Abstract
In the budding yeast Saccharomyces cerevisiae, the structure and function of telomeres are maintained by binding proteins, such as Cdc13-Stn1-Ten1 (CST), Yku, and the telomerase complex. Like CST and Yku, telomerase also plays a role in telomere protection or capping. Unlike CST and Yku, however, the underlying molecular mechanism of telomerase-mediated telomere protection remains unclear. In this study, we employed both the CDC13-EST1 fusion gene and the separation-of-function allele est1-D514A to elucidate that Est1 provided a telomere protection pathway that was independent of both the CST and Yku pathways. Est1's ability to convert single-stranded telomeric DNA into a G quadruplex was required for telomerase-mediated telomere protection function. Additionally, Est1 maintained the integrity of telomeres by suppressing the recombination of subtelomeric Y′ elements. Our results demonstrate that one major functional role that Est1 brings to the telomerase complex is the capping or protection of telomeres.
Telomeres are protein-DNA complexes found at the ends of linear chromosomes in eukaryotes. Their complete replication facilitates continuous cell division and prevents cells from undergoing senescence (33, 50). Telomeric DNA typically consists of simple repetitive double-stranded region and a protruding single-stranded G-rich 3′ overhang (60). In most eukaryotes, telomeric DNA is maintained either through the telomerase pathway (20) or through a homologous recombination pathway (30, 56). In the budding yeast Saccharomyces cerevisiae, each chromosome usually includes ∼300 ± 75 bp of C1-3A/TG1-3 telomeric DNA as well as Y′ and X telomere-associated repeats (62).
The yeast telomerase holoenzyme is composed of the catalytic reverse transcriptase Est2, the RNA template Tlc1, and the regulatory subunits Est1 and Est3 (27, 31). Est2 and Tlc1 together form the telomerase core enzyme that exerts nucleotide addition activity in vitro (29, 49). Est1 and Est3 are required for telomerase activity in vivo, and deletion of either subunit causes telomere shortening and cellular senescence (27, 28). Est1 is expressed in S phase of the cell cycle and binds specifically to single-stranded TG DNA (38, 59) and a bulged stem-loop in Tlc1 RNA to associate with the telomerase core enzyme (47, 51). Additionally, Est1 recruits telomerase to telomeres via interaction with the single-stranded telomeric DNA binding protein Cdc13 (8, 43). Furthermore, Est1 can convert single-stranded telomeric DNA into a G quadruplex to activate telomerase (63). Est3 also associates with telomerase, but its precise function is not well understood (23, 26, 61).
When an EST gene is absent, all of the telomerase-deficient cells show progressive telomere shortening and most of the cells are subjected to cell cycle arrest at the G2/M-phase boundary and cell death (31). A small number of “survivor” cells are able to overcome senescence by utilizing a RAD52-dependent homologous recombination pathway to maintain telomeres (30, 56). Type I survivors have tandem arrays of the subtelomeric Y′ element, whereas type II survivors have very long terminal tracts of C1-3A/TG1-3 DNA (30, 56). Since the telomere ends share similarities with double-strand breaks (DSBs), there is the potential for telomeres to be viewed as DNA damage, thereby triggering cell cycle arrest and/or repair using the two recombination-dependent survivor pathways described above. Telomeres do not, however, elicit DNA damage signaling, presumably because telomeres are protected by specific binding proteins that help distinguish themselves from normal DNA damage. For example, Cdc13, Stn1, and Ten1 form a complex (CST complex) that “protects” or “caps” telomeric DNA from being seen as DNA damage (17, 18, 37, 44). The Yku complex, which contains the YKU70 and YKU80 gene products, is also required for telomere protection (4, 11). Like the cdc13-1, stn1-13, and ten1-31 mutants, yku70Δ and yku80Δ cells accumulate long single-stranded G-rich overhangs and have growth defects when the cells are cultured at a higher than normal restrictive temperature of 36°C (17-19, 32).
Telomere shortening caused by telomerase deletion in S. cerevisiae increases the amount of single-stranded telomere DNA (21) and triggers a DNA damage response (35). In Candida albicans, the deletion of either EST2 or TER1 also increases the amount of telomeric single-stranded DNA (ssDNA) (22). Additionally, in S. cerevisiae, the combinatorial deletion of YKU and any telomerase component, such as EST1, EST2, EST3, or TLC1, generates a synthetic lethal phenotype (19, 36, 41). Together, these lines of evidence indicate that in addition to elongating telomeric DNA, telomerase also participates in telomere “capping” or “protection” via a molecular mechanism that has not yet been elucidated. In this article, we provide experimental evidence that demonstrates that loss of telomere protection is the source of the synthetic lethal phenotype of EST1 and YKU double mutant cells. Additionally, we demonstrate that the G-quadruplex promotion activity of Est1 plays an essential role in telomere protection. Finally, we show that the deprotection of telomeres that results from mutation of EST1 leads to subtelomeric Y′-element amplification, suggesting Est1 also plays a role in promoting the telomerase pathway by simultaneously suppressing telomere recombination.
MATERIALS AND METHODS
Plasmids and strains.
Yasumasa Tsukamoto, Daniel Gottschling, Virginia Zakian, and Victoria Lundblad kindly provided CEN plasmids pRS316-EST1, pRS316-EST2, pRS415-CDC13-EST2 (pVL1107), pRS415-CDC13-EST1 (pVL1091), pDBL-EST1, pDBL-CDC13, pRS315-YKU80, pRS315-YKU70, and pRS315-yku80-135i. We constructed CEN plasmids pRS316-CDC13-EST2 and pRS317-CDC-EST2 by inserting CDC13-EST2 into the SalI and NotI sites of the pRS316 and pRS317 vectors, respectively. To construct the point mutation alleles of est1-D514A or est1-60, we performed oligonucleotide-directed point mutagenesis in the plasmids pRS315-EST1, pDBL-EST1, and pVL1091. Information about the strains used in this study is provided in Table 1 (14, 48).
TABLE 1.
Genotypes of the yeast strains used in this study
| Strain | Relevant genotype | Referencea |
|---|---|---|
| Wild type | ||
| YPH499 | MATaura3-52 lys2-801_amber ade2-101_ochre trp1-Δ63 his3-Δ200 leu2-Δ1 | 44 |
| YPH501 | MATa/α ura3-52/ura3-52 lys2-801_amber/lys2-801_amber ade2-101_ochre/ade2-101_ochre trp1-Δ63/trp1-Δ63 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 | 44 |
| cdc13-1 mutants | 14 | |
| TXJ002 | YPH501 EST1/est1Δ::TRP1 KU70/ku70Δ::HIS3/CEN pRS316-EST1/CEN pRS315-EST1 | |
| TXJ003 | YPH501 EST1/est1Δ::TRP1 KU80/ku80Δ::HIS3/CEN pRS316-EST1/CEN pRS315 | |
| TXJ004 | YPH501 EST1/est1Δ::TRP1 KU80/ku80Δ::HIS3/CEN pRS316-EST1/CEN pRS315-EST1 | |
| TXJ005 | YPH501 CDC13/cdc13Δ::ADE2 EST1/est1Δ::TRP1 KU70/ku70Δ::HIS3/CEN pRS316-EST1/CEN pRS415-CDC13-EST2 | |
| TXJ006 | YPH501 CDC13/cdc13Δ::ADE2 EST1/est1Δ::TRP1 KU80/ku80Δ::HIS3/CEN pRS316-EST1/CEN pRS415-CDC13-EST2 | |
| TXJ007 | YPH501 CDC13/cdc13Δ::ADE2 EST1/est1Δ::TRP1 KU80/ku80Δ::HIS3/CEN pRS316-CDC13-EST2/CEN pRS315-ku80-135i | |
| TXJ008 | YPH499 ku70Δ::HIS3 | |
| TXJ009 | YPH499 ku80Δ::HIS3 | |
| TXJ010 | YPH499 ku80Δ::HIS3 est1Δ::TRP1/CEN pRS316-EST1/CEN pRS315-ku80-135i | |
| TXJ011 | YPH499 ku80Δ::HIS3/CEN pRS315-ku80-135i | |
| TXJ012 | YPH499 ku70Δ::HIS3 est1Δ::TRP1 exo1Δ::ADE2/CEN pRS316-EST1 | |
| TXJ013 | YPH499 ku80Δ::HIS3 est2Δ::TRP1/CEN pRS316-EST2 | |
| TXJ014 | YPH501 ku80Δ::HIS3 est1Δ::TRP1 cdc13Δ::ADE2/CEN pRS316-EST1/CEN pRS317-CDC13-EST2/CEN pRS315-EST1 | |
| TXJ015 | YPH501 ku80Δ::HIS3 est1Δ::TRP1 cdc13Δ::ADE2/CEN pRS316-EST1/CEN pRS317-CDC13-EST2/CEN pRS315-est1-D514A | |
| TXJ016 | YPH499 ku80Δ::HIS3 est1Δ::TRP1 cdc13Δ::ADE2/CEN pRS316-EST1/CEN pRS317-CDC13-EST2/CEN pRS317-est1-60 | |
| TXJ017 | YPH501 EST1/est1Δ::TRP1/CEN pRS316-EST1 | |
| TXJ018 | YPH501 EST1/est1Δ::TRP1/CEN pRS316-est1-D514A | |
| TXJ019 | YPH501 EST1/est1Δ::TRP1 RAD50/rad50Δ::HIS3/CEN pRS316-EST1 | |
| TXJ020 | YPH501 EST1/est1Δ::TRP1 RAD50/rad50Δ::HIS3/CEN pRS316-est1-D514A | |
| TXJ021 | YPH501 EST1/est1Δ::TRP1 RAD51/rad51Δ::HIS3/CEN pRS316-EST1 | |
| TXJ022 | YPH501 EST1/est1Δ::TRP1 RAD51/rad51Δ::HIS3/CEN pRS316-est1-D514A |
Unless otherwise indicated with a reference number, all strains were constructed in this study.
Telomere Southern blot.
The genomic DNA was prepared from yeast cells and digested with XhoI. The telomere Southern blot was performed using the TG1-3 probe as previously described (7, 34, 57).
Growth dotting assay on plates.
A single colony for the indicated yeast strains was inoculated into 3 ml yeast extract-peptone-dextrose (YEPD) or selective medium at 30°C to reach saturation and then diluted to an optical density at 600 nm (OD600) of 0.2 and kept in culture for about 4 h until the culture reached an OD600 of 0.35. Cells were plated at 5-fold serial dilutions onto 5′-fluoroorotic acid (5′-FOA) or a selective medium plate for 2 to 3 days at 30°C or 37°C before being photographed.
Single-stranded DNA measurements.
Single-stranded DNA was measured as described previously (3). Cells were harvest at an OD600 of 0.7. Cells were treated with Zymolyase (2.5 μg/μl), which was followed by genomic DNA purification by Qiagen Genomic-tip (catalog no. 10223).
RESULTS
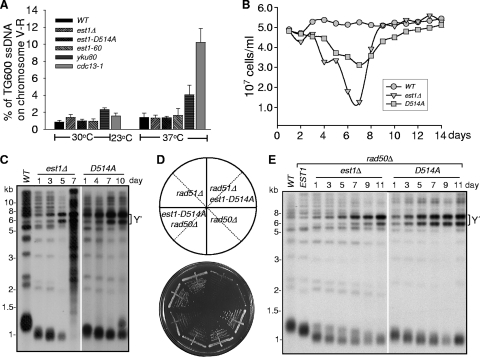
The est1Δ ykuΔ double deletion is synthetic lethal in cells with longer than normal telomeres.
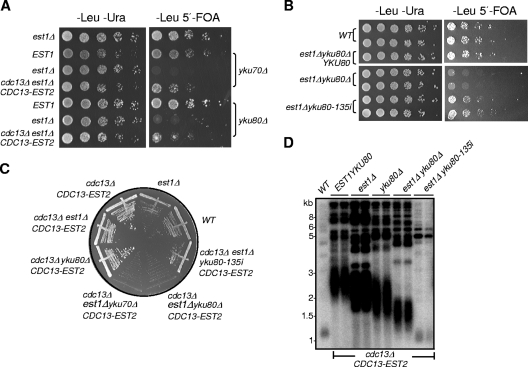
Previous genetic studies revealed that combinatorial mutations of Yku and telomerase components cause synthetic lethality/sickness (9, 19, 36, 41) (Fig. 1 A). One hypothesis to explain the synthetic lethal phenotype is that inactivation of both Yku and telomerase exacerbates the telomere shortening rate and accelerates senescence, thereby manifesting a phenotype of synthetic lethality. To test this hypothesis, we took advantage of a CDC13-EST2 fusion gene whose expression produced very long telomeres when Est1 was present and maintained wild-type (WT) telomere length and cell viability when Est1 was absent (8, 63). We constructed two diploid strains in the YPH501 background, namely, TXJ005 and TXJ006 (Table 1), in which one chromosomal copy of CDC13, EST1, and YKU70 (TXJ005) or YKU80 (TXJ006) was deleted. Then we performed tetrad dissection to obtain different mutant haploid strains, including the cdc13Δ/pRS415-CDC13-EST2, cdc13Δ est1Δ/pRS316-EST1/pRS415-CDC13-EST2, cdc13Δ yku80Δ/pRS415-CDC13-EST2, and cdc13Δ yku80Δ est1Δ/pRS316-EST1/pRS415-CDC13-EST2 (or cdc13Δ yku70Δ est1Δ/pRS316-EST1/pRS415-CDC13-EST2) strains. Due to the presence of EST1 and the pRS415-CDC13-EST2 plasmids in these diploid cells, the telomeres were gradually elongated along consecutive passages and became super-long after several restreaks (data not shown). The loss of the pRS316-EST1 covering plasmid for these experiments was achieved by 5′-FOA selection against cells harboring the URA3 selectable marker on the plasmid (Fig. 1A). The colonies grown on the 5′-FOA plate were immediately restreaked on a YEPD plate (Fig. 1C) or collected and subjected to Southern blot analysis (Fig. 1D). After eviction of the covering EST1 plasmid by 5′-FOA selection, the est1Δ yku70Δ and est1Δ yku80Δ cells exhibited markedly reduced viability compared with that of wild-type cells and the expression of the Cdc13-Est2 fusion protein could only improve their growth slightly (Fig. 1A). Further serial streak plating of the cdc13Δ est1Δ ykuΔ/pRS415-CDC13-EST2 cells uncovered a more pronounced lethal-like phenotype (Fig. 1C). As a control, we also constructed the strains TXJ007 and TXJ010 (Table 1), which contained pRS315-ku80-135i. After 5′-FOA selection or tetrad dissection, we obtained ku80Δ est1Δ/pRS315-yku80-135i and cdc13Δ ku80Δ est1Δ/pRS315-yku-135i/pRS316-CDC13-EST2 haploid cells. They were viable and did not exhibit a synthetic lethal phenotype (Fig. 1B and C), likely because the yku80-135i mutation causes a loss of Yku80's interaction with Tlc1 RNA but does not affect its function in telomere capping (2, 52). Southern blot analysis revealed that the extremely long telomeres in the cells expressing the Cdc13-Est2 fusion protein were not shortened to wild-type length upon eviction of the covering EST1 plasmid in the single-deletion YKU80 or EST1 strains or double-deletion YKU80 and EST1 strains, and telomere length in cdc13Δ est1Δ yku-135i/pRS316-CDC13-EST2 cells was comparable to that of wild-type telomeres (Fig. 1D). These results suggested that the synthetic lethality of YKU and EST1 mutation was not likely caused by short telomeres, and Cdc13-Est2 fusion protein could not bypass the requirement for Est1 when Yku was absent.
FIG. 1.
The est1Δ ykuΔ cells with longer than normal telomeres are synthetic lethal. (A) Growth analysis of cdc13Δ est1Δ yku70Δ or cdc13Δ est1Δ yku80Δ mutants in the presence of Cdc13-Est2 fusion expression. As indicated, the cdc13Δ est1Δ yku70Δ (or yku80Δ)/pRS415-CDC13-EST2/pRS316-EST1 cells were 5-fold serially diluted and spotted at 30°C on Leu− Ura− medium (left panel) to select for the presence of both the pRS415-CDC13-EST2 and pRS316-EST1 plasmids and on the Leu− 5′-FOA+ medium (right panel) to select for eviction of the pRS316-EST1 plasmid. The parallel transformation and growth analysis were done with pRS315-EST1 and pRS315 vector alone for controls. (B) Growth analysis of est1Δ yku80-135i cells. est1Δ yku80Δ/pRS316-EST1 cells were generated by tetrad dissection of diploid strain TXJ004 (Table 1) and were transformed with pRS315-yku80-135i. Five-fold dilutions were spotted at 30°C on Leu− Ura− medium (left panel) to select for the presence of both pRS315-yku80-135i and pRS316-EST1 plasmids and on Leu− 5′-FOA+ medium (right panel) to select for eviction of pRS316-EST1 plasmid. The parallel transformation and growth analysis were done with pRS315-YKU80 and pRS315 vectors for controls. (C) Streak plate growth analysis of isogenic strains generated in panel A and the est1Δ yku80-135i/pRS316-CDC13-EST2 cells (by tetrad dissection from diploid strain TXJ007 shown in Table 1) on YEPD medium at 30°C. (D) Analysis of telomere length of the indicated isogenic strains shown in panel C. Genomic DNA of two independent colonies of isogenic strains was subjected to telomere Southern blot analysis with a TG1-3 telomeric probe (34).
The amount of single-stranded telomeric DNA is increased in est1Δ ykuΔ cells.
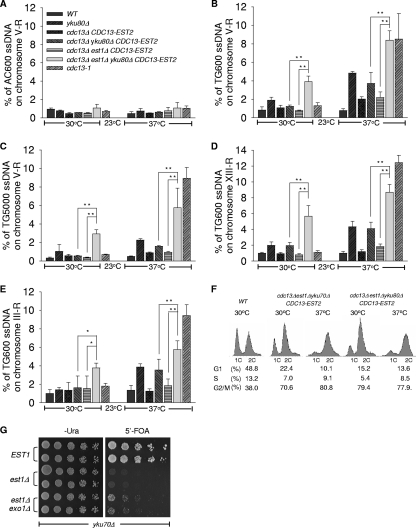
Since the cdc13Δ yku80Δ est1Δ/pRS415-CDC13-EST2 cells displayed a synthetic lethal phenotype even with longer than normal telomeres (Fig. 1D), it was possible that the synthetic lethality of the EST1 and YKU double deletion was attributable to the deprotection of telomeres. One consequence of telomere deprotection that has previously been observed in yku, cdc13, stn1, and ten1 mutants is the increased formation of an irregular single-stranded G-rich overhang (6, 17-19). We therefore performed a quantification amplification of single-stranded DNA (QAOS) assay to examine the level of single-stranded telomeric DNA at the subtelomeric loci (3). We chose to examine the extent of single-stranded telomeric DNA that is present in both wild-type and mutant cells approximately 600 bp and/or 5,000 bp from the right telomere end of chromosomes III, V, and XIII (Fig. 2 A to E). The right arm telomere of chromosome V or XIII is usually silenced, while the right arm telomere of chromosome III is usually transcribed and not silenced (42). As expected, we barely detected C-rich single-stranded telomeric DNA in all the cells examined (Fig. 2A). Also as expected, the amount of G-rich single-stranded telomeric DNA was increased in the yku80Δ cells either with or without CDC13-EST2 fusion expression when cultured at the nonpermissive 37°C temperature. A significant increase of G-rich single-stranded telomeric DNA was detected in cdc13Δ yku80Δ est1Δ/pRS415-CDC13-EST2 cells at both 30°C and 37°C and the increase was comparable to that of cdc13-1 mutant (Fig. 2B to E). Parallel fluorescence-activated cell sorter (FACS) analysis revealed that over 70% of the cells deficient in both Yku and Est1 were arrested in G2/M phase at both 30 and 37°C (Fig. 2F). Consistent with a previous report (1, 32), inactivation of Exo1, a 5′→3′ exonuclease that is responsible for C-stand resection and G-tail production, partially rescued the synthetic lethality of YKU and EST1 double deletion (Fig. 2G). Together these data suggested that the EST1 deletion in a yku mutant background led to a further increase in the amount of single-stranded telomeric DNA, and this consequently triggered the cells to arrest at the G2/M-phase boundary.
FIG. 2.
The est1Δ ykuΔ cells have increased single-stranded telomeric DNA. (A to E) QAOS analysis with yku80Δ mutants. Strains generated from 5′-FOA selection described in the legend to Fig. 1A and the wild-type strain were cultured at 30°C or 37°C for 4 h. cdc13-1 mutant strains cultured at 23 or 37°C were used as a positive control. Genomic DNA was extracted by the Zymolyase digestion method and monitored by QAOS analysis as described previously (3). The error bars represent the standard deviation (SD) from three independent experiments. **, P < 0.01; *, P < 0.05. (A) The relative amount of single-stranded CA DNA from the genomic locus 600 bp from the right-arm telomere in chromosome V (V-R) was determined. (B, D, and E) The relative amounts of single-stranded TG DNA from the genomic locus 600 bp from the right-arm telomeres in chromosome V (V-R) (B), chromosome XIII (XIII-R) (D), and chromosome III (III-R) (E) were determined. (C) The relative amount of single-stranded TG DNA from the genomic locus 5,000 bp from the telomere in the chromosome V right arm was determined. (F) FACS analysis with est1Δ ykuΔ mutant cells. Wild-type (WT) cells and the mutant strains indicated were cultured in selective liquid medium at 30 or 37°C for 4 h, and their DNA content was examined by FACS analysis. The percentages of cells in the G1, S, and G2/M phases are indicated at the bottom. (G) Growth analysis of est1Δ yku70Δ exo1Δ cells. est1Δ yku70Δ exo1Δ cells and est1Δ yku70Δ cells carrying plasmid pRS316-EST1 were plated at 30°C on medium selecting for the presence of pRS316-EST1 (Ura−) and on medium that selected for the eviction of covering EST1 plasmid (5′-FOA), respectively.
Telomere-tethered Est1 rescues the temperature sensitivity of the YKU deletion mutant in a manner that is independent of Est2.
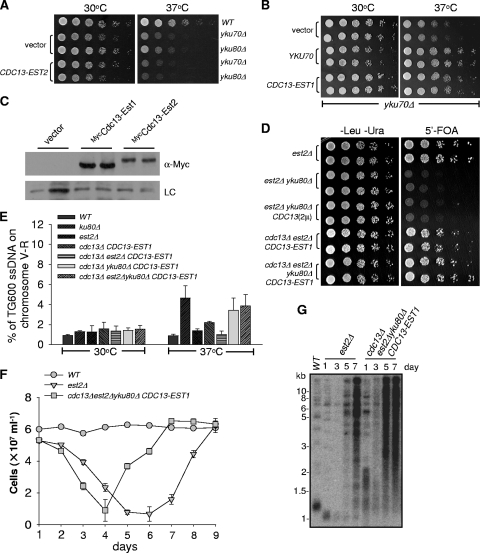
The Est2/Tlc1 complex associates with telomeres in both G1 phase, via a Yku-Tlc1 interaction, and in S phase, via a Cdc13-Est1-Tlc1 interaction (5, 8, 13). Est1 is expressed and associated with telomeres in S phase (38, 53) when the telomeric G-overhang reaches more than 30 nucleotides (60). Since Est1 and Est2 are normally dependent on each other for their association with telomeric DNA in S phase (5), it was not clear if both the telomerase core enzyme of Est2/Tlc1 and the accessory subunit of Est1 contributed to telomere protection directly. We therefore examined temperature sensitivity of Yku-deficient cells when Est2 or Est1 was artificially tethered to telomeres via fusion with Cdc13. Anchoring of Est2 to telomeres via a Cdc13-Est2 fusion protein could not rescue the temperature sensitivity of yku70Δ or yku80Δ cells (Fig. 3 A). Conversely, tethering Est1 to telomeres via a Cdc13-Est1 fusion (8) alleviated the temperature sensitivity phenotype of the yku70Δ mutant (Fig. 3B), suggesting that the physical presence of Est1 at telomeres compensated for the absence of Yku. Therefore, Est1 likely played an important role in telomerase complex-mediated telomere protection. Western blot analysis revealed that the expression level of mycCdc13-Est1 was higher than that of mycCdc13-Est2 (Fig. 3C), so we could not rule out the possibility that the telomere protection of Est1 is attributed to the higher expression of Cdc13-Est1 fusion protein.
FIG. 3.
Targeting Est1 to telomeres suppresses synthetic lethality of YKU80 and EST2 double deletion. (A) Growth analyses of the yku70Δ or yku80Δ mutants. The pRS415-CDC13-EST2 or pRS415 plasmid was transformed into the yku70Δ or yku80Δ cells. Five-fold serial dilutions were spotted on Leu− medium to select for the presence of plasmid at 30°C and 37°C, respectively. (B) Growth analyses of the yku70Δ mutant. The plasmid pRS415-CDC13-EST1 (CDC13-EST1) was transformed into the yku70Δ cells. Five-fold dilutions were spotted on the medium (Leu−) at 30 and 37°C, respectively. The parallel transformation and growth analyses were done with pRS315-YKU70 and pRS315 vector alone for controls. (C) Western blot analyses of Cdc13-Est1 and Cdc13-Est2 expression. pRS415 vector, pRS415-Myc-CDC13-EST1, or pRS415-Myc-CDC13-EST2 plasmid was transformed into YPH499 cells, and the total cell lysate was subjected to Western blot analyses with anti-Myc antibody (α-Myc) (upper panel). A nonspecific band shown in the same film was set as loading control (LC) (lower panel). (D) Growth analyses of the est2Δ yku80Δ cells in the presence of CDC13-EST1 fusion expression. The est2Δ/pRS316-EST2/pRS415, yku80Δ est2Δ/pRS316-EST2/pRS415, yku80Δ est2Δ/pRS316-EST2/pDBL-CDC13 (63), est2Δ cdc13Δ/pRS316-EST2/pRS415-CDC13-EST1, and yku80Δ est2Δ cdc13Δ/pRS316-EST2/pRS415-CDC13-EST1 haploid cells were generated from tetrad dissection. Five-fold serial dilutions were spotted at 30°C on Leu− Ura− medium to select for the presence of both pRS415-CDC13-EST1 (or pRS415) and pRS316-EST2 plasmids (left panel) and on Leu− 5′-FOA+ medium (right panel) that selected for eviction of pRS316-EST2 plasmid. (E) QAOS analysis of the est2Δ yku80Δ mutant. Strains generated from the 5′-FOA plate described in panel D were cultured at 30 or 37°C as described. Genomic DNA was extracted by the Zymolyase digestion method and monitored by QAOS analysis. The single-stranded DNA of the TG strand at the genomic locus of 600 bp from the right telomere in chromosome V was measured (3), and the error bars represent the standard deviation (SD) from three independent experiments. (F) Liquid cell viability assay for the est2Δ yku80Δ mutant. est2Δ and est2Δ yku80Δ/pRS415-CDC13-EST1 cells were grown in selective medium to saturation, and each culture was diluted to an OD600 of 0.05 every 24 h. The total cell number was monitored. The error bars represent the results from two independent clones. (G) Analysis of telomere length of est2Δ yku80/pRS415-CDC13-EST1 cells. est2Δ and cdc13Δ est2Δ yku80Δ/pRS415-CDC13-EST1 haploid cells were passaged in liquid medium for the indicated time in days and collected for telomere Southern blot analysis using a TG1-3 telomeric probe.
To address whether the protective role of Est1 at telomeres requires the simultaneous presence of Est2, we examined synthetic lethality of a YKU and EST2 double-deletion mutant when Est1 was tethered to telomeres via the Cdc13-Est1 fusion protein. Interestingly, telomere-targeted Cdc13-Est1 protein, but not the Cdc13 protein alone, sufficiently suppressed the synthetic lethality of the YKU80 and EST2 double deletion (Fig. 3D). Accordingly, the single-stranded G tails in the cdc13Δ est2Δ yku80Δ/pRS415-CDC13-EST1 cells was comparable to that in yku80Δ or cdc13Δ yku80Δ/pRS415-CDC13-EST1 cells (Fig. 3E). The continuous passage of cells in liquid media revealed that cdc13Δ est2Δ yku80Δ/pRS415-CDC13-EST1 cells senesced after about 50 generations, and a small number of survivors eventually began repopulating the culture (Fig. 3F). As expected, the telomeres gradually shortened and TG recombination took place to generate the survivors (Fig. 3G) (15). Taken together, these results strongly supported the idea that Est1 directly participated in telomere protection.
The G-quadruplex-promoting activity of Est1 contributes to telomere protection.
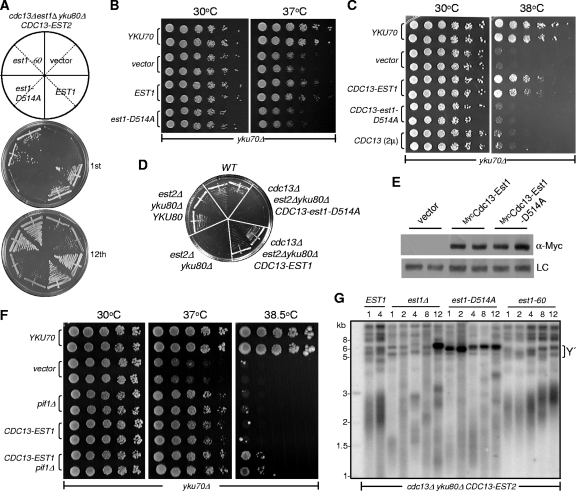
Because Est1 interacts with both Tlc1 and Cdc13 to help recruit telomerase (43, 47, 51) and is able to convert single-stranded DNA into a G quadruplex (63), we wondered which activity of Est1 was more important to its telomere protection function. We therefore employed the est1-60 and est1-D514A separation-of-function mutants. Cells with either mutation exhibit a senescence phenotype because of gradually shortening telomeres (40, 63). The Est1-60 mutant protein does not interact with Cdc13 and is defective in telomerase recruitment (40). The Est1-D514A mutant protein can interact with both Tlc1 and Cdc13 to recruit telomerase; however, it cannot convert single-stranded telomeric DNA into a G quadruplex (63). We examined the synthetic lethality of YKU with these two separation-of-function EST1 mutants in the cdc13Δ/pRS415-CDC13-EST2 fusion background. When serially restreaked on plates, cells bearing est1-60 and yku80Δ mutations grew as robust as EST1 yku80Δ cells, presumably because the deficiency of telomerase recruitment in the est1-60 mutant was bypassed by CDC13-EST2 fusion (Fig. 4 A) (8). In contrast, cells with both the est1-D514A and yku80Δ mutations appeared to be very sick and barely produced colonies in the first several restreaks (Fig. 4A). When these “sick” cells were continuously passaged on solid medium, their growth ability was gradually recovered to a reasonable level at later restreaks: e.g., the 12th restreak (Fig. 4A). To confirm this result, we also examined other EST1 mutations that were previously shown to be deficient in quadruplex-promoting activity and strains harboring either the est1-D510A or the est1-D521A allele showed synthetic lethality with a YKU deletion (data not shown) (9, 63). Additionally, overexpression or targeting of wild-type Est1, but not Est1-D514A, alleviated the temperature sensitivity of the yku70Δ mutant (Fig. 4B and C) (10), and overexpression Cdc13 alone was not able to improve the growth of the yku70Δ mutant (Fig. 4C). Moreover, cdc13Δ est2Δ yku80Δ cells expressing Cdc13-Est1-D514A fusion protein could not produce clones after eviction of the covering EST2 plasmid on the 5′-FOA plate (Fig. 4D), indicating that targeting Est1-D514A to telomeres by fusing it to CDC13 would not rescue the synthetic lethality of EST2 and YKU double deletion. Western blot analysis showed that the expression levels of mycCdc13-Est1-D514A and mycCdc13-Est1 were comparable (Fig. 4E). These results indicated that the telomeric DNA G-quadruplex-promoting activity of Est1 is directly involved in telomere protection.
FIG. 4.
Telomere protection of Est1 depends on its G-quadruplex promotion activity. (A) Growth analysis of est1-D514A and est1-60 mutants in the est1Δ yku80Δ/pRS317-CDC13-EST2 background. A cdc13Δ est1Δ yku80Δ strain carrying both pRS316-EST1 and pRS317-CDC13-EST2 plasmids was obtained from a tetrad dissection and was transformed with pRS315-EST1, pRS315-est1-D514A, pRS315-est1-60, or pRS315 empty vector as indicated. After eviction of pRS316-EST1 plasmid by 5′-FOA selection, two independent clones were sequentially restreaked on selective Leu− plates at 30°C. The number of passages for each streak plate is indicated below. (B) Growth analysis of Est1-D514A overexpression in yku70Δ cells. The plasmid pRS315-yku70, pDBL-EST1 (63), pDBL-est1-D514A or an empty vector was transformed into yku70Δ cells. Five-fold serial dilutions were spotted on Leu− medium and grown at 30 or 37°C as indicated. (C) Growth analysis of the yku70Δ mutant. Plasmid pRS415-CDC13-EST1 (CDC13-EST1), pRS415-CDC13-est1-D514A (CDC13-est1-D514A), or pDBL-CDC13 (CDC13) was transformed into the yku70Δ cells. Five-fold dilutions were spotted on Leu− medium and grown at 30 or 38°C as indicated. The parallel transformation and growth analyses were done with pRS315-YKU70 and pRS415 vector alone for controls. (D) Growth analyses of est2Δ yku80Δ cells in the presence of the CDC13-EST1 or CDC13-est1-D514A fusion gene. est2Δ yku80Δ/pRS316-EST2 cells were generated by tetrad dissection and then transformed with pRS415-CDC13-EST1 or pRS415-CDC13-est1-D514A. The parallel transformation and growth analyses were done with pRS315-YKU80 and pRS415 vector alone for controls. Cells were streaked on the 5′-FOA medium to evict the pRS316-EST2 plasmid. (E) Western blot analyses of Cdc13-Est1 and Cdc13-Est1-D514A expression. pRS415 empty vector, pRS415-Myc-CDC13-EST1, or pRS415-Myc-CDC13-EST1-D514A plasmid was transformed into YPH499 cells, and the total cell lysate was subjected to Western blot analyses with anti-Myc antibody (α-Myc) (upper panel). A nonspecific band shown in the same film was set as a loading control (LC) (lower panel). (F) Growth analysis of the yku70Δ mutant. Plasmid pRS415-CDC13-EST1 was transformed into the yku70Δ or yku70Δ pif1Δ cells. Five-fold dilutions were spotted on Leu− medium and grown at 30, 37, or 38.5°C as indicated. The parallel transformation and growth analyses were done with pRS315-YKU70 and pRS415 vector alone for controls. (G) Telomere Southern blot analysis of est1 mutants in the cdc13Δ yku80Δ est1Δ/pRS317-CDC13-EST2 background. The isogenic strains shown in panel A were restreaked on plates, and genomic DNA was isolated from the 1st, 2nd, 4th, 8th, and 12th restreaks, as labeled at the top of the blot. DNA was digested by XhoI and subjected to Southern blot analysis with a TG1-3 probe. The subtelomeric Y′ signal is indicated on the right.
Previous studies have shown that Pif1 family helicases could unwind G-quadruplex structures (45, 46). In order to address whether the hypothesized telomere G-quadruplex structure facilitated telomere protection, we deleted PIF1 in yku70Δ cells because we assumed that inactivation of Pif1 helicase would stabilize the G quadruplex (45). A temperature sensitivity assay showed that deletion of PIF1 partially rescued the growth ability of yku70Δ cells at 37°C (Fig. 4F) (58). Additionally, deletion of PIF1 could improve the growth of yku70Δ/pRS415-CDC13-EST1 cells at 38.5°C (Fig. 4F). These results were in agreement with the argument that the telomere protection function of Est1 is attributed to its G-quadruplex promotion activity. However, we could not rule out the possibility that the deletion of PIF1 might enhance the occupancy of telomerase on telomere ends and contribute to telomere protection (58).
Telomere Southern blot analysis of the cells sequentially passaged in liquid medium revealed that the Cdc13-Est2 fusion protein generated extremely long telomeres, and they were maintained in the cdc13Δ yku80Δ est1-60/pRS415-CDC13-EST2 cells (Fig. 4G). However, prominent Y′ amplification appeared in the cdc13Δ yku80Δ est1-D514A/pRS415-CDC13-EST2 and cdc13Δ yku80Δ est1Δ/pRS415-CDC13-EST2 survivor-like cells. Notably, the Y′ amplification occurred abruptly in the early passages of the cdc13Δ yku80Δ est1-D514A/pRS415-CDC13-EST2 mutant (Fig. 4G). These results indicate that defective protection of the single-stranded TG DNA by the dysfunctional Est1-D514A mutation promoted Y′ recombination to repair telomeres with exposed single-stranded DNA.
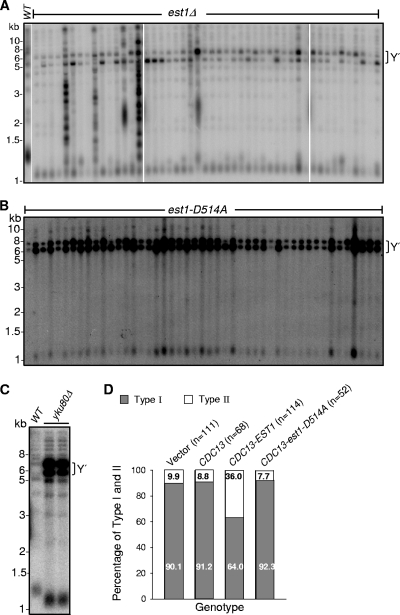
Y′ amplification occurs frequently in the est1-D514A mutant background.
In order to investigate further how the lack of G-quadruplex-promoting activity of Est1 affected Y′ recombination, we obtained an est1-D514A mutant strain from a tetrad dissection. This est1-D514A mutant strain did not contain the CDC13-EST2 fusion plasmid. The telomere single-stranded DNA in est1-D514A mutant strain was examined and compared with that in est1Δ, est1-60, yku80Δ, and cdc13-1 cells. The result in Fig. 5 A showed that no significant single-stranded DNA was accumulated in est1-D514A cells (Fig. 5A). The mutant cells were grown in liquid medium, and their growth potential and telomere phenotypes were also examined. Interestingly, the est1-D514A mutant strain did not exhibit a senescence curve that was typical of an est1Δ mutant (24) (Fig. 5B). A telomere blot analysis showed that Y′ amplification occurred much more dominantly and efficiently in the est1-D514A mutants than in the est1Δ cells (Fig. 5C). Accordingly, deletion of RAD51 in the est1-D514A background, which is normally required for Y′ recombination, but not for TG tract recombination (25), caused a phenotype that was nearly lethal (Fig. 5D). Conversely, deletion of RAD50 in the est1-D514A background, which is required for TG tract recombination but not Y′ recombination and amplification (25), did not affect survivor formation (Fig. 5D) and Rad51-mediated Y′ amplification effectively took place to repair telomeres (Fig. 5E). To validate that est1-D514A mutant cells amplified Y′ efficiently to escape senescence, we randomly selected 50 colonies that were derived from est1Δ (Fig. 6 A) or est1-D514A (Fig. 6B) spores after three serial restreaks on solid medium. A telomere Southern blot revealed that all of the colonies derived from est1-D514A spores displayed shortened TG tracts and Y′ amplification, resembling the phenotypes of type I survivors (Fig. 6B). Consistent with a previous report (12), inactivation of Yku resulted in short but stable telomeres as well as Y′ amplification (Fig. 6C). Taken together, we concluded that as in the yku mutant, Y′ amplification functions as a telomere repair mechanism that is activated in the est1-D514A mutant.
FIG. 5.
Y′ amplification takes place in the est1-D514A mutant to repair deprotected telomeres. (A) QAOS analysis of the est1 mutant as described in the legend to Fig. 2B. est1Δ, est1-D514A, and est1-60 haploid strains were generated from tetrad dissection. Wild-type and yku80Δ cells cultured at 30 or 37°C or the cdc13-1 mutant cells cultured at 23 or 37°C were used as negative or positive controls. (B) Cell viability assay for the est1 mutants. Dissected est1Δ spores carrying empty vector, pRS316-EST1, or pRS316-est1-D514A plasmid were grown in selective medium until the culture reached growth saturation, and each culture was diluted to an OD600 of ∼0.05 every day (24 h) for 14 days. The total cell number was determined after each indicated day. (C) Analysis of the telomere length of the est1 mutant strains. Genomic DNA was isolated from the yeast cells in panel B, digested with XhoI, and subjected to Southern blot analysis with a TG probe. The numbers at the top indicate the number of days the cells were grown. (D) Growth analysis of rad50Δ est1-D514A and rad51Δ est1-D514A mutants. est1Δ rad50Δ or est1Δ rad51Δ cells carrying either the pRS316-EST1 or the pRS316-est1-D514A mutant plasmid were grown repeatedly in Ura− medium until survivors were generated, and the 6th restreak is shown. (E) Telomere Southern blot analysis of rad50Δ est1-D514A and rad50Δ est1Δ mutant strains. Genomic DNA isolated from the indicated isogenic strains was digested with XhoI and subjected to Southern blot analysis using a TG probe. The numbers at the top indicate the numbers of days the cells were passaged.
FIG. 6.
Targeting Est1 to telomeres inhibits type I survivor formation. (A and B) Haploid est1Δ cells with pRS316 (A) or pRS316-est1-D514A (B) plasmid were passaged on solid medium to generate survivors. Fifty independent clones were randomly picked, and their DNA was subjected to a telomere Southern blot analysis to examine the survivor types. The telomere phenotype of 46 clones is shown. (C) Telomere Southern blot analysis of yku80Δ cells. Genomic DNA from two independent clones of the yku80Δ strain cultured at 30°C was subjected to telomere Southern blot analysis using a TG1-3 probe. (D) The est2Δ strain with pRS415, pRS314-CDC13, pRS415-CDC13-EST1 vector, or pRS415-CDC13-est1-D514A was passaged by serial streakouts on solid medium to generate survivors, and DNA isolated from the indicated independent clones was subjected to telomere Southern blot analysis (data not shown). The survivor rate of est2Δ strain cells with pRS415, pRS314-CDC13, pRS415-CDC13-EST1 vector, or pRS415-CDC13-est1-D514A plasmid was calculated according to the telomere Southern blot analysis.
Est1 normally inhibits Y′ amplification.
In any of the estΔ mutants, about 90% of postsenescent survivors maintain telomeres by Y′ amplification to form type I survivors, and about 10% of postsenescent survivors maintain telomeres by TG recombination to form type II survivors (55). If Est1 normally inhibits Y′ amplification, physically tethering Est1 to telomeres via a Cdc13-Est1 fusion should decrease the frequency of type I survivor formation in telomerase null cells. To test this hypothesis, we prepared est2Δ cells and immediately transformed the pRS415-CDC13-EST1, pRS415-CDC13-est1-D514A, pRS314-CDC13, or pRS415 plasmid into est2Δ cells, respectively. Due to the lack of Est2, cells expressing Cdc13-Est1, Cdc13-Est1-D514A fusion protein, or Cdc13 alone underwent senescence and survivors were recovered after the third serial restreak on the solid medium. More than 50 colonies for each strain were subsequently isolated, and telomeres of individual colonies were analyzed by Southern blotting. The percentage of survivor types is summarized in Fig. 6D. Consistent with a previous report, est2Δ postsenescent cells produced 9.9 and 90.1% of type II and type I survivors, respectively (55). In contrast, est2Δ/pRS415-CDC13-EST1 postsenescent cells gave rise to 36.0 and 64.0% of type II and type I survivors, respectively, indicating that the telomere-tethered Est1 increased the rate of type II survivor formation and decreased the rate of type I survivor formation. In a manner that paralleled est2Δ cells, est2Δ/pRS314-CDC13 postsenescent cells yielded 8.8 and 91.2% of type II and type I survivors, respectively, and est2Δ/pRS415-CDC13-est1-D514A postsenescent cells yielded 7.7 and 92.3% of type II and type I survivors, respectively. These results support a model in which the telomere association of a functional Est1 inhibits Y′ recombination and the G-quadruplex-promoting activity of Est1 is required for this inhibitory role.
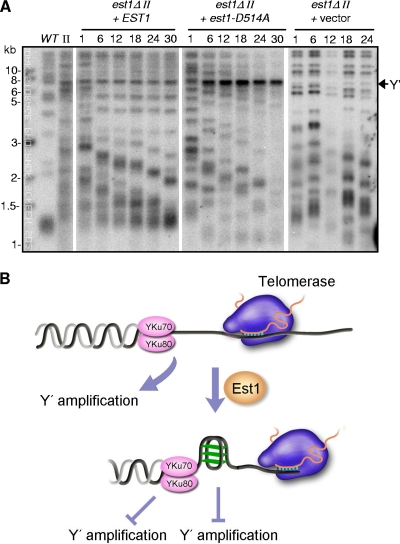
Telomerase and Rad52-dependent recombination are the respective primary and alternative pathways used for telomere elongation in yeast. Reintroduction of telomerase into type II survivor cells gradually restored a wild-type telomere Southern blot pattern (7, 56). To validate further that the lack of G-quadruplex-promoting activity of Est1 might increase Y′ recombination-mediated telomere repair, we reintroduced either wild-type EST1 or mutant est1-D514A into type II survivor cells that were derived originally from est1Δ cells. As expected, reintroduction of wild-type EST1 gradually restored the wild-type telomere Southern blot pattern (Fig. 7 A). In contrast, reintroduction of an est1-D514A mutant, but not an empty vector, promoted Y′-element amplification and trimmed down the TG tracts to a length shorter than that of the wild type (Fig. 7A). Because only an inactive telomerase was assembled at telomeres in the est1-D514A mutant (63), the type II survivors derived from est1Δ cells likely transformed into type I cells in the presence of the Est1-D514A mutant protein. These results further support our conclusion that the G-quadruplex-promoting activity of Est1 inhibits Y′ amplification (Fig. 7B).
FIG. 7.
Reexpression of Est1-D514A mutant protein switches the est1Δ-derived type II survivor to a type I survivor. (A) Telomere Southern blot analysis of type II survivors that express Est1-D514A mutant protein. est1Δ-derived type II survivors transformed with pRS315-EST1, pRS315-est1-D514A, or pRS315 plasmid were passaged on Leu− medium. Genomic DNA from the indicated restreaks (at the top) was digested by XhoI and subjected to Southern blot analysis using a TG1-3 probe. (B) Model of Est1's functions in telomere protection. In the presence of Est1, G tail could be converted into G quadruplex. The higher-order DNA structure on one hand avoids telomere being recognized by recombination factors and on the other hand activates telomerase.
DISCUSSION
As the natural ends of chromosomes, telomeres have to be shielded by various associated proteins to distinguish themselves from undesired DNA double-strand breaks. Telomere protection appears to involve at least three independent pathways, namely, the CST, Yku, and telomerase pathways. A defect in one pathway usually doesn't cause lethality; however, a combinatorial deficiency in two protection pathways leads to cell death. For example, both ykuΔ est1Δ (or est2Δ or est3Δ) and cdc13-1 ykuΔ mutant cells are unviable (19, 36, 41).
The telomerase holoenzyme is made up of the core enzyme complex of Est2/Tlc1 and the accessory subunits Est1 and Est3. A previous study suggested the telomerase core enzyme was involved in telomere protection (22). Since a lack of Est2 or Tlc1 reduces Est1-telomere association, it remained a possibility that the telomere-protective role of telomerase core enzyme is indirect and is instead mediated by the Est1 subunit (5). We favor this model because the tethering of Est2 to telomeres via fusion with Cdc13 did not suppress either the temperature sensitivity of yku cells (Fig. 3A) or the synthetic lethality of the ykuΔ est1Δ cells (Fig. 1A), whereas the tethering of Est1 to telomeres by fusion with Cdc13 rescued both the temperature sensitivity of yku cells and the synthetic lethality of ykuΔ est2Δ cells (Fig. 3). These observations support a model in which Est1 directly participates in telomere protection.
Overexpression of Est1-D514A, which possesses telomerase recruitment activity, could not rescue the temperature sensitivity of a yku mutant (Fig. 4). In addition, when the Cdc13-Est2 fusion protein was present, synthetic lethality was observed in a yku and est1-D514A double mutant, but not in a yku and est1-60 double mutant (Fig. 4). Moreover, inactivation of Pif1 helicase, a G-quadruplex resolver (45, 46), could partially rescue the temperature sensitivity of ykuΔ cells and increase the viability of yku70Δ/pRS415-CDC13-EST1 cells at 38.5°C (Fig. 4F). Therefore, the G-quadruplex-promoting, but not telomerase recruitment, activity of Est1 plays a complementary role in the single-stranded DNA protection that is missing in a yku mutant. The abrogation of the temperature sensitivity phenotype of the yku mutants via the overexpression of either Est2 or Tlc1 was therefore likely attributable to an increase of Est1 at telomeres due to an overabundance of Est2 and/or Tlc1 at the telomeres. Since the precise role of Est3 in telomere maintenance is poorly understood, we could not rule out the possibility that Est3 is required for telomere protection through a mechanism that employs Est1.
In wild-type cells, because telomerase maintains normal telomere length and capping factors like the Yku and CST complexes guard the single-stranded G tails, Y′ amplification and TG tract recombination are extremely rare events. When a telomere protection factor is absent or dysfunctional, the exposure of single-stranded TG DNA could be recognized as a DNA lesion and subsequently become a favorable substrate for homologous recombination (Fig. 7B). In yku mutant cells, although short and stable telomeres are maintained by telomerase, the deprotected single-stranded TG DNA appears to be repaired through Y′ amplification (Fig. 6C) (12). In the cdc13-1 mutant, a small portion of cells could survive at a nonpermissive temperature of 37°C with recombination-repaired telomeres (16). Likewise, in est1-D514 cells, the Est1-D514A mutant protein can help to assemble telomerase at telomeres, but cannot convert single-stranded TG DNA into a G quadruplex (63). The deprotected single-stranded TG DNA in the est1-D514 cells was repaired by Y′ amplification but not TG tract recombination, thereby allowing cells to escape senescence (Fig. 5). Interestingly, it is unclear why the est1Δ and est1-D514A cells behaved differently in activating telomere recombination (Fig. 5B and C). In est1Δ cells, both Y′ recombination and TG tract recombination seem to be difficult to initiate because the survivor formation rate is extremely low (30). In contrast, in est1-D514A cells Y′ recombination occurred much more efficiently, and type I survivors arose much earlier (Fig. 5B and C). One explanation was that in the est1Δ cells, the absence of telomerase might result in telomere recruitment of the factors that are involved in both homologous recombination (Y′ or TG tract recombination) and nonhomologous end joining (NHEJ) (30, 39, 56), competing for the same substrate to make any event less efficient, while in the est1-D514A mutant, on one hand, telomere-associated inactive telomerase inhibited both TG tract recombination and NHEJ, and on the other hand, the lack of G-quadruplex formation promoted Y′ recombination (Fig. 5 to 7). Therefore, both Yku and Est1 protect single-stranded TG DNA from Y′ recombination, which serves as a means of repairing deprotected telomeric DNA (Fig. 7B).
Because Est1 is expressed and associates with telomeric DNA via interaction with Cdc13 in S phase (8, 38, 40, 43), it is likely that Est1 exerts its protection function during telomere elongation in S phase. However, since the expression of the CDC13-EST1 fusion gene was under the promoter of CDC13, it remains possible that due to an intrinsic telomere association of Cdc13 (54), the Cdc13-Est1 fusion protein has targeted Est1 to telomeres during all the phases of cell cycle. The constant association of Est1 with telomeres may compensate for the loss of Yku protection in the YKU null mutant (Fig. 3B). We propose a model where upon C-strand degradation by exonuclease (i.e., ExoI) (1, 32), Est1 binds to and converts single-stranded TG DNA into a G quadruplex (63). This transformation of G-rich DNA on one hand facilitates the activation of telomerase (63) and on the other hand prevents single-stranded TG DNA from being recognized by recombination machinery (Fig. 7B). Therefore, in addition to activating telomerase, Est1 likely functions as the primary factor in the telomerase pathway that provides the function of telomere protection (Fig. 7B).
Acknowledgments
We are grateful to Yasumasa Tsukamoto, Daniel Gottschling, Virginia Zakian, and Victoria Lundblad for providing the plasmids and yeast strains. We thank Brian A. Lenzmeier for critical reading of our manuscript.
This work is supported by grants from the Natural Science Foundation of China (NSFC30630018 and 90919027) and the Ministry of Science and Technology (2007CB914502).
Footnotes
Published ahead of print on 10 January 2011.
REFERENCES
- 1.Bertuch, A. A., and V. Lundblad. 2004. EXO1 contributes to telomere maintenance in both telomerase-proficient and telomerase-deficient Saccharomyces cerevisiae. Genetics 166:1651-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertuch, A. A., and V. Lundblad. 2003. Which end: dissecting Ku's function at telomeres and double-strand breaks. Genes Dev. 17:2347-2350. [DOI] [PubMed] [Google Scholar]
- 3.Booth, C., E. Griffith, G. Brady, and D. Lydall. 2001. Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res. 29:4414-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulton, S. J., and S. P. Jackson. 1996. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 24:4639-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, A., J. B. Boule, and V. A. Zakian. 2008. Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet. 4:e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandra, A., T. R. Hughes, C. I. Nugent, and V. Lundblad. 2001. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 15:404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, X. F., F. L. Meng, and J. Q. Zhou. 2009. Telomere recombination accelerates cellular aging in Saccharomyces cerevisiae. PLoS Genet. 5:e1000535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117-120. [DOI] [PubMed] [Google Scholar]
- 9.Evans, S. K., and V. Lundblad. 2002. The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics 162:1101-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, S. K., M. L. Sistrunk, C. I. Nugent, and V. Lundblad. 1998. Telomerase, Ku, and telomeric silencing in Saccharomyces cerevisiae. Chromosoma 107:352-358. [DOI] [PubMed] [Google Scholar]
- 11.Feldmann, H., and E. L. Winnacker. 1993. A putative homologue of the human autoantigen Ku from Saccharomyces cerevisiae. J. Biol. Chem. 268:12895-12900. [PubMed] [Google Scholar]
- 12.Fellerhoff, B., F. Eckardt-Schupp, and A. A. Friedl. 2000. Subtelomeric repeat amplification is associated with growth at elevated temperature in yku70 mutants of Saccharomyces cerevisiae. Genetics 154:1039-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher, T. S., and V. A. Zakian. 2005. Ku: a multifunctional protein involved in telomere maintenance. DNA Repair (Amst.) 4:1215-1226. [DOI] [PubMed] [Google Scholar]
- 14.Garvik, B., M. Carson, and L. Hartwell. 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15:6128-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grandin, N., and M. Charbonneau. 2009. Telomerase- and Rad52-independent immortalization of budding yeast by an inherited-long-telomere pathway of telomeric repeat amplification. Mol. Cell. Biol. 29:965-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grandin, N., C. Damon, and M. Charbonneau. 2001. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J. 20:6127-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grandin, N., C. Damon, and M. Charbonneau. 2001. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 20:1173-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grandin, N., S. I. Reed, and M. Charbonneau. 1997. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 11:512-527. [DOI] [PubMed] [Google Scholar]
- 19.Gravel, S., M. Larrivee, P. Labrecque, and R. J. Wellinger. 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280:741-744. [DOI] [PubMed] [Google Scholar]
- 20.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405-413. [DOI] [PubMed] [Google Scholar]
- 21.Hackett, J. A., and C. W. Greider. 2003. End resection initiates genomic instability in the absence of telomerase. Mol. Cell. Biol. 23:8450-8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu, M., et al. 2007. Telomerase core components protect Candida telomeres from aberrant overhang accumulation. Proc. Natl. Acad. Sci. U. S. A. 104:11682-11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes, T. R., S. K. Evans, R. G. Weilbaecher, and V. Lundblad. 2000. The Est3 protein is a subunit of yeast telomerase. Curr. Biol. 10:809-812. [DOI] [PubMed] [Google Scholar]
- 24.Joseph, I. S., et al. 2010. An mre11 mutation that promotes telomere recombination and an efficient bypass of senescence. Genetics 185:761-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:1431-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, J., E. K. Mandell, T. M. Tucey, D. K. Morris, and V. Lundblad. 2008. The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat. Struct. Mol. Biol. 15:990-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lendvay, T. S., D. K. Morris, J. Sah, B. Balasubramanian, and V. Lundblad. 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144:1399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lingner, J., T. R. Cech, T. R. Hughes, and V. Lundblad. 1997. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. U. S. A. 94:11190-11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lingner, J., et al. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 30.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 31.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633-643. [DOI] [PubMed] [Google Scholar]
- 32.Maringele, L., and D. Lydall. 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 16:1919-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McEachern, M. J., A. Krauskopf, and E. H. Blackburn. 2000. Telomeres and their control. Annu. Rev. Genet. 34:331-358. [DOI] [PubMed] [Google Scholar]
- 34.Meng, F. L., et al. 2009. Sua5p a single-stranded telomeric DNA-binding protein facilitates telomere replication. EMBO J. 28:1466-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nautiyal, S., J. L. DeRisi, and E. H. Blackburn. 2002. The genome-wide expression response to telomerase deletion in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 99:9316-9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nugent, C. I., et al. 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8:657-660. [DOI] [PubMed] [Google Scholar]
- 37.Nugent, C. I., T. R. Hughes, N. F. Lue, and V. Lundblad. 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274:249-252. [DOI] [PubMed] [Google Scholar]
- 38.Osterhage, J. L., J. M. Talley, and K. L. Friedman. 2006. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 13:720-728. [DOI] [PubMed] [Google Scholar]
- 39.Pardo, B., and S. Marcand. 2005. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 24:3117-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 41.Polotnianka, R. M., J. Li, and A. J. Lustig. 1998. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 8:831-834. [DOI] [PubMed] [Google Scholar]
- 42.Pryde, F. E., and E. J. Louis. 1999. Limitations of silencing at native yeast telomeres. EMBO J. 18:2538-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi, H., and V. A. Zakian. 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 14:1777-1788. [PMC free article] [PubMed] [Google Scholar]
- 44.Qian, W., et al. 2009. Ten1p promotes the telomeric DNA-binding activity of Cdc13p: implication for its function in telomere length regulation. Cell Res. 19:849-863. [DOI] [PubMed] [Google Scholar]
- 45.Ribeyre, C., et al. 2009. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 5:e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanders, C. M. 2010. Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem. J. 430:119-128. [DOI] [PubMed] [Google Scholar]
- 47.Seto, A. G., A. J. Livengood, Y. Tzfati, E. H. Blackburn, and T. R. Cech. 2002. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 16:2800-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 50.Smogorzewska, A., and T. de Lange. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73:177-208. [DOI] [PubMed] [Google Scholar]
- 51.Steiner, B. R., K. Hidaka, and B. Futcher. 1996. Association of the Est1 protein with telomerase activity in yeast. Proc. Natl. Acad. Sci. U. S. A. 93:2817-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stellwagen, A. E., Z. W. Haimberger, J. R. Veatch, and D. E. Gottschling. 2003. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 17:2384-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taggart, A. K., S. C. Teng, and V. A. Zakian. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297:1023-1026. [DOI] [PubMed] [Google Scholar]
- 54.Taggart, A. K., and V. A. Zakian. 2003. Telomerase: what are the Est proteins doing? Curr. Opin. Cell Biol. 15:275-280. [DOI] [PubMed] [Google Scholar]
- 55.Teng, S. C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6:947-952. [DOI] [PubMed] [Google Scholar]
- 56.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsukamoto, Y., C. Mitsuoka, M. Terasawa, H. Ogawa, and T. Ogawa. 2005. Xrs2p regulates Mre11p translocation to the nucleus and plays a role in telomere elongation and meiotic recombination. Mol. Biol. Cell 16:597-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vega, L. R., et al. 2007. Sensitivity of yeast strains with long G-tails to levels of telomere-bound telomerase. PLoS Genet. 3:e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Virta-Pearlman, V., D. K. Morris, and V. Lundblad. 1996. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 10:3094-3104. [DOI] [PubMed] [Google Scholar]
- 60.Wellinger, R. J., A. J. Wolf, and V. A. Zakian. 1993. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell 72:51-60. [DOI] [PubMed] [Google Scholar]
- 61.Yu, E. Y., F. Wang, M. Lei, and N. F. Lue. 2008. A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat. Struct. Mol. Biol. 15:985-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zakian, V. A. 1996. Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet. 30:141-172. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, M. L., et al. 2010. Yeast telomerase subunit Est1p has guanine quadruplex-promoting activity that is required for telomere elongation. Nat. Struct. Mol. Biol. 17:202-209. [DOI] [PubMed] [Google Scholar]