (S)-to-alanine (A) substitutions introduced in the following experiments are shown in boldface. (D) Phos-tag and standard WB analysis of wild-type and mutant HP1αs. WCLs were prepared from NIH 3T3 cells transiently expressing FLAG-tagged wild-type HP1α or HP1α with N-terminal (N-term: S11-14A) mutations, hinge (Hinge: S92, 93, 95, 97A) mutations, or both (N-term + hinge). Each FLAG-HP1α with an SAP-treated control was analyzed by phos-tag WB using an anti-FLAG antibody. The phosphorylation state of HP1α represented by each protein band was deduced from the distance of migration level of each band (Rf) and is indicated to the right of the phos-tag WB panel. (E) Phosphorylation state of FLAG-HP1α with each single amino acid substitution in the N-terminal region. The phosphorylation patterns of the indicated FLAG-HP1α species were analyzed by phos-tag WB using a higher concentration (80 μM) of phos-tag acrylamide. (F) Phosphorylation state of FLAG-HP1α with each single amino acid substitution in the hinge region. (G) The numbers of phosphorylated peptides identified by liquid chromatography (LC)-MS/MS analysis of affinity-purified HP1α. (H) Summary of the phosphorylation sites of mouse HP1α deduced from the experiments described above.
To examine HP1α phosphorylation in detail, we used Phos-tag (26). In the presence of manganese(II) ion (Mn2+), Phos-tag forms a complex with two Mn2+ ions and selectively traps R-OPO32− (R = protein, peptide, DNA, etc.). Therefore, the addition of this chemical to a PAGE gel slows down the migration of phosphorylated proteins in SDS-PAGE. The degree of the band shift depends on the number of phosphate groups on a single protein molecule and the site of phosphorylation within a protein. The endogenous HP1α in NIH 3T3 cells was detected as at least two distinct bands by the use of phos-tag-PAGE, and both bands shifted to a faster-migrating form after SAP treatment (Fig. 2A, phos-tag), indicating that the majority of the HP1α was phosphorylated at one or more sites.
We next examined whether the phosphorylation pattern of HP1α changes according to cell cycle stage. NIH 3T3 cells were grown under asynchronous (control), serum-starved (G0-arrest), or nocodazole-treated (G2/M-arrest) conditions. G2/M-arrested cells were further enriched by the mitotic shake-off method, and then the phosphorylation state of HP1α in these cells was analyzed by phos-tag-PAGE (Fig. 2B). HP1α showed a major band that was consistently present irrespective of the cell cycle stage in all the cell populations (Fig. 2B, arrowhead), and slower-migrating minor bands appeared with different degrees of intensity, depending on the cell cycle stage (Fig. 2B, asterisk). All the HP1α species shifted to the faster-migrating form after SAP treatment (Fig. 2B, +SAP). Since the unphosphorylated form of HP1α was hardly detected in the examined cell cycle stages, it appeared that HP1α was constitutively phosphorylated at one or more sites in vivo. The intensities and numbers of slower-migrating bands were relatively low in G0-phase-arrested and asynchronous cells but became prominent in the G2/M-phase-arrested cells, indicating that these bands represented “metaphase-associated” phosphorylation. These results agree with a previous observation from a study of human cells showing HP1α to be more heavily labeled with radioactive phosphate in G2/M-phase-arrested cells than in asynchronous cells (34).
Determination of the phosphorylation sites of mouse HP1α.
To determine the phosphorylation sites of HP1α, we cloned mouse HP1α cDNA and introduced amino acid substitutions for the candidate serine residues that were predicted by several phosphorylation site prediction programs (Fig. 2C, indicated in boldface; see also Materials and Methods). The wild-type (WT) or mutant HP1α was expressed with the FLAG tag in NIH 3T3 cells, and the phosphorylation patterns found under asynchronous conditions were examined by phos-tag-PAGE. Wild-type HP1α (HP1αWT) was detected as two distinct bands, both of which shifted to a faster-migrating band after SAP treatment (Fig. 2D, WT). The overall band patterns and relative migration levels were similar to those of endogenous HP1α (Fig. 2A).
Amino acid substitutions in the N-terminal region altered the migration levels of both the faster- and slower-migrating bands (Fig. 2D, N-term). On the other hand, mutations in the hinge region led to the specific disappearance of the slower-migrating band (Fig. 2D, Hinge). Combining both mutations resulted in a shift of all the species to the unphosphorylated level (Fig. 2D, N-term + hinge). These results suggest that the faster-migrating HP1α contains phosphorylated serine residues in its N terminus, whereas the slower-migrating HP1α contains the metaphase-associated phosphorylation in its hinge region in addition to the N-terminal phosphorylation.
To further characterize the phosphorylation sites, we created HP1α mutants carrying a single point mutation in the N-terminal (S11A, S12A, S13A, or S14A) or hinge (S92A, S93A, S95A, or S97A) region. Three N-terminal point mutations, S11A, S12A, and S13A, only slightly changed the overall migration pattern of HP1α (Fig. 2E, asterisks). In contrast, the S14A mutation caused a dramatic shift of the band, reaching the level of the unphosphorylated control (Fig. 2E [S14A, arrowheads]) and indicating that S14 is the key N-terminal phosphorylation site. Although the presence of phosphorylation at S11, S12, or S13 has not been clearly demonstrated by the mutation studies, mass spectrometry analysis of affinity-purified HP1α enabled us to identify peptides containing phosphorylation(s) at these residues in addition to S14 phosphorylation (Fig. 2G and data not shown). Together, these results suggested that the N-terminal serine residues (S11-S14) are multiply phosphorylated in vivo and that S14 phosphorylation is a prerequisite for or enhances the phosphorylation of its neighboring serine residues.
With regard to the hinge phosphorylation, the S93A mutation caused the loss of the slower-migrating HP1α (Fig. 2F), suggesting that S93 is the major metaphase-associated phosphorylation site. In addition to S93, S95 and S97 may play a role in the control of HP1α's phosphorylation in vivo, since there were slight but obvious shifts in the bands of the S95A and S97A mutants compared with WT results (Fig. 2F) and phosphorylation at those serine residues was detected by mass spectrometry analysis (Fig. 2G). Taken together, the results indicated that N-terminal phosphorylation contributes to the major band shift of HP1α whereas S93 in the hinge region is the major mitotic phosphorylation site in vivo (Fig. 2H).
The N-terminal phosphorylation of HP1α enhances its binding to H3K9me3.
The N-terminal phosphorylation for the HP1α in different mouse and human cells was widely observed (Fig. 3A). In addition, the corresponding serine residues lie close to the conserved CD (Fig. 3B). We therefore hypothesized that the N-terminal phosphorylation could affect the interaction between the CD and H3K9me3. To test this directly, we introduced N-terminal phosphorylation in vitro and examined the binding affinity of the phosphorylated HP1α for H3K9me3.
FIG. 3.
N-terminal phosphorylation of HP1α enhances the interaction between HP1αCD and H3K9me3. (A) Phos-tag WB analysis of the HP1α phosphorylation state in various mouse (NIH 3T3 and MEF) and human (U2OS and HeLa) cell lines. SAP-treated WCLs were used as unphosphorylated controls. The same anti-HP1α antibody was used to detect human and mouse HP1α. (B) Schematic diagram showing HP1α with the amino acid sequences of the N-terminal phosphorylation sites (top) and the CK2 consensus sequences (bottom). Serine residues that could be phosphorylated are underlined, and the surrounding acidic amino acid residues are shown in red. (C) In vitro kinase activity of CK2 for the recombinant HP1α. 6×His-tagged HP1αWT, HP1αS14A, and HP1αS11-14A were treated with recombinant CK2, and the phosphorylation states were analyzed by phos-tag SDS-PAGE, followed by Coomassie staining. (D) LC-MS/MS analysis of CK2-treated full-length HP1α (HP1αFL) and HP1α chromodomain (HP1αCD). (E) In vitro pulldown assays with N-terminally phosphorylated HP1α. Bacterially produced 6×His-HP1α (WT and S11-14A) was phosphorylated using recombinant CK2 in vitro. The CK2-phosphorylated- and control HP1αs were subjected to pulldown assays with the H3unmod. and H3K9me3 peptides as described for Fig. 1B. (F) Binding of the HP1-CD to fluorescein-labeled H3 peptides as measured by fluorescence anisotropy. Curves represent the best fit to data as described in Materials and Methods. Binding assays were performed for the H3K9me3 peptide with HP1αCD, CK2-treated HP1αCD (phospho-HP1αCD), HP1βCD, and HP1γCD and for the unmodified H3 peptide with HP1αCD and phospho-HP1αCD. Error bars represent the ranges for the standard deviations of the results of three independent measurements. (G) The dissociation constants (Kd) measured by fluorescence anisotropy.
Since the N-terminal serine residues closely resemble the consensus target sequence of casein kinase II (CK2) (Fig. 3B) (40), we first examined whether the N-terminal serine residues of HP1α could be phosphorylated by CK2 in vitro (see Materials and Methods). The incubation of recombinant HP1α (HP1αWT) with CK2 led to a clear band shift, and two distinct bands (major and minor) were detected using phos-tag-PAGE (Fig. 3C, WT and CK2+). The shift of the major band was completely abolished when the S11-14A mutation was introduced into the recombinant HP1α (Fig. 3C, S11-14A), showing that the N-terminal serine residues of HP1α were phosphorylated by CK2 in vitro. The presence of the CK2-mediated phosphorylation at the N-terminal serine residues was also confirmed by MS/MS analysis (Fig. 3D and data not shown). This analysis further revealed that S97 phosphorylation mainly contributed to the minor band shift, which is fully consistent with the fact that S97 is located within a consensus CK2 target sequence (Fig. 2C). While elucidation of the in vivo involvement of CK2 in HP1α phosphorylation remains elusive, we used this in vitro phosphorylated HP1α for further analyses.
Unphosphorylated recombinant HP1α bound to H3K9me3 with a moderate affinity and bound negligibly to the unmodified H3 (Fig. 1B and 3E). Notably, the CK2-mediated phosphorylation strongly improved the binding of HP1α to H3K9me3 (Fig. 3E, WT+CK2) to a level similar to that of the rHP1α purified from NIH 3T3 cells (see Fig. 3E, rHP1α [NIH 3T3]). CK2 treatment had little or no effect on the binding affinity of the HP1αS11-14A mutant, confirming that the N-terminal phosphorylation, but not the minor hinge phosphorylation(s), promoted the H3K9me3 binding of HP1α. Intriguingly, the CK2-mediated phosphorylation also increased the affinity of HP1α for unmodified H3 (Fig. 3E, WT+CK2). Although the levels differed, weak binding to unmodified H3 was also observed for the HP1α purified from NIH 3T3 cells (Fig. 1D and 3E). This implied a distinct role of HP1α's N-terminal phosphorylation in its binding to histone H3 (see below).
To confirm the effect of the N-terminal phosphorylation, we monitored the affinity of CK2-phosphorylated HP1αCD for the H3K9me3 peptide by fluorescence anisotropy measurements (18) (Fig. 3F). By MS/MS analysis, we confirmed that the N-terminal phosphorylation pattern of the CK2-phosphorylated HP1αCD was similar to that of CK2-phosphorylated full-length HP1α (Fig. 3D). As seen with the results of the pulldown assays, the unphosphorylated HP1αCD had an affinity for the H3K9me3 peptide 4 to 6 times lower than that of HP1βCD or HP1γCD (Fig. 3F and G). CK2-mediated N-terminal phosphorylation increased the affinity of the HP1αCD for the H3K9me3 peptide to a level comparable to that of the other HP1 CDs (Fig. 3F and G, Phos-HP1αCD). Taken together, these results indicated that the N-terminal phosphorylation of HP1α enhanced the binding of HP1α to H3K9me3. Note that the increased affinity of phosphorylated HP1α for unmodified H3 was also confirmed in this experiment (Fig. 3F), although the affinity was weak, and it was difficult to obtain a reliable Kd value.
The N-terminal phosphorylation and the CD make distinct contributions to HP1α's binding to H3K9me3.
The N-terminal serine residues of HP1α that undergo phosphorylation are located next to a cluster of acidic amino acids (composed of amino acids E and D), and additional acidic amino acids are present in the corresponding region of HP1β and HP1γ (Fig. 4A). To test whether phosphorylated serine plays a role similar to that of the acidic amino acids, we replaced the N-terminal serine residues (S11-14) of recombinant HP1α with glutamic acid (E) and tested its binding affinity for H3K9me3. Although the S11-14E mutation indeed increased HP1α's affinity for H3K9me3 compared with the unphosphorylated HP1αWT, HP1αS11-14E did not perfectly mimic the phosphorylated HP1αWT (Fig. 4B; compare WT + CK2 and S11-14E). A clear difference was also seen in its binding to the unmodified H3 tail. These results suggested that the phosphorylated serine and acidic amino acids are functionally related but that they have distinct impacts on HP1α's interaction with the histone H3 tail.
FIG. 4.
HP1α's N-terminal phosphorylation and CD make distinct contributions to its H3K9me3 binding. (A) N-terminal amino acid sequences of the mouse HP1 isoforms (Mm_HP1α, Mm_HP1β, and Mm_HP1γ), Drosophila HP1a (Dm_HP1), and S. pombe Swi6 (Sp_Swi6). Acidic amino acids E and D are shown in red, and the phosphorylatable serine residues are shown in blue. The gray box indicates the amino acid residues of the conserved CD. (B) In vitro pulldown assays of mutant HP1αs. Mutant HP1αs (S11-14A and S11-14E) were prepared, and their levels of binding capacity for the H3unmod. and H3K9me3 peptides were compared to those of wild-type HP1α with or without N-terminal phosphorylation. (C) Amino acid sequences of the mouse HP1 chromodomains. Residues that are conserved in HP1β and HP1γ, but not in HP1α, and that could change the amino acid's properties are indicated by boxes and a different color as follows: red, basic amino acids; blue, acidic amino acids; green, polar amino acids. The positions of secondary structure elements are indicated by green arrows (β-strands) and an orange cylinder (α-helix). (D) Schematic drawing of mutant HP1 proteins: HP1α with the CD mimicking HP1βCD (HP1α-CDβ*) and HP1β containing the CD mimicking HP1αCD (HP1β-CDα*). Black and gray boxes represent the conserved CD and CSD, respectively. The positions of amino acids mutated in this study are indicated. (E) In vitro pulldown assays of mutant HP1s. Mutant HP1αs (HP1α-CDβ* and HP1β-CDα*) were prepared, and their levels of binding capacity for the H3unmod. and H3K9me3 peptides were compared to those of wild-type HP1.
We next examined how HP1β and HP1γ achieve their strong binding to H3K9me3 without the contribution of the N-terminal phosphorylation. In comparing the amino acid sequences of the HP1-CDs (Fig. 4C), we introduced a combination of amino acid substitutions to produce a mutant HP1α with the CD that mimics HP1βCD (HP1α-CDβ*) (Fig. 4D) and tested its binding to H3K9me3 (Fig. 4E). The mutant HP1α-CDβ* showed stronger binding to H3K9me3 than wild-type HP1α (Fig. 4E). On the other hand, a mutant HP1β with the CD mimicking HP1αCD (HP1β-CDα*) showed weaker binding to H3K9me3 than wild-type HP1β (Fig. 4E). These mutations, however, had little effect on the binding of the HP1s to unmodified H3. Together, these results suggest that the binding of HP1α to H3K9me3 is mediated by distinct contributions of both the N-terminal phosphorylation (or acidic residues, to some extent) and the CD.
The N-terminal phosphorylation of HP1α enhances its binding to the histone H3 tail.
To confirm the effect of HP1α's N-terminal phosphorylation on its H3K9me binding, we purified the unphosphorylatable mutant HP1αS14A from NIH 3T3 cells by the use of a 6×His tag (Fig. 5A) and compared its ability to bind to H3K9me3 with that of the N-terminally phosphorylated HP1αWT (Fig. 5B). As seen with the results of in vitro phosphorylation (Fig. 3E), the HP1αS14A showed weaker binding to H3K9me3 compared with the HP1αWT (Fig. 5B, H3K9me3). In addition, HP1αS14A exhibited a lower affinity for the unmodified H3 (Fig. 5B, H3unmod.). This finding confirmed the results obtained using in vitro-phosphorylated HP1α (Fig. 3E) and raised the possibility that the N-terminal phosphorylation somehow facilitates HP1α's interaction with the histone H3 tail rather than simply increasing its affinity for H3K9me3.
FIG. 5.
N-terminal mutation influences the initial binding, but not the stability, of the HP1α-H3K9me3 interaction. (A) Silver staining image of the immunopurified 6×His-HP1α. 6×His-tagged HP1αWT and mutant HP1αS14A were transiently expressed in NIH 3T3 cells and purified using anti-His-tag agarose. The immunopurified proteins were resolved by 8 to 15% SDS-PAGE and visualized by silver staining. (B) In vitro pulldown assays of immunopurified HP1α with histone H3 peptides. The amount of immunopurified HP1α was estimated by comparison with bacterially expressed recombinant HP1α, and ∼1-pmol-equivalent 6×His-HP1α purified from NIH 3T3 cells was examined in a pulldown assay with H3unmod. and H3K9me3 peptides as described for Fig. 1B. (C) Diagram showing the positions of the amino acid substitutions introduced into the histone H3 peptide. (D) In vitro pulldown assays of immunopurified HP1α with mutant histone H3 peptides. H3K9me3 or K9-unmodified H3 peptide containing amino acid substitutions at K14 and K18 was examined. (E) Quantitative pulldown assays of in vivo-phosphorylated HP1αWT and the unphosphorylatable S14A mutant with H3K9me3 peptide. Recombinant 6×His-HP1α proteins (1 pmol) were incubated with increasing molar excesses of H3K9me3-peptide beads. Input and bound fractions were analyzed by immunoblotting using an anti-HP1α antibody. (F) Quantitative elution assays of wild-type and mutant HP1α. 6×His-HP1αWT and 6×His-HP1αS14A were first bound to H3K9me3 beads, and then the stability of the binding was tested in the presence of increasing amounts of free H3K9me3 peptide.
The previously solved structure of the HP1β CD complexed with a histone H3 peptide (amino acids 1 to 13) implies that the region just N terminal to the CD lies close to the histone H3 tail, somewhere C terminal to H3K9 (23, 37). Histone H3 contains several positively charged residues in its distal tail region (Fig. 5C); hence, it is possible that the phosphorylated serine residues of HP1α enhance its interaction with the distal tail region of H3. Based on this idea, we investigated the effect of amino acid substitutions in the H3 tail on its interaction with HP1α.
Mutant H3K9me3 peptides (amino acids 1 to 20), in which lysine 14 and lysine 18 were replaced with alanine (A) or glutamine (Q [mimicking acetylated lysine]) (Fig. 5C), were tested for their ability to interact with the HP1αWT or HP1αS14A by a pulldown assay (Fig. 5D). The amino acid substitutions reduced the interaction with both the HP1αWT and HP1αS14A, but they impaired more severely the H3K9me3 peptide's interaction with N-terminally phosphorylated HP1αWT, as was apparent from the increased and decreased levels of the unbound (U) and bound (B) fractions, respectively (Fig. 5D, HP1αWT and HP1αS14A). We also performed a similar assay using K9-unmodified H3 peptide with the K-to-Q substitutions. The binding of HP1αWT to unmodified H3 peptide was partially perturbed by the substitution, suggesting that K14 and K18 of histone H3 contributed to the interaction with N-terminally phosphorylated HP1αWT. Since HP1αWT maintained residual binding activity against the mutated H3 tail, another tail region(s) may also contribute to the interaction. These data suggest that the phosphorylated N-terminal serine residues and possibly the adjacent acidic amino acid residues of HP1α facilitate its interaction with the H3 tail in concert with methylated K9, presumably by interacting with the distal tail region of H3.
The N-terminal phosphorylation of HP1α enhances its initial binding to H3K9me3.
To investigate further the role of the N-terminal phosphorylation of HP1α, we analyzed HP1α's binding to H3K9me3 by a titration assay (Fig. 5E). HP1αWT and HP1αS14A were assayed with various concentrations of H3K9me3. As seen with the results of the simple pulldown assay, HP1αWT could stably interact with the H3K9me3 peptide at a lower peptide concentration than HP1αS14A (Fig. 5E), suggesting that N-terminal phosphorylation enhances the initial binding of HP1α to H3K9me3.
We next examined the stability of HP1α's binding to H3K9me3 by a competition assay (Fig. 5F). In this experiment, the wild-type or mutant HP1α was first incubated with H3K9me3-cross-linked beads, and the binding stability was examined by adding an excess amount of free H3K9me3 peptide. Both wild-type HP1α and mutant HP1α stably bound H3K9me3, even in the presence of a large molar excess of competitor peptide (Fig. 5F, ×100). The N-terminal phosphorylation appeared to weakly enhance the release of HP1α from H3K9me3 beads (Fig. 5F, lane ×1,000). These results indicate that, although HP1α's N-terminal phosphorylation plays a role in its initial binding to H3K9me, it might be dispensable for maintaining the stable interaction between the CD and H3K9me3.
HP1α's N-terminal phosphorylation is required for its proper heterochromatic localization.
To gain insight into the physiological roles of HP1α phosphorylation, wild-type or mutant HP1α was transiently expressed as an EGFP fusion protein in NIH 3T3 cells, and its nuclear localization was examined by fluorescence microscopy. As previously observed (20, 45), EGFP-fused wild-type HP1α showed punctate nuclear signals and largely colocalized with the DAPI-dense heterochromatic regions (Fig. 6A, WT). This localization pattern of EGFP-HP1α is consistent with that of endogenous HP1α detected by specific antibodies (data not shown) (34). The S14A mutation in the N-terminal phosphorylation sites, however, resulted in a marked decrease in the cell population with clear heterochromatic localization (Fig. 6A, S14A).
FIG. 6.
Disruption of HP1α N-terminal phosphorylation leads to defects in the heterochromatic localization of HP1α. (A) Examples of images of NIH 3T3 cells transfected with EGFP-fused wild-type or mutant (S14A) HP1α. Cells were fixed 24 h after transfection, counterstained with DAPI to visualize nuclei, and observed under a fluorescence microscope. Enlarged images of the cells indicated by white arrows are shown as insets. (B) Representative fluorescence images and line plot profiles of fluorescent intensity showing the three major nuclear localization patterns of EGFP-HP1α. (C) The percentages of cells showing the three nuclear localization patterns for EGFP-HP1α expression as described for panel B. For this analysis, the cells were costained with an anti-phosphohistone H3 antibody, and all the cells with positive results were excluded from the scoring. (D) HP1αWT, but not mutant HP1αS14A, stably bound to the chromatin-enriched nuclear fraction. NIH 3T3 cells expressing wild-type or mutant (S14A) HP1α were transfected with control siRNA or siRNA directed against HP1α, and whole-cell lysate (W), soluble supernatant (S), and chromatin-enriched pellet (P) fractions were obtained. The proteins in each fraction were resolved by SDS-PAGE and analyzed by immunoblotting. (E) Coimmunoprecipitation of histone H3 nucleosomes with FLAG-HP1α. Whole-cell lysates of NIH 3T3 cells stably expressing FLAG-HP1αWT or FLAG-HP1αS11-14A were treated with micrococcal nuclease, and FLAG-HP1α and associated histone H3 nucleosomes were precipitated using anti-FLAG M2 beads. The precipitated fractions were analyzed by immunoblotting using the antibodies against the histone H3 C terminus, H3K9me3, or H3K4me3.
To quantify the heterochromatic localization, we categorized the HP1α localization into three patterns, strong heterochromatic (strong), weak heterochromatic (weak), and diffused localization (Fig. 6B; see also Materials and Methods for details of the categorization), and the number of cells that fell into each category was scored (Fig. 6C). Since mammalian HP1 delocalizes from the chromatin during the G2/M phase of the cell cycle (18, 22), phospho-H3S10-positive cells were excluded from the scoring (data not shown). When wild-type HP1α was expressed, more than 90% of the cells showed strong heterochromatic localization (Fig. 6C). In contrast, this percentage was markedly reduced in the cell populations expressing HP1α with N-terminal mutations (S14A, 42.9%; S11-14A, 49.4%), and nearly half of the cells in these populations exhibited weak heterochromatic or diffused localization of the mutant HP1α species (Fig. 6C). These observations suggested that phosphorylation of the N-terminal serine residues is required for the proper heterochromatic localization of HP1α. Since reduced heterochromatic localization was observed for HP1α with the S11-13A mutation (Fig. 6C), not only S14 phosphorylation but also neighboring phosphorylation(s) of the other N-terminal serine residues are likely to contribute to targeting of HP1α to heterochromatin.
Although we mapped S93 as the major metaphase-specific phosphorylation site, HP1α with a hinge mutation (S93A or S93E) did not have a noticeable effect on HP1α's heterochromatic localization in our mitotic cell-excluding scoring (data not shown). In addition, these mutant HP1αs did not show any obvious defect in localization, even in mitotic cells (data not shown). Further studies are needed to clarify the physiological role of this S93 phosphorylation in HP1α function.
To verify the role of N-terminal phosphorylation in the chromatin binding of HP1α, we performed a chromatin fractionation assay. NIH 3T3 cells expressing FLAG-tagged wild-type or mutant HP1α were fractionated to obtain a soluble fraction and chromatin-enriched pellet, and the chromatin-bound HP1α was analyzed by Western blotting (Fig. 6D). In this assay, nearly 90% of the endogenous HP1α and more than half of the exogenously expressed HP1αWT were detected in the chromatin-bound fraction (P) (Fig. 6D [WT, control]). In contrast, most of the mutant HP1αS14A was present in the soluble fraction (S), and only a minor portion was detected in the chromatin-bound fraction (P) (Fig. 6D [S14A, control]). The HP1αS14A appeared to bind to chromatin via an interaction with endogenous HP1α, because the depletion of endogenous HP1α clearly abolished the chromatin-bound mutant HP1α (Fig. 6D [siHP1α, S14A]).
We further tested whether HP1αWT bound more efficiently to H3K9me3-containing nucleosomes than HP1αS11-14A in vivo by an immunoprecipitation assay (Fig. 6E). Approximately three times more histone H3 was coimmunoprecipitated with FLAG-HP1αWT as with FLAG-HP1αS11-14A, and K9-trimethylted H3 was preferentially enriched in the precipitated fraction compared with K4-trimethylated H3 (Fig. 6E). Together, these results further confirmed that HP1α's N-terminal phosphorylation has an important role in its chromatin- and histone H3-binding activity.
Expression of mutant HP1α leads to increased chromosomal instability.
We next examined the roles of HP1α phosphorylation in chromosomal stability, since it has been reported that HP1 homologues contribute to genome stability in a diverse range of eukaryotic cells (2, 7, 13, 15). We generated NIH 3T3-derived clones that stably expressed either FLAG-tagged HP1αWT or HP1αS11-14A (Fig. 7A) and examined their chromosome configurations. Among the stable clones expressing HP1αWT, most cells showed a normal chromosome configuration (Fig. 7B). Although a minor population of cells (8 to 13%) contained some sort of chromosomal abnormality, this proportion was comparable to that of the parental NIH 3T3 cells (13.1%) (Fig. 7F). In contrast, a much higher proportion (18 to 68%) of cells exhibited aberrant chromosomes in the stable clones expressing mutant HP1αS11-14A (Fig. 7F). Moreover, these HP1αS11-14A-expressing cells often contained multiple abnormal chromosomes with various aberrations (Fig. 7C to E), in contrast to the parental NIH 3T3 or HP1αWT-expressing cells, which usually contained only a single abnormal chromosome when they harbored such a defect (data not shown). These results indicate that the N-terminal phosphorylation of HP1α is necessary for the maintenance of genome integrity. Considering that the unphosphorylatable mutant HP1α showed defects in heterochromatin localization, it is possible that the mutant HP1α formed a heterodimer with endogenous HP1α or other HP1s and thereby hampered their function at centromeric or telomeric heterochromatin.
FIG. 7.
Prolonged expression of unphosphorylatable mutant HP1α leads to increased genome instability. (A) Western blot analysis of NIH 3T3-derived clones stably expressing FLAG-HP1αWT or FLAG-HP1αS11-14A. (B) Metaphase chromosome spread of FLAG-HP1αWT-expressing NIH 3T3 cells, showing the normal situation. (C to E) Metaphase chromosomes of FLAG-HP1αS11-14A-expressing NIH 3T3 cells exhibited a variety of chromosomal aberrations, including diplochromosomes (C), a circular chromosome (D), and long fused chromosomes (E). White arrowheads indicate aberrant chromosomes. (F) Percentages of cells containing aberrant chromosomes. Metaphase chromosomes of NIH 3T3-derived cell lines expressing FLAG-HP1αWT or FLAG-HP1αS11-14A were examined under a microscope. The dotted line indicates the percentage of parental NIH 3T3 cells exhibiting abnormal chromosome configurations. (G) Proposed model for the role of N-terminal phosphorylation in the targeting of HP1α. We propose that the N-terminal phosphorylation on HP1α facilitates its initial binding to K9-methylated histone H3 and chromatin. Once it is localized, HP1α's CD stabilizes the interaction.
DISCUSSION
In this report, we demonstrate that the N-terminal phosphorylation of HP1α enhances its binding to H3K9me and is essential for proper interaction between HP1α and heterochromatin. Our detailed biochemical analyses further revealed that the N-terminal phosphorylation promotes the initial binding of HP1α to H3K9me3. This study provides novel insights into how the CD function is modulated by accompanying posttranslational modifications.
The disruption of HP1α's N-terminal phosphorylation considerably reduced its affinity for H3K9me and impaired its targeting to heterochromatin. The phosphorylated HP1α appeared to interact with the histone H3 tail in a manner independent, at least to a certain degree, of H3K9me3, suggesting that the N-terminal phosphorylation induces and/or secures the interaction between HP1α and histone H3 by providing a binding site other than the CD. This idea is supported by earlier studies suggesting that the mere binding of HP1-CD to methyl H3K9 may not be sufficient to achieve a stable or rapid interaction between HP1α and chromatin (43) and proposing a number of additional modules for the HP1-chromatin interaction, including the interaction of HP1 with RNA (31), auxiliary factors (14), or the globular domain of histone H3 (36) or the involvement of the hinge and CSD (6, 32). The N-terminal phosphorylation of HP1α thus provides another layer of regulation for the HP1-chromatin interaction.
Our peptide competition data implied that the N-terminal phosphorylation is more important for the “initial” contact of HP1α with the histone H3 tail and that it might contribute little to the stabilization of the HP1α-H3K9me3 interaction (Fig. 5E and F). The phosphorylated N terminus of HP1α might act as a “hook” to bring the CD and methyl H3K9 into close proximity for their rapid interaction (Fig. 7G). From the previously solved structure of the HP1-CD complexed with H3K9me peptide (23, 37), the N-terminal region of HP1α is assumed to interact with the histone H3 tail somewhere C terminal to H3K9, and our data support this idea (Fig. 5D). Alternatively, the N-terminal phosphorylation may influence the structure of the CD. Indeed, circular dichroism spectra obtained from the phosphorylated HP1αCD and unphosphorylated HP1αCD suggested that their secondary structures are very similar but not identical (data not shown). It would be very interesting to determine the three-dimensional structure of the phosphorylated HP1α N terminus complexed with H3K9me peptide and investigate how the N-terminal phosphorylation affects the HP1α-chromatin interaction.
The N-terminal phosphorylation sites closely resemble the consensus target sequence of CK2 (Fig. 3B), and we demonstrated here that they could be phosphorylated by CK2 in vitro (Fig. 3C). As shown in previous reports from Drosophila HP1a and S. pombe Swi6 studies (42, 47), it is likely that CK2 is the kinase responsible for the N-terminal phosphorylation of HP1α in mouse and human cells. However, we could not obtain direct evidence for the involvement of CK2 in the N-terminal phosphorylation of HP1α: 4,5,6,7-tetrabromobenzotriazole (TBB), a potent CK2-specific inhibitor, did not clearly block the phosphorylation of endogenous HP1α, and knockdown of CK2α/α′ catalytic subunits did not have a marked effect on the HP1α phosphorylation (data not shown). Thus, although CK2 is the most likely kinase candidate for the N-terminal phosphorylation of HP1α, it remains possible that some other kinase(s) is responsible for it in vivo.
Phosphorylation is one of the most common mechanisms for regulating the localization and functions of HP1 family proteins. For example, HP1γ's euchromatic localization and transcriptional activator functions coincide with the phosphorylation of its hinge region (29). In this study, we showed that HP1α undergoes similar phosphorylation in its hinge region, but this phosphorylation occurs in a cell cycle-dependent manner and does not have obvious effects on HP1α's nuclear localization (data not shown). The role of the hinge phosphorylation, therefore, may differ depending on the HP1 isoform or homologue.
In studies of Drosophila, it has been demonstrated that an N-terminal serine (S15) of HP1a is phosphorylated by CK2 and that this phosphorylation plays a role in heterochromatin-mediated silencing (47, 48). However, it remains unresolved how S15 contributes to HP1a's chromatin targeting function. Since S15 also lies close to the CD (Fig. 4A), it is likely that its phosphorylation plays a similar role to promote the binding of Drosophila HP1a. Indeed, CK2 treatment increased the affinity of Drosophila HP1a for H3K9me3 in a manner similar to that observed for mouse HP1α (data not shown). Thus, the phosphorylation-mediated regulation of CD function appears to be conserved in evolutionarily distant species. In S. pombe, Swi6, one of the HP1 family proteins, also undergoes CK2-mediated phosphorylation in vivo (42). In that case, however, the phosphorylation does not appear to alter the chromatin-binding activity of Swi6 but rather modulates the interaction between Swi6 and other trans-acting factors.
Our data suggest that the primary role of the N-terminal phosphorylation is the efficient targeting of HP1α to H3K9me-enriched heterochromatic regions. The initial targeting step appears to be critical to the HP1α's function in vivo. The unphosphorylatable mutant HP1α showed defects in heterochromatic localization (Fig. 6), and the overexpression of this mutant caused chromosomal abnormalities as a dominant effect (Fig. 7). It was recently reported that the expression state of HP1α, but not of HP1β or HP1γ, is tightly linked with tumorigenesis (7). Although we have not yet identified a situation in which the N-terminal region of HP1α becomes dephosphorylated in vivo, it is possible that the deregulation of HP1α phosphorylation further accelerates chromosomal instability. Exploring the regulatory mechanisms of HP1α phosphorylation may elucidate the functional relationship between HP1 proteins and chromosome integrity.
Acknowledgments
We thank D. Goto, N. Suka, M. Oki, A. Hayashi, and M. Sadaie for critical reading, other fellow laboratory members at the RIKEN Center for Developmental Biology (CDB) for helpful discussions, and S. Seno for excellent secretarial work.
None of us has a financial interest related to this work.
This research was supported by Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 18 January 2011.
REFERENCES
- 1.Aasland, R., and A. F. Stewart. 1995. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 23:3168-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aucott, R., et al. 2008. HP1-beta is required for development of the cerebral neocortex and neuromuscular junctions. J. Cell Biol. 183:597-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayoub, N., A. D. Jeyasekharan, J. A. Bernal, and A. R. Venkitaraman. 2008. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature 453:682-686. [DOI] [PubMed] [Google Scholar]
- 4.Bannister, A. J., et al. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 5.Brasher, S. V., et al. 2000. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 19:1587-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheutin, T., et al. 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299:721-725. [DOI] [PubMed] [Google Scholar]
- 7.De Koning, L., et al. 2009. Heterochromatin protein 1alpha: a hallmark of cell proliferation relevant to clinical oncology. EMBO Mol. Med. 1:178-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinant, C., and M. S. Luijsterburg. 2009. The emerging role of HP1 in the DNA damage response. Mol. Cell. Biol. 29:6335-6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eissenberg, J. C., and S. C. Elgin. 2000. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 10:204-210. [DOI] [PubMed] [Google Scholar]
- 10.Eissenberg, J. C., Y. W. Ge, and T. Hartnett. 1994. Increased phosphorylation of HP1, a heterochromatin-associated protein of Drosophila, is correlated with heterochromatin assembly. J. Biol. Chem. 269:21315-21321. [PubMed] [Google Scholar]
- 11.Eissenberg, J. C., et al. 1990. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 87:9923-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eissenberg, J. C., G. D. Morris, G. Reuter, and T. Hartnett. 1992. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics 131:345-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekwall, K., et al. 1995. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 269:1429-1431. [DOI] [PubMed] [Google Scholar]
- 14.Eskeland, R., A. Eberharter, and A. Imhof. 2007. HP1 binding to chromatin methylated at H3K9 is enhanced by auxiliary factors. Mol. Cell. Biol. 27:453-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fanti, L., G. Giovinazzo, M. Berloco, and S. Pimpinelli. 1998. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell 2:527-538. [DOI] [PubMed] [Google Scholar]
- 16.Festenstein, R., et al. 2003. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science 299:719-721. [DOI] [PubMed] [Google Scholar]
- 17.Festenstein, R., et al. 1999. Heterochromatin protein 1 modifies mammalian PEV in a dose- and chromosomal-context-dependent manner. Nat. Genet. 23:457-461. [DOI] [PubMed] [Google Scholar]
- 18.Fischle, W., et al. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438:1116-1122. [DOI] [PubMed] [Google Scholar]
- 19.Grewal, S. I., and S. Jia. 2007. Heterochromatin revisited. Nat. Rev. Genet. 8:35-46. [DOI] [PubMed] [Google Scholar]
- 20.Hayakawa, T., T. Haraguchi, H. Masumoto, and Y. Hiraoka. 2003. Cell cycle behavior of human HP1 subtypes: distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase. J. Cell Sci. 116:3327-3338. [DOI] [PubMed] [Google Scholar]
- 21.Hiragami, K., and R. Festenstein. 2005. Heterochromatin protein 1: a pervasive controlling influence. Cell. Mol. Life Sci. 62:2711-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirota, T., J. J. Lipp, B. H. Toh, and J. M. Peters. 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438:1176-1180. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs, S. A., and S. Khorasanizadeh. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295:2080-2083. [DOI] [PubMed] [Google Scholar]
- 24.James, T. C., et al. 1989. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol. 50:170-180. [PubMed] [Google Scholar]
- 25.James, T. C., and S. C. Elgin. 1986. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 6:3862-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinoshita, E., E. Kinoshita-Kikuta, K. Takiyama, and T. Koike. 2006. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell Proteomics 5:749-757. [DOI] [PubMed] [Google Scholar]
- 27.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 28.LeRoy, G., et al. 2009. Heterochromatin protein 1 is extensively decorated with histone code-like post-translational modifications. Mol. Cell Proteomics 8:2432-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lomberk, G., D. Bensi, M. E. Fernandez-Zapico, and R. Urrutia. 2006. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat. Cell Biol. 8:407-415. [DOI] [PubMed] [Google Scholar]
- 30.Maison, C., and G. Almouzni. 2004. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5:296-304. [DOI] [PubMed] [Google Scholar]
- 31.Maison, C., et al. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30:329-334. [DOI] [PubMed] [Google Scholar]
- 32.Meehan, R. R., C. F. Kao, and S. Pennings. 2003. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. EMBO J. 22:3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meshorer, E., et al. 2006. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell 10:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minc, E., Y. Allory, H. J. Worman, J. C. Courvalin, and B. Buendia. 1999. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma 108:220-234. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen, A. L., et al. 2001. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell 7:729-739. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen, P. R., et al. 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416:103-107. [DOI] [PubMed] [Google Scholar]
- 38.Olsen, J. V., et al. 2006. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127:635-648. [DOI] [PubMed] [Google Scholar]
- 39.Paro, R., and D. S. Hogness. 1991. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl. Acad. Sci. U. S. A. 88:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinna, L. A. 2002. Protein kinase CK2: a challenge to canons. J. Cell Sci. 115:3873-3878. [DOI] [PubMed] [Google Scholar]
- 41.Sadaie, M., K. Shinmyozu, and J. Nakayama. 2008. A conserved SET domain methyltransferase, Set11, modifies ribosomal protein Rpl12 in fission yeast. J. Biol. Chem. 283:7185-7195. [DOI] [PubMed] [Google Scholar]
- 42.Shimada, A., et al. 2009. Phosphorylation of Swi6/HP1 regulates transcriptional gene silencing at heterochromatin. Genes Dev. 23:18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh, P. B., and S. D. Georgatos. 2002. HP1: facts, open questions, and speculation. J. Struct. Biol. 140:10-16. [DOI] [PubMed] [Google Scholar]
- 44.Smothers, J. F., and S. Henikoff. 2000. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol. 10:27-30. [DOI] [PubMed] [Google Scholar]
- 45.Sugimoto, K., H. Tasaka, and M. Dotsu. 2001. Molecular behavior in living mitotic cells of human centromere heterochromatin protein HPLalpha ectopically expressed as a fusion to red fluorescent protein. Cell Struct. Funct. 26:705-718. [DOI] [PubMed] [Google Scholar]
- 46.Thiru, A., et al. 2004. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 23:489-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao, T., and J. C. Eissenberg. 1999. Phosphorylation of heterochromatin protein 1 by casein kinase II is required for efficient heterochromatin binding in Drosophila. J. Biol. Chem. 274:15095-15100. [DOI] [PubMed] [Google Scholar]
- 48.Zhao, T., T. Heyduk, and J. C. Eissenberg. 2001. Phosphorylation site mutations in heterochromatin protein 1 (HP1) reduce or eliminate silencing activity. J. Biol. Chem. 276:9512-9518. [DOI] [PubMed] [Google Scholar]









