Abstract
Polo-like kinases (Plks) are characterized by the presence of a specific domain, known as the polo box (PBD), involved in protein-protein interactions. Plk1 to Plk4 are involved in centrosome biology as well as the regulation of mitosis, cytokinesis, and cell cycle checkpoints in response to genotoxic stress. We have analyzed here the new member of the vertebrate family, Plk5, a protein that lacks the kinase domain in humans. Plk5 does not seem to have a role in cell cycle progression; in fact, it is downregulated in proliferating cells and accumulates in quiescent cells. This protein is mostly expressed in the brain of both mice and humans, and it modulates the formation of neuritic processes upon stimulation of the brain-derived neurotrophic factor (BDNF)/nerve growth factor (NGF)-Ras pathway in neurons. The human PLK5 gene is significantly silenced in astrocytoma and glioblastoma multiforme by promoter hypermethylation, suggesting a tumor suppressor function for this gene. Indeed, overexpression of Plk5 has potent apoptotic effects in these tumor cells. Thus, Plk5 seems to have evolved as a kinase-deficient PBD-containing protein with nervous system-specific functions and tumor suppressor activity in brain cancer.
Polo was originally identified in Drosophila as a mutant with mitotic and meiotic defects in the microtubule spindle (51). This mutation was later mapped to a serine-threonine protein kinase specifically concentrated in dividing cells (32). Polo-like kinases (Plks) are found in all eukaryotes and are characterized by a highly conserved kinase domain and one or two C-terminal polo box domains (PBDs). These include the Plk1 subfamily, containing Drosophila polo and mammalian Plk1; the SAK subfamily, containing Drosophila SAK and mammalian Plk4, both of which are widely distributed across the eukaryotes; and the Plk2 subfamily, containing vertebrate Plk2 and Plk3 and also including homologs from echinoderms (4, 8, 10, 35). The PBD participates in subcellular localization and partner interaction (16, 54). The founding member of the family, Plk1, localizes to the cytoplasm and centrosomes in interphase and concentrates to the kinetochores and the cytokinesis bridge during cell division. This protein has major functions in centrosome maturation, mitotic entry, and cytokinesis (4, 8, 40, 53).
The other members of the family are less studied. Plk4 (Sak) is a critical regulator required for centriole duplication both in Drosophila and in mammals (9, 21). Plk2 (also known as Snk) localizes to the centrosome and may also participate in centrosome biology and S-phase checkpoints (37). Plk3 (Fnk or Prk) activity peaks in G1 and localizes to the nucleolus in interphase. This protein may function in S-phase entry (64), and it is activated in response to replicative stress and genotoxic insults leading to apoptosis in a p53-dependent manner (56, 61, 62). The three Plk subfamilies have distinct functions and operate in multiple cell types. Their expression is regulated differentially in cells and tissues and in response to several cellular processes and stimuli (59). Whereas Plk1 and Plk4 are found only in dividing cells, Plk2 and Plk3 are also expressed in neurons and other, nondividing differentiated cells (47, 55). Plk2 and perhaps Plk3 seem to have a crucial function in modulating synaptic plasticity in neuronal dendritic spines through the phosphorylation of the spindle-associated protein SPAR (3, 46). Plk2 also phosphorylates the neuronal alpha-synuclein, a phospho-protein accumulated in several neurological diseases, such as Parkinson's and dementia with Lewy bodies (23).
Given the critical roles of Plk1 during cell division and its frequent overexpression in human tumors (15), it has been considered a cancer therapeutic target (34, 50). Several small-molecule Plk1 inhibitors that compete with ATP at the kinase domain are in clinical trials (39), and efforts are ongoing to identify and validate small molecules that bind to the PBD to inhibit the interaction of Plk1 with its partners (39, 42). These efforts, however, must take into account that inhibition of Plk2, Plk3, or Plk4 may lead to tumor development, as these less-known Plks may function as tumor suppressors in specific cell types (28, 39, 52, 63).
The mammalian genome contains a fifth member of the Plk family, Plk5, initially described as a putative protein encoded by a pseudogene (12) and recently linked to DNA damage (2). Although mouse cells express a full-length Plk5 similar in size to Plk2 or Plk3, human cells express a shorter PLK5 form in which the kinase domain is disrupted due to a stop codon in exon 6, which is followed by an in-frame ATG codon immediately downstream, in the boundary between exons 6 and 7. However, both the murine (long) and human (short) forms display similar cellular functions, and the kinase domain of the murine protein seems to be inactive in kinase assays. Plk5 is specifically expressed in the eye and the brain as well as the ovary in the mouse. Interference with Plk5 expression in PC12 cells and primary hippocampus neuroblasts results in reduced axon growth and neurite formation upon stimulation with neuronal growth factors, suggesting a role for Plk5 in the proper formation of neurite processes. Accordingly, the human PLK5 (hPLK5) protein is detected in the cytoplasm of neurons and glia. Interestingly, PLK5 is dramatically downregulated in human brain tumors, and its expression is inactivated by hypermethylation of the PLK5 promoter region. Reexpression of PLK5 in glioblastoma multiforme (GBM) cells results in the induction of apoptosis, suggesting a therapeutic potential of this protein.
MATERIALS AND METHODS
Plk5 sequences, cDNA constructs, and mutagenesis.
All gene and protein sequences were taken from Ensembl (http://www.ensembl.org/) or Uniprot (http://www.uniprot.org/). Sequence alignment and comparison were performed using Clustal2 (http://www.ebi.ac.uk/Tools/clustalw2/). The mouse Plk5 (mPlk5; NM_183152) and human PLK5 (NR_026557) cDNAs were obtained from the Mammalian Gene Collection Consortium (http://mgc.nci.nih.gov/). Each cDNA was verified by sequencing and subsequently subcloned and fused either to enhanced green fluorescent protein (EGFP) (pDEST3.1-GFP) or to a triple c-myc tandem tag (pcDNA3.1/3× myc-C from Invitrogen) N-terminal to the mouse Plk5 (IMAGE Consortium cDNA Collection; clone 6489647) or to the predicted human PLK5 short form (IMAGE:40000565). mPlk5-ΔN was generated by PCR amplification of the mouse Plk5 cDNA from position 900 to the final stop codon. Point mutations in the cDNA were performed using the QuikChange site-directed mutagenesis kit (Stratagene) and verified by DNA sequencing. The human PLK1-PBD (residues 326 to 603), PLK2-PBD (residues 394 to 677), and PLK3-PBD (residues 372 to 646) were subcloned and fused to EGFP (pDEST3.1-GFP) for expression experiments.
Cell culture and transfection.
NIH 3T3 mouse fibroblasts, B16F10 mouse melanoma cells, HeLa human cervical carcinoma cells, U2OS human osteosarcoma cells, 293T human kidney cells, and the glioblastoma-derived cell lines (U251MG, U87MG, U373MG, LN18, A172) were grown in petri dishes at 37°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. PC12 rat pheochromocytoma cells were cultured in gelatin-coated petri dishes with DMEM supplemented with 10% horse serum and 5% fetal bovine serum. Mouse embryonic fibroblasts (MEFs) derived from p53-null embryos were kindly gifted from Manuel Serrano's laboratory (CNIO). Cell cycle synchronization was performed by different means. G1/S synchronization was achieved by double thymidine block. Cells were incubated with 2.5 mΜ thymidine (Sigma) for 16 h, released into fresh medium for 10 h, and incubated again in thymidine for another 12 h. Cells were then released in fresh medium, and nocodazole 0.2 μM (Sigma) was added 5 h after release. Samples were taken each hour up to 9 h. Quiescent cells were obtained by serum starvation for at least 48 h and released in fresh medium with 10% serum, and samples were taken at the time points indicated in Fig. 2F. Cell transfection was performed using Lipofectamine 2000 reagent (Invitrogen) by following the manufacturer's recommendations. Stable transfectants were achieved by incubating transfected cells with either 400 μg/ml of G418 (Sigma) or 200 μg/ml hygromycin (Invitrogen) for at least 3 weeks, changing the medium every 2 or 3 days. Individual clones were isolated with cloning rings. For RNA interference, several vectors expressing short hairpin RNAs (shRNAs) against Plk5 were generated. Three shRNAs for the mouse, rat, and human Plk5 genes were designed using the GenScript algorithm and cloned into the pRNAT-U6.3/Hygro plasmid (GenScript USA Inc.) (see the supplemental material). Focus assays, colony formation, and soft agar cultures were performed as described previously (41, 49).
Cell cycle and apoptosis profiling analysis was performed by flow cytometry using FACSCanto (BD). For cell cycle analysis, cells were trypsinized, fixed in 1% formaldehyde in phosphate-buffered saline (PBS) for 10 min, and permeabilized in cold 70% ethanol. DNA was stained with propidium iodide (Sigma-Aldrich) supplemented with RNase (100 μg/ml) for 30 min. For the apoptosis analysis, cells were trypsinized and stained with the annexin V-allophycocyanin (APC) apoptosis kit (BD Pharmingen) by following the manufacturer's recommendations. All collected data were further analyzed using FACSDiva software (BD).
Northern blot analysis.
Northern blot analysis was performed using a commercial membrane with 22 different mouse tissue mRNAs (Zyagen). A 350-bp probe of the mouse Plk5 cDNA (from residue 1322 to 1671, overlapping exons 12 to 15) was generated by PCR amplification. The probe was radioactive labeled by using the Rediprime II kit from Amersham, and the membrane was incubated with the probe for 2 h at 65°C in a rocking oven. After incubation, the membrane was washed three times in 2× SSC buffer (SSC 20× stock solution consists of 3 M sodium chloride and 300 mM trisodium citrate, adjusted to pH 7.0 with HCl) for 10 min at room temperature, with a final wash in 1× SSC plus 0.1% SDS for 10 min at 65°C. Finally, the membrane signal was retrieved using a Storm 820 phosphorimager scanner (Amersham).
Antibodies.
Rabbit polyclonal antibodies were raised using specific peptides for the mouse Plk5 protein (Sigma-Genosys) located at the N-terminal segment (mPlk5-NT) (N-PRGGAGRLRLRGKV-C) and the linker region between the kinase domain and the PBD (mPlk5-LK) (N-PDHMEAGNEERDPLC-C). Similarly, two antibodies were generated against two peptides of the human PLK5 protein: one was located at the putative N-terminal segment (hPLK5-NT) (N-RDPGSGRVYRRKGL-C), and the other was located at a linker region between the kinase domain and the PBD (hPLK5-LK) (N-YQNIREGHYPEPAH-C) (Genosphere Biotechnologies). The resulting sera were affinity purified against the antigenic peptides and stored at −20°C in 50% glycerol in PBS. Other commercial antibodies used were anti-α-tubulin antibody (mouse monoclonal DM1a from Sigma), anti-cyclin D antibody (mouse monoclonal DCS-6 from LabVision), anti-p21Cip1 antibody (rabbit polyclonal from Santa Cruz), anti-c-myc antibody (mouse monoclonal 9E10 from ATCC), anti-GFP antibody (mouse monoclonal 7.1 plus 13.1 from Roche), anti-actin antibody (mouse monoclonal AC-40 from Sigma), anti-EphA2 (mouse monoclonal 371805 from R&D Systems), and anti-phospho-histone H3-S10 antibody (rabbit polyclonal from Upstate).
Protein detection.
For Western blot analysis, cells were scraped from petri dishes, washed in PBS, and resuspended for 30 min in radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris HCl [pH 7.5], 150 mM NaCl, 0.5% NP-40, and 10% glycerol) supplemented with protease inhibitor cocktail (Calbiochem), phosphatase inhibitor cocktail (Calbiochem), 0.5 mM dithiothreitol (DTT), and 50 units/ml of benzonase. Protein extraction from mouse tissues was performed by grinding the tissue in liquid nitrogen using a ceramic mortar. The resulting powder was lysed using RIPA buffer for 30 min, and lysates were clarified by centrifugation for 20 min at 16,000 × g. Protein extracts from human tissues were obtained from frozen tissue sections in optimal cutting temperature (OCT) embedding media (Tissue Tek). Extracts were frozen in dry ice and stored at −80°C. Lysates were then separated in 4 to 12% Bis-Tris Criterion XT gels (Bio-Rad) and transferred onto a nitrocellulose membrane (Bio-Rad). Transfer efficiency was checked by red Ponceau S staining, and the blot was then blocked with 5% nonfat milk in PBS-Tween (PBS plus 0.05% Tween 20) for 1 h at room temperature. After being blocked, the blot was incubated with primary antibodies at 4°C overnight. Thereafter, the blots were washed in PBS-Tween and incubated with either anti-rabbit or anti-mouse antibodies (Alexa Fluor 680 labeled from Invitrogen) for 45 min at room temperature. Finally, the fluorescent signal was detected by using the Odyssey infrared imaging system (Li-Cor Biosciences).
For immunohistochemistry, 5-μm sections were taken from human normal or tumor samples and put on glass slides. Slides were deparaffinized, rehydrated, and immersed in 10 mM citrate solution for antigen retrieval. Slides were washed in water, blocked in a 1:10 dilution of normal goat serum (Vector Labs), and incubated with primary antibodies. Slides were then incubated with secondary biotinylated antibodies followed by signal development with an immunoperoxidase reagent (ABC-HRP; Vector Labs) and DAB (Sigma). Sections were lightly counterstained with hematoxylin and analyzed by light microscopy using a Nikon microscope. Human glioblastoma and brain samples were selected from the archives of the Virgen de la Salud Hospital (Toledo, Spain) between 2001 and 2004. Informed consent was required from patients according to the policies of the ethical committee of the hospital. Tissue macroarrays were constructed by getting 1-mm-thick cylinders from formalin-fixed and paraffin-embedded tissue blocks from 157 different glioblastoma samples. Five nontumor controls (four normal brain tissue samples and one tonsil tissue sample) were included in each array. Hematoxylin and eosin-stained full sections from each donor block were used for morphological selections of the representative areas of each tumor.
Kinase assays.
Protein lysates from 293T cells transfected with different constructs of c-myc-tagged Plk5 cDNA or Plk1 cDNA (0.5 to 1 mg of protein) were incubated for 2 h at 4°C in a total volume of 200 μl with 10 μl of anti-c-myc antibody agarose beads (Santa Cruz). One-third of the immunoprecipitates was collected by centrifugation, washed in RIPA lysis buffer, and subjected to SDS-PAGE and immunoblot analysis. The remnant two-thirds of the immunoprecipitation were used for the kinase activity assay. The immunoprecipitates were washed in RIPA buffer three times, with an extra wash in lysis buffer supplemented with 0.5 M NaCl and 5 mM DTT to remove possible nonspecific bound kinases. Then, beads were washed in kinase buffer (10 mM HEPES-NaOH [pH 7.4], 150 mM NaCl, 10 mM MgCl2, and 1 mM EGTA) supplemented with 0.5 mM DTT and phosphatase inhibitor cocktail (Calbiochem). The kinase reaction was initiated by the addition to the beads of 0.2 mM ATP, 2 mCi [32P]ATP (Amersham) in 10 μl of kinase buffer, and either 3 μg of myelin basic protein (MBP) or 3 μg of histone H1 as the substrate. After incubation for 30 min at 30°C, the reaction was stopped with Laemmli sample buffer and analyzed by SDS-PAGE, and signal was obtained by using a Storm 820 phosphorimager scanner (Amersham).
Culture and transfection of neuroblasts and neurite outgrowth assays.
Differentiation and neurite outgrowth were induced in PC12 cells either by incubating cells with neuronal growth factor (NGF) (Sigma) as described previously (19) or by expressing oncogenic Ras (20). After treatment, PC12 cells were kept in low serum medium (1% fetal bovine serum plus 0.5% horse serum) for 5 to 7 days, renewing the medium every 2 days. After the treatment, cells were fixed in 4% formaldehyde and stained with HCS CellMask blue stain (Invitrogen) and propidium iodide for nuclear staining. Neurite length quantification was performed using a specific Metamorph script (Molecular Imaging).
Cultures of primary hippocampal neurons were prepared from E18 rat embryos as described previously (7, 24). Dissociated neurons were seeded on coverslips or dishes coated with poly-l-lysine (1 mg/ml) in plating minimal essential medium (MEM; supplemented with 0.6% glucose and 10% horse serum) for 4 to 6 h until neurons attached. The medium was then replaced by MEM containing N2 supplement (Invitrogen, Carlsbad, CA). Cultures were incubated at 37°C in a humidified atmosphere containing 5% CO2. Transfection of neurons in suspension was done using the Nucleofector device (Amaxa, Gaithersburg, MD) according to the manufacturer's instructions. Transfection of neurons seeded over poly-l-lysine-coated coverslips was done in serum-free medium with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). Hippocampal neurons were treated at day 1 in vitro (DIV1) with 50 ng/ml brain-derived neurotrophic factor (BDNF), and their morphology was analyzed at DIV3. To analyze the neuronal morphology, shRNA-transfected cells were chosen based on GFP expression. Axonal processes were traced, and their lengths were measured using the semiautomatic plug-in NeuronJ from the ImageJ software (National Institutes of Health). Axons were defined as neurites with a length at least twice that of the cell body and always longer than 100 μm at DIV3.
Immunofluorescence and video time-lapse microscopy.
Cells transfected with EGFP-Plk5 cDNA were grown on coverslips for 20 h after transfection, fixed in 4% formaldehyde in PBS for 10 min, and permeabilized in cold (−20°C) methanol. Cells were then incubated with 20 mM glycine in PBS for 10 min, followed by an additional incubation with 10% calf serum in PBST (PBS plus Triton X-100 [0.03%]) for 20 min. Primary antibodies were diluted in PBST and incubated for 1 h in a humid chamber. Samples were washed three times in PBST and then incubated with the corresponding Alexa dye-tagged secondary antibodies (Molecular Probes). DNA was counterstained with 4′,6-diamidine-2-phenylindole (DAPI; 2 μg/ml). Finally, cells were mounted with Mowiol (Sigma-Aldrich) and analyzed with a Leica SP5 confocal scanning microscope. For in vivo video time- lapse, cells stably transfected with EGFP-Plk5 cDNA were grown on glass-bottomed 35-mm plates (Matek). After 20 h of expression, petri dishes were directly mounted onto a Leica SP5 confocal scanning microscope equipped with an argon laser. The growing medium was kept at 37°C and 5% CO2 in a sealed chamber. The 488-nm laser and a 63× Plan Apo lens with a 1.4 numerical aperture (NA) were used in imaging experiments, and cells were filmed for 12 h.
Methylation of CpG islands.
The specific oligonucleotides against the CpG island of the human PLK5 gene (see the supplemental material) were designed using Methprimer (31). Methylation-specific PCR and bisulfite sequencing of normal and tumor samples were performed essentially as reported previously (11).
Statistical analysis.
Error bars and P values were obtained upon three different assays in each experiment. P values were obtained using the t test or one-way analysis of variance (ANOVA) with a Bonferroni posttest in order to compare individual points (GraphPad software).
RESULTS
Plk5 is an antiproliferative Plk family member.
The murine Plk5 gene contains 15 exons, as does Plk3, and has clear orthologs in all mammals, including humans (2) (see Fig. S1 in the supplemental material). Plk5 contains the key catalytic and ATP-binding residues but has unusual substitutions in certain regions, including the activation loop, which lacks the key activatory phosphorylation site (T210 in human Plk1) conserved as serine or threonine in all other Plks. Plk5 contains a clear polo box domain (PBD1) and has weaker sequence similarity in the PBD2 region, which lacks some key residues involved in substrate recognition in Plk1 to Plk3 (see Fig. S1 in the supplemental material), suggesting relevant differences in partner binding. Despite being of more recent origin than Plk2 and Plk3, Plk5 is more divergent in sequence: Plk2 and Plk3 share 68% identity in their kinase domains, 50% overall, while they are just 50% identical (kinase domain) and 37 to 41% identical (overall) to Plk5.
In humans, the corresponding PLK5 locus (ENSG00000185988; SgK384ps) has been considered a pseudogene since there is an in-frame stop codon in exon 6 disrupting the kinase domain (2, 12, 36). This stop codon is present in all human samples in public databases and in samples we have analyzed by genomic PCR in either tumor samples or healthy individuals (see Fig. S2 in the supplemental material). However, the human PLK5 contains a conserved ATG in frame in the boundary between exons 6 and 7, and it therefore has the potential to encode a short (about 36-kDa) protein containing the final fragment of the kinase domain, the PBDs, and the linker between these two domains. All human cDNAs skip the DNA region corresponding to mouse exon 13, which encodes the lesser conserved part of PBD1, although the genomic sequence appears to still be under evolutionary constraint and so may be expressed under some conditions. All other primate genomes sequenced have gaps within the Plk5 coding sequence. None have the frameshift or stop codon within the human sequence, but the chimp sequence has a distinct frameshift within the kinase domain confirmed by four individual reads.
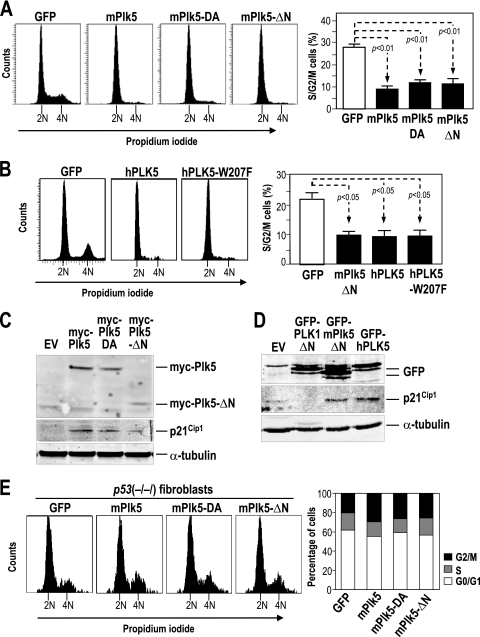
We first tested the effects of Plk5 on the cell cycle by overexpressing the mouse Plk5 cDNA in NIH 3T3 cells. Plk5 induces a significant arrest in G1 and the subsequent reduction in S-phase and G2/M cells (Fig. 1A). This arrest is independent of kinase activity since expression of a D172A mutant (Plk5-DA) (in which the predicted catalytic residue Asp172 has been mutated to alanine) or the N terminus-deleted mutant (Plk5-ΔN) results in a similar arrest. The predicted human PLK5 is also efficient in inducing G1 arrest (Fig. 1B), whereas overexpression of the PBD region of Plk1, Plk2, or Plk3 (constructs similar to Plk5-ΔN or PLK5) results in no or minor differences in G1 arrest (17, 22, 24) (see Fig. S3 in the supplemental material). Since the human PLK5 is predicted to lack the kinase domain, we mutated the conserved tryptophan 207, a residue involved in substrate recognition in PLK1 (18, 29). As shown in Fig. 1B, the human PLK5-W207F mutant is similarly efficient in inducing G1 arrest in these cells. Similar results were found using the corresponding mutant in the mouse sequence (Plk5-W421F; data not shown). These data indicate that the C-terminal region present in the human sequence is sufficient for inducing G1 arrest and that this function does not require the conserved tryptophan in the PBD.
FIG. 1.
Overexpression of Plk5 results in cell cycle arrest. (A) Overexpression of mouse wild-type or mutant Plk5 constructs results in G0/G1 arrest in NIH 3T3 cells as measured by propidium iodide staining for DNA content. (B) The shorter human PLK5 protein and the PBD mutant hPLK5-W207F result in G1 arrest in NIH 3T3 cells to an extent similar to that of the mPlk5-ΔN mutant. (C) The protein levels of myc-Plk5 were measured by immunodetection with antibodies against the myc epitope. Transfection of the Plk5 wild-type or mutant forms results in the upregulation of the cell cycle inhibitor p21Cip1 to an extent comparable with the expression levels of exogenous Plk5. EV, empty vector. (D) Both the human PLK5 and the N terminus deletion mutant of mouse Plk5 result in p21Cip1 induction in NIH 3T3 cells. On the other hand, expression of the PLK1-ΔN mutant does not lead to p21Cip1 upregulation. α-Tubulin was used as a control for protein loading in these assays. (E) Overexpression of wild-type or mutant EGFP-Plk5 does not result in G1 arrest in p53-null fibroblasts as measured by DNA content in EGFP-positive cells. Histograms represent means ± standard errors of the means (SEM).
The antiproliferative effect of the murine or human Plk5 proteins is accompanied by a clear induction of p21Cip1 in these fibroblasts (Fig. 1C and D), suggesting a role for the p53 pathway in these proliferative arrests. These results were obtained using both GFP- and myc-tagged proteins, thus excluding the remote possibility that these tags may modify the function of Plk5. In fact, overexpression of the murine Plk5 or the mutant Plk5-DA or Plk5-ΔN has no significant effect on the cell cycle of p53-null fibroblasts (Fig. 1E). These data suggest a p53-dependent G1 arrest specifically mediated by Plk5 overexpression in primary fibroblasts.
Murine Plk5 accumulates during quiescence and displays limited kinase activity.
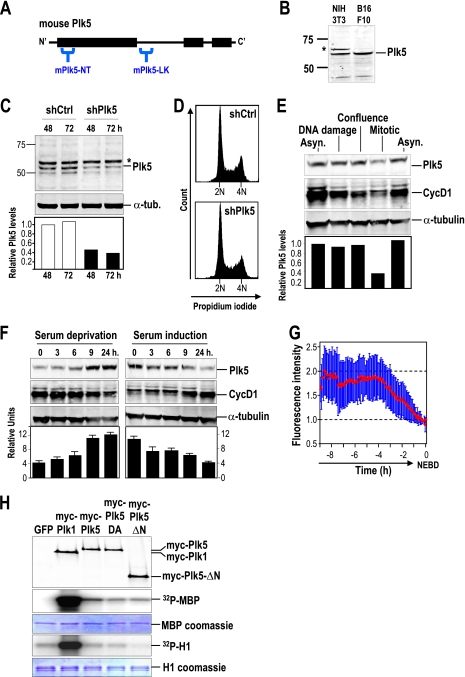
To test the presence of Plk5 proteins in mouse cells, we generated antibodies targeting specific amino acid sequences at the N-terminal region of the murine Plk5 kinase domain (mPlk5-NT) or at the linker region (mPlk5-LK) (Fig. 2A). As a first validation for the antibodies, both mPlk5-NT and mPlk5-LK are able to recognize the EGFP-mPlk5 fusion transiently transfected in 293T cells but do not cross-react with EGFP-Plk1 (see Fig. S4 in the supplemental material). These antibodies are able to detect the endogenous mPlk5 protein in several cell lines, including NIH 3T3 fibroblasts and B16F10 melanoma cells (Fig. 2B). To validate specificity of the antibodies toward the endogenous mPlk5, we tested NIH 3T3 cells treated with specific short hairpin RNAs (shRNAs) against the mouse Plk5 sequence. These shRNAs provoke a significant reduction in Plk5 protein levels 48 h and 72 h after transfection (Fig. 2C). Despite this effect on Plk5 protein levels, treatment with these shRNAs did not result in any alteration in the cell cycle profile or cell proliferation of NIH 3T3 cells (Fig. 2D).
FIG. 2.
The mouse Plk5 is preferentially expressed in quiescent cells and has no detectable kinase activity. (A) Schematic representation of the mPlk5-NT (N terminus) and mPlk5-LK (linker) epitopes selected for generating antibodies against murine Plk5. (B) The mPlk5-NT antibody detects an endogenous protein of about 65 to 68 kDa in NIH 3T3 fibroblasts and B16F10 melanoma cells. An additional upper strong band is found in NIH 3T3 fibroblasts (asterisk). (C) Immunodetection of endogenous mPlk5 in NIH 3T3 cells transfected with vectors expressing short hairpin RNAs (shRNAs) against Plk5 (shPlk5) or scrambled sequences (shCtrl). The additional band (asterisk) found frequently in these cells is not modulated by these shRNAs, suggesting that it is an unspecific band. Protein levels of Plk5 were quantified and normalized to α-tubulin. (D) Expression of shRNAs against Plk5 does not result in major alterations in DNA content profile in these cells. (E) Quantification of Plk5 protein levels in different cellular conditions, such as asynchronous cultures (Asyn.), DNA damage induced by doxorubicin, full confluence during 48 h, or mitosis (nocodazole arrested). Cyclin D1 was used as an additional control for some of these culture conditions. The relative mPlk5 levels were normalized to α-tubulin. (F) mPlk5 is induced during the exit from the cell cycle upon serum starvation, and its protein levels decrease during the entry into the cell cycle after serum stimulation. In panels E and F, α-tubulin was used as a loading control and cyclin D1 was used as an additional control for some of these culture conditions. Histograms represent means ± SEM. (G) mPlk5 was stably expressed in U2OS cells as a GFP fusion protein, and fluorescence was recorded by video microscopy. Green fluorescence of asynchronously growing cells (20 cells per assay) in two different assays was quantified and plotted at different time points before nuclear envelope breakdown (NEBD), showing that GFP-Plk5 expression levels decrease in the G2/M transition. The red line shows the fluorescence average intensity of all the recorded cells. Bars represent the standard deviations of the recorded fluorescence intensity. (H) Mouse Plk1 and Plk5, as well as Plk5 mutant isoforms, were immunoprecipitated using antibodies against the myc epitope or the endogenous proteins (not shown) and assayed for kinase activity on the myelin basic protein (MBP) or histone H1. The immunoprecipitation of the kinase was measured with antibodies against myc (top), whereas the loading of MBP or H1 was measured by Coomassie blue staining.
Using the mPlk5-NT antibody, we detected decreased levels of Plk5 in mitotic cells compared to those in asynchronous or confluent NIH 3T3 cultures (Fig. 2E). When NIH 3T3 cells were treated with doxorubicin to induce DNA damage, Plk5 protein levels were similar to those of asynchronous cultures (Fig. 2E). To further analyze the response of endogenous Plk5 levels, we cultured NIH 3T3 cells in the presence of reduced amounts of mitogenic signals (0.5% serum). As shown in Fig. 2F, serum deprivation induces the accumulation of Plk5 in about 9 h, and this level is maintained for 24 h after removal of serum. Restimulation of these cells with 10% serum results in downregulation of Plk5 in 9 to 24 h. We then next evaluated whether Plk5 levels fluctuate during cell cycle progression. Synchronization of cells in S-phase with thymidine shows a clear downregulation of Plk5 within 4 h of release from the thymidine block, at a time when cells exit from S-phase and enter into G2/M as detected by cell cytometry (see Fig. S5 in the supplemental material). Overall, these results suggest specific downregulation of Plk5 in the later stages of the cell cycle, in agreement with the downregulation of exogenous Plk5 during G2/M when expressed as a stable GFP fusion protein, as shown by video microscopy (Fig. 2G).
The absence of the typical activation residue in the T-loop of Plk5 (see Fig. S1 in the supplemental material) suggests some differences in the regulation of Plk5 activity. In fact, when tested in a typical kinase assay from immunoprecipitated proteins, wild-type or mutant mouse Plk5 only induces a residual signal on the myelin basic protein (MBP) and background activity on histone H1, two general substrates of many kinases (Fig. 2H). Plk1, however, displays a robust kinase activity on both substrates. Immunoprecipitated Plk5 tagged with GFP also failed to phosphorylate these substrates, while GFP-tagged Plk1 provided a significant signal (data not shown), suggesting that these different tags have no effect on the overall kinase activity of either Plk1 or Plk5.
Plk5 suppresses malignant transformation by Ras oncogenes.
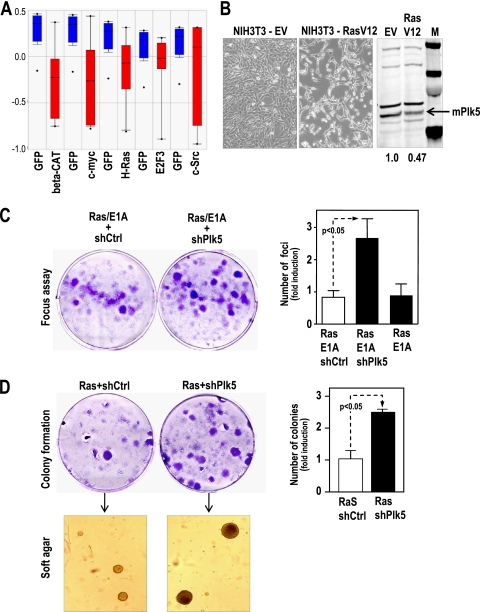
The fact that Plk5 is downregulated in proliferating cells suggests it may be repressed by mitogenic signals or oncogenes. In fact, both the human and mouse Plk5 genes are repressed at the mRNA levels after transfection of primary human cells with oncogenes such as Ras, c-myc, and others (Fig. 3A and B). We also tested whether the endogenous Plk5 may counteract the proliferative effect of oncogenes. Downregulation of endogenous Plk5 by specific shRNAs cooperates with Ras and E1A oncogenes in cell proliferation and malignant transformation in mouse fibroblasts, as measured by the focus assay as well as the formation of transformed colonies that are able to grow in soft agar (Fig. 3C and D). These results indicate that the endogenous Plk5 counteracts the proliferative effect of mitogenic pathways and may function as a tumor suppressor in vitro.
FIG. 3.
Plk5 functions as a tumor suppressor in mouse fibroblasts. (A) hPLK5 mRNA is downregulated upon cell transformation with different oncogenes (data from Oncomine). (B) Oncogenic Ras represses Plk5 expression in NIH 3T3 cells. These cells were transfected with oncogenic H-RasV12 or the empty vector, and the Plk5 protein levels were assessed by immunodetection. (C) Downregulation of mPlk5 by specific shRNAs (shPlk5) cooperates with Ras and E1A oncogenes in cell transformation in a classical focus assay in primary MEFs. (D) shPlk5 NIH 3T3 cells generate more colonies upon transformation with the Ras oncogene. These colonies are transformed, as they are able to grow in soft agar. Histograms represent means ± SEM.
Plk5 is expressed in differentiated cells, such as neurons and glia, both in mice and in humans.
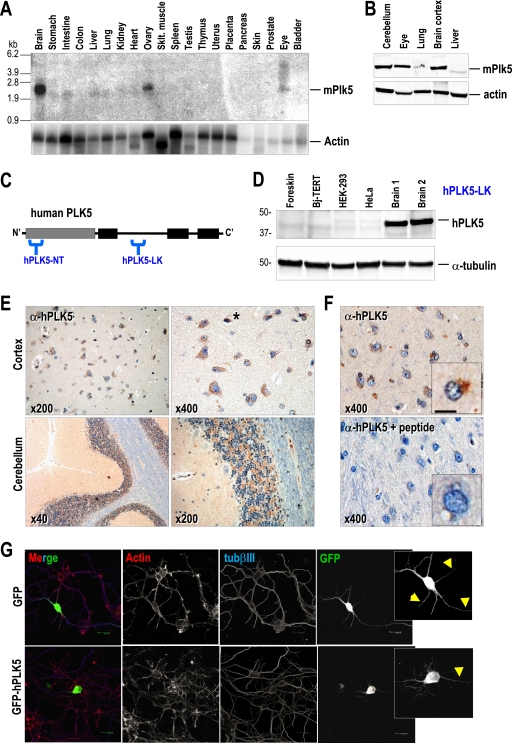
We next tested the expression of Plk5 in several mouse tissues. As depicted in Fig. 4A, the Plk5 mRNA is highly expressed in differentiated tissues, such as the brain, ovary, or eye, but is not detectable in proliferating tissues, such as the colon, spleen, placenta, etc. This expression pattern was confirmed at the protein level, indicating significant levels of Plk5 in the murine central nervous system (cerebellum and brain cortex) as well as in the eye (Fig. 4B). These expression data are also confirmed by available massive cDNA microarray analysis or in situ hybridization in the mouse embryo or young mice (see Fig. S6 in the supplemental material).
FIG. 4.
Mouse and human Plk5 proteins are preferentially expressed in brain cells. (A) Northern blot of mouse tissues indicating a preferential expression of the murine Plk5 mRNA in brain, eye, and ovary. (B) Immunodetection of mPlk5 protein in different tissues, showing the preferential expression in the nervous system. (C) Schematic representation of the predicted hPLK5-NT (N terminus) and hPLK5-LK (linker) epitopes selected for generating antibodies against human PLK5. Note that the hPLK5-NT antibody has been designed against an epitope not present in the predicted 36-kDa short form. (D) The hPLK5-LK antibody is able to detect a band that runs at a position slightly higher than that of the predicted molecular size (values in kilodaltons at left). This protein is present in two different samples from human brain tissue, but it is barely detectable in different cell lines, including primary human foreskin fibroblasts. (E) Detection of PLK5 in human brain cortex and cerebellum sections by immunohistochemistry. PLK5 (brown signal) is present mostly in the cytoplasm of neurons and glial cells in the cortex and granular cells in the cerebellum. Eventual nuclear staining is also observed in some of these cells (asterisk), although it is not evident with this technique. (F) Preabsorption with the antigenic hPLK5-LK peptide used to generate the antibody eliminates the signal in the brain, suggesting the specificity of the hPLK5-LK antibody in these samples. (G) When expressed in neuroblasts as a GFP fusion protein, the human PLK5 is mostly confined to the somatic and proximal neuritic levels (arrowheads), localizing to the cytoplasm and eventually to the nucleus. In contrast, soluble GFP localizes all over the cell, including the nucleus, cytoplasm, and neuritic processes.
We next analyzed whether human cells also express PLK5 at the protein level by generating two different antibodies. The hPLK5-LK antibody was generated against amino acid sequences in the region homologous to the linker of the murine protein (Fig. 4C). An additional antibody was generated using the predicted amino acid sequences at the N-terminal region (hPLK5-NT). The hPLK5-LK antibody clearly recognizes the human PLK5 protein when overexpressed as a GFP fusion protein, and this signal is significantly decreased by PLK5-specific shRNAs (see Fig. S7 in the supplemental material). Interestingly, the hPLK5-LK antibody recognizes a short, endogenous, ∼40-kDa protein in brain tissue extracts, in agreement with the predicted sequence originated from the ATG codon at the boundary between exons 6 and 7. Human PLK5 is highly expressed in brain but not in other cell types, such as primary fibroblasts or tumoral HeLa or HEK293T cells (Fig. 4D). On the other hand, hPLK5-NT (designed against the human N terminus) detects the long Plk5 isoforms in mouse tissues, but it is not able to detect any peptide in human cells (data not shown).
To further analyze in detail the expression of the human PLK5 protein, we used the hPLK5-LK antibody in immunohistochemistry of a tissue macroarray containing a variety of human tissues. As represented in Fig. 4E, hPLK5 displays a characteristic pattern of expression, with polarized localization in neurons and glial cells. This staining is specific, and it is lost when the primary antibody is previously preabsorbed with the peptide used for its generation (Fig. 4F). The cytoplasmic localization observed by this antibody is in agreement with the cytoplasmic signal detected with the human PLK5-GFP fusion constructs in stable cell lines (see Fig. S8 in the supplemental material) as well as in primary neuroblasts (Fig. 4G). In addition to these cells in the central nervous system, hPLK5 seems to be expressed in other highly differentiated cells, such as cells of the serous acini in the parotid gland, distal and proximal tubules of the kidney, tubules of the seminal gland, Kupffer cells and some hepatocytes in the liver, and some cells in the germinal center of lymph nodes (see Fig. S9 in the supplemental material).
Plk5 is required for axonal growth in neuroblasts.
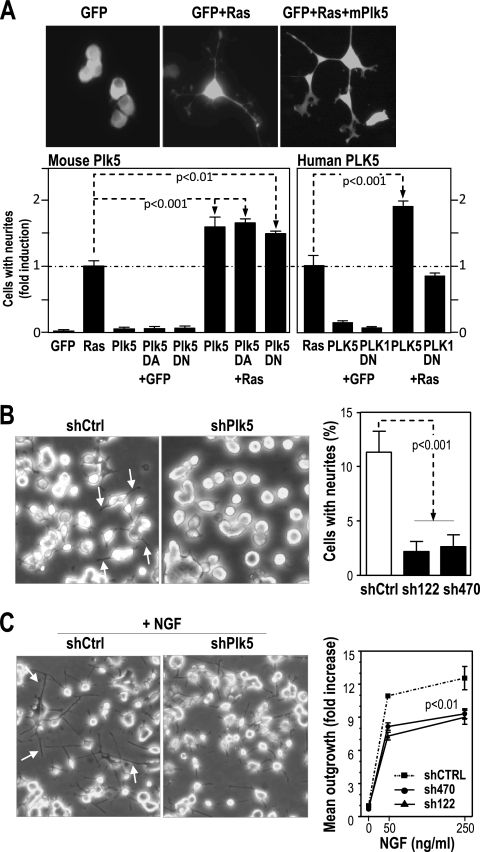
Since both mouse and human Plk5 proteins seem to be preferentially expressed in the central nervous system, we next tested the effect of modulating Plk5 levels on neuronal function. We first used rat pheochromocytoma PC12 cells, which can be differentiated into neuron-like cells in vitro by either Ras oncogenes or nerve growth factor (NGF). Overexpression of either mouse or human Plk5 per se does not induce cell differentiation (measured by the formation of neurites) but cooperates with Ras in this assay (Fig. 5A). Overexpression of mPlk5-D172A or mPlk5-ΔN resulted in similar effects, suggesting that Plk5 synergizes with Ras in neurite formation in a kinase-independent manner. Interestingly, the human PLK5 construct also resulted in a similar cooperation, whereas the PBD region of PLK1 did not have any effect in this assays (Fig. 5A). We also tested the effect of the loss of the endogenous Plk5 on neurite formation. Rat PC12 cells were stably transfected with different shRNAs against the rat Plk5 transcript (shPlk5). In general, shPlk5 transfected cells display a significantly reduced number of neurites when cultured in rich medium with serum (Fig. 5B). In addition, NGF-induced neurite outgrowth was reduced in these cultures in the presence of the two different shRNAs against Plk5, suggesting that this protein is required for maintaining neurites or for de novo formation of these cellular extensions in these assays (Fig. 5C).
FIG. 5.
Plk5 modulates the outgrowth of neurites. (A) Overexpression of mouse and human Plk5 cDNA synergizes with Ras in neurite formation in PC12 cells. PC12 cells were cotransfected with GFP (to label cells and cellular extensions) plus H-Ras and wild-type or mutant Plk5 mouse isoforms and human PLK5. The ratio of cells with neurite emissions was scored. The Plk1-PBD fragment was used as a control. (B) PC12 cells were stably transfected with shRNAs against rat Plk5 (shPlk5) or scrambled sequences (shCtrl). The number of cells with neurites (arrows) was scored in clones expressing two different shRNAs, sh122 and sh470, against Plk5. Representative microphotographs of shCtrl and shPlk5(sh122) are also shown. (C) PC12 cells were treated with different doses (50 and 250 ng/ml) of NGF, and the outgrowth of neurites was quantified and represented as the fold increase versus nontreated cells. Representative microphotographs of shCtrl and shPlk5(sh122) cultures treated with 250 ng/ml are shown. Data are represented as means ± SEM.
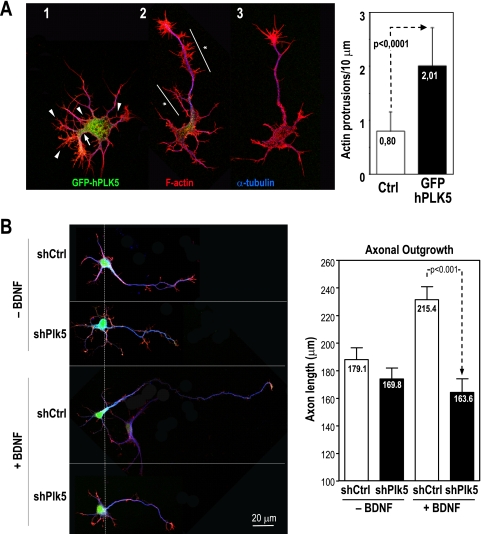
To analyze the relevance of Plk5 in primary neuroblasts, we next investigated the effect of Plk5 overexpression or inhibition in primary cells isolated from embryonic rat hippocampus. Overexpression of human PLK5 induces the formation of abundant F-actin-rich protrusions that are much less abundant in neuroblasts negative for the GFP-Plk5 construct (Fig. 6A). These Plk5-overexpressing neurons present a distorted phenotype with an abnormally thick neuritic initial segment at the basis of the neurites and an increased abundance of thin actin-positive protrusions. Transfection of Plk5 shRNAs into these neuroblasts does not result in major effects in the existing neuron extensions. However, knockdown of Plk5 prevents the growth of axons induced by brain-derived neurotrophic factor (BDNF) (Fig. 6B). Whereas BDNF induces a significant growth of the axons, increasing their lengths by 20%, no axonal growth is observed in the presence of the shRNAs against Plk5, thus suggesting that Plk5 is involved in neuron differentiation and axonal growth induced by growth factors such as NGF or BDNF.
FIG. 6.
Plk5 modulates cellular morphology in primary neuroblasts. (A) A GFP-hPLK5 fusion protein was overexpressed for 48 h in primary neuroblasts from rat hippocampus. High expression levels of GFP-hPLK5 (neuron 1, green) preclude normal axon development, inducing abnormally thick hillocks at the basis of the neurites (arrow) and abundant thin F-actin-positive protrusions (arrowheads). In the case of medium/low expression levels (neuron 2, green), axons develop normal length, but in contrast to control nontransfected neurons (neuron 3), they display axon tracts highly enriched in F-actin-positive protrusions (asterisks). In the graph, the numbers of protrusions per 10 μm of axon length are compared between controls and GFP-hPLK5-transfected neurons. (B) Expression of shRNAs against Plk5 (shPlk5) or scrambled sequences (shCtrl) (green signal) does not have a major impact on the morphology of nontreated primary neuroblasts. However, whereas the control cultures respond to stimulation with BDNF by elongating their axons, this response is impaired in cells in which Plk5 is knocked down. Green reflects shRNA expression, red indicates F-actin staining, and blue shows α-tubulin staining. Results are means plus standard deviations of results from three independent experiments (25 cells/experiment).
PLK5 is downregulated in human brain tumors by promoter hypermethylation.
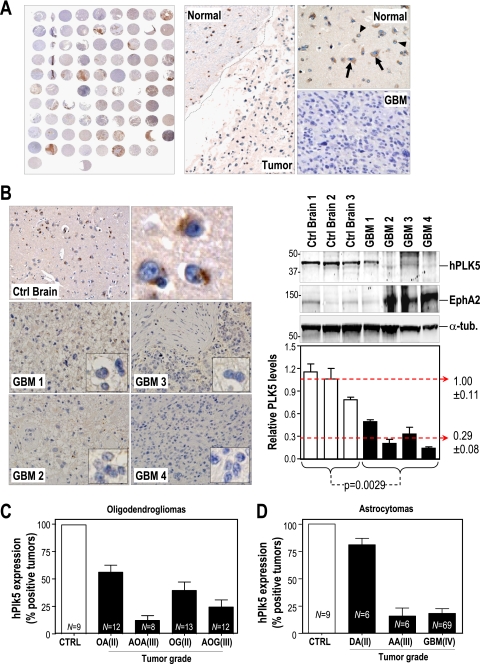
The presence of Plk5 in the central nervous system and its possible function in modulating neuron differentiation prompted us to analyze the expression of hPLK5 in human brain tumors. Our preliminary data using cDNA expression profiles suggested that hPLK5 is indeed downregulated at the mRNA level in human primary brain tumors, including oligodendrogliomas, oligoastrocytomas, and astrocytomas of different grades (see Fig. S10 in the supplemental material). To test PLK5 protein levels in these tumors, we used the hPLK5-LK antibody for immunohistochemistry in a panel of 144 gliomas of different grades. As represented in Fig. 7A, hPLK5 is readily detectable in the normal brain controls in this array, whereas this specific signal is reduced in tumor cells. We also analyzed specific tumors and normal brain samples by immunoblotting, confirming that hPLK5 protein levels are severely decreased in advanced tumors, such as glioblastoma multiforme (GBM) samples both in paraffin sections and in protein lysates (Fig. 7B). Whereas hPLK5 is detected in neurons and glia cells of control brains (an average of 95% of control cells are positive), most tumors are negative or present reduced expression of hPLK5 staining (an average of 25 to 35% of positive cells in astrocytomas and 30 to 55% of positive cells in oligodendrogliomas). Higher-grade tumors tend to show a stronger reduction in PLK5 transcripts. Thus, whereas about 80% of grade II astrocytomas are positive for hPLK5, only 15 to 20% of advanced tumors (grade III and IV) maintain this signal (Fig. 7C and D; see also Fig. S10 in the supplemental material). PLK5 and PLK2 are the only Plk family members downregulated in these tumors, whereas PLK1 is upregulated and PLK3 and PLK4 display only minor changes in expression (see Fig. S10).
FIG. 7.
PLK5 is downregulated in human brain tumors. (A) Human PLK5 expression is evaluated in 144 different gliomas by immunohistochemistry (IHC) on tissue microarrays. In some of these tumors, hPLK5 (brown signal; arrows) is expressed in normal areas but not in the tumor mass. (B) The reduction in hPLK5 expression is demonstrated by IHC and Western blot analysis with similar results. Three different control brains and four different glioblastomas (GBM) are represented. The glioblastoma marker EphA2 is overexpressed in three out of the four glioblastomas, but it is expressed at low levels in control brains. The relative hPLK5 protein levels were normalized versus the levels of α-tubulin. (C) Quantification of hPLK5 expression in oligodendrogliomas sorted by tumor type and by malignant grade. OA, oligoastrocytoma (grade II); AOA, anaplastic oligoastrocytoma (grade III); OG, oligodendroglioma (grade II); AOG, anaplastic oligodendroglioma (grade III). (D) Quantification of hPLK5 expression in astrocytomas sorted by tumor type and by malignant grade. DA, diffuse astrocytoma (grade II); AA, anaplastic astrocytoma (grade III); GBM, glioblastoma multiforme (grade IV). Cases with cytoplasmic staining of hPLK5 in more than 10% of the cells were scored as a positive tumor in this analysis. However, whereas the average number of positive cells was close to 90% in normal samples, this average ranged from 10% to 60% in the positive tumors, indicating that PLK5 levels in the tumors never reached the protein levels found in normal samples. Error bars represent SEM.
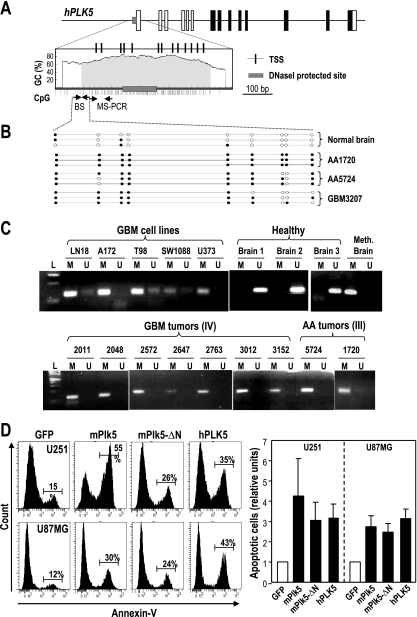
We then studied the mechanism that silences human PLK5 in brain tumors. Plk5 genes contain a CpG island, close to the transcription factor binding site both in mouse (not shown) and in humans (Fig. 8A), that is susceptible to changes in methylation. Whereas most CpG dinucleotides are not methylated in control brains, we have detected a significant methylation of CpG dinucleotides in DNA isolated from human astrocytoma and GBM. Indeed, 16 out of 18 tumors tested by either methylation-specific PCR (MS-PCR) or DNA sequencing after bisulfite modification showed a significant methylation of the human PLK5 promoter region. Similar data were obtained when analyzing cell lines derived from glioblastomas (Fig. 8B and C). Thus, the observed tumor-associated reduction in hPLK5 protein levels is likely due to epigenetic alteration of the PLK5 gene, at least in brain tumors.
FIG. 8.
Human PLK5 is silenced by epigenetic means, and its reexpression results in apoptotic cell death of brain tumor cells. (A) Schematic representation of the CpG island upstream of the human PLK5 gene. The positions of the oligonucleotides used for methylation-specific (MS)-PCR or DNA sequencing after bisulfite modification are shown as small arrows. The predicted transcription start sites (TSS) and the region protected upon DNase I treatment are also shown to indicate the overlap between these regulatory regions and the CpG island. (B) Methylation analysis after bisulfite sequencing (BS) of a fragment of the CpG island in normal brain tissue and three representative high-grade glioblastomas. Empty circles represent nonmethylated cytosines, whereas closed circles indicate methylation at these positions. (C) Methylation of the hPLK5 CpG island in different glioblastoma (GBM) cell lines and primary tumors and primary astrocytomas (AA) as detected by MS-PCR. Three unmethylated (U) healthy brain samples are shown as controls. The methylated (M) band is detected in the healthy brain after in vitro sample methylation, but not before treatment. On the other hand, the unmethylated band is no longer detected after this treatment. Most tumor cells display a strong preference for the M bands. (D) U251MG and U87MG glioblastoma cells were transfected with GFP or the GFP fusions of the wild-type mouse Plk5, the mPlk5-ΔN mutant, and human PLK5. Apoptosis was scored by annexin V staining and fluorescence-activated cell sorting (FACS) analysis. Data represent means ± SEM.
We next tested whether the reexpression of hPLK5 has any effect in these tumor cells. We observed that several GBM cell lines, such as A172, T98, SW1088, U373MG, or LN18, contain methylated PLK5 promoters (Fig. 8C). Overexpression of hPLK5 in these cell lines (Fig. 8D; see also Fig. S11 in the supplemental material) results in a dramatic apoptotic response as measured by annexin V staining. Overexpression of the mouse wild-type Plk5 or the Plk5-ΔN mutant, which mimics the truncated human protein, has a similar apoptotic effect. Similarly, the conserved W207 in the PBD is not required for the apoptotic effect in GBM cells (see Fig. S3 in the supplemental material). The effect of the other Plk family members is less dramatic, although overexpression of Plk1 and Plk2 also results in a moderate increase in apoptosis, at least in the p53-null U373MG cell line (see Fig. S3). All together, these data suggest that PLK5 silencing through epigenetic means may help brain tumors to escape from apoptosis.
DISCUSSION
Polo kinases are considered central modules in cell cycle regulation and have attracted much attention due to their therapeutic potential (4, 34, 39, 50). Most research efforts have been focused on the analysis of Plk1 as an essential protein in mitotic entry and cytokinesis (8, 40, 53) and, recently, on the relevant role of Plk4 in centriole biology and mitotic fidelity (9, 21, 27, 28, 43).
Whereas Plk1 and Plk4 orthologs are present in most eukaryotes, the Plk2 subfamily is first clearly seen in deuterostomes, duplicates to form Plk2 and Plk3 in vertebrates, and gives rise to Plk5 in mammals (see Fig. S1 in the supplemental material). Plk2 seems to share some functions with Plk1 and Plk4 in centriole function (13, 58), while Plk3 localizes either to the nucleolus or to the microtubules around the nuclear membrane and may be involved in several cellular processes, including S-phase progression and cell cycle checkpoints, Golgi fragmentation, cytokinesis, and apoptosis (5, 6, 14, 33, 44, 57, 60, 62). Mouse Plk5 is highly similar to Plk2 and Plk3 in sequence, and it may share similar functions in DNA damage, as recently reported (2). However, whereas both Plk2 and Plk3 are induced by mitogens, mouse Plk5 is downregulated in serum-stimulated cells and accumulates upon serum deprivation (Fig. 2). Downregulation during G2/M is likely to be due to protein degradation, as it is also observed in exogenous GFP-Plk5 proteins (Fig. 2G; see also Fig. S5 in the supplemental material). Proteasome-dependent degradation has also been observed for Plk3 (1).
The most striking feature of the human PLK5 gene is the presence of a stop codon in exon 6, close to the end of the kinase domain, and the presence of a new ATG in-frame codon in the boundary between exons 6 and 7. This stop codon is not observed in any primate sequence, suggesting a significant functional shift in this gene within the human lineage. Surprisingly, while we cannot detect the expression of an N-terminal-truncated protein, we do see strong expression of a product likely (by size and epitope) to initiate just downstream of the stop codon. Thus, as predicted from the sequence and also reported in reference 2, the human gene encodes a smaller, 40-kDa protein that lacks the kinase domain but contains the linker sequences and the PBD, which is in agreement with our results using specific antibodies against the N terminus or the linker of the predicted proteins. This downstream ATG is conserved in all Plk5 orthologs and in the other Plk family members, although we have not detected shorter Plk5 proteins in mouse extracts. All Plk5 orthologs lack an activation loop threonine, which is the activatory residue on Plk1 and conserved in all other Plks. In addition, the mouse Plk5 protein fails to efficiently phosphorylate typical kinase substrates, such as histone H1 or MBP, suggesting that Plk5 proteins may have lost some or all kinase catalytic activity and function in a PBD-dependent manner. Indeed, mouse Plk5 not only lacks the activatory threonine at the T-loop but also has a divergence in the highly conserved DFG motif present in the ATP binding pocket of almost all kinase domains of the kinome (see Fig. S1 in the supplemental material). This is carried further in humans, where only a truncated form lacking the kinase domain is expressed. The apparent deletion of the kinase domain in the human protein may be a logical extension of this trend. Several kinases are known to lack catalytic function and instead serve as scaffolds or kinase substrates. About 45 kinases lack at least one of the catalytic sites required for the kinase function (36). An almost identical set of pseudokinases is predicted in mice, supporting a conserved noncatalytic function for many mammalian kinases (12).
Both Plk2 and Plk3 participate in neuron structure and synaptic plasticity, and the conserved PBD is involved in this neuronal function (26). Plk2 is also required for plasticity of hippocampal neurons during epileptiform activity and chronically elevated activity (46, 48). The present study demonstrates that Plk5 is expressed mostly in the adult murine and human brain and may modulate neurite formation in established cell lines and primary neurons. In addition, Plk5 participates in the formation of neuritic processes in response to NGF/BDNF or Ras. The formation of these processes is not observed upon interference with the endogenous Plk5 transcript, suggesting a relevant role for Plk5 in this pathway. As suggested for Plk2 and Plk3, this function is also likely to be dependent on the PBD, as human PLK5 also modulates neurite formation (Fig. 5 and 6) and does not contain the kinase domain. The PBD is a relevant domain that confers specificity in partner binding (54). Thus, it is tempting to speculate that Plk5 has evolved as a kinase-deficient protein, where the PBD may function in protein-protein interaction without the subsequent phosphorylation. Similar gene duplication and subfunctionalization or neofunctionalization have been suggested for other kinase family members, such as VRK3 (45), a duplicate gene that loses catalytic activity but maintains a modification of the regulatory site, similar to the PBD here. Whether the Plk5 PBD recognizes specific substrates or may compete for common partners with other Plk family members is currently unknown. However, the fact that Plk5 lacks relevant residues involved in protein interaction in PBD2 (H538/K450 in Plk1) and the dispensability of the conserved tryptophan in PBD1 suggests that Plk5 may use different mechanisms than those reported for Plk1, as suggested for PLK2 and PLK3 (54).
Human PLK5 is significantly downregulated in brain tumors at both the mRNA and the protein level. In fact, the PLK5 gene is frequently silenced by promoter hypermethylation in astrocytomas and glioblastoma multiforme, the most common and most aggressive type of primary brain tumor, accounting for 52% of all primary brain tumor cases and 20% of all intracranial tumors. This epigenetic silencing suggests the relevance of this protein in either maintaining the differentiated phenotype or triggering cell death in neurons, glia, or their progenitors. Loss of heterozygosity (LOH) for human chromosome 19p13.3, where PLK5 is located, is frequently found in several human cancers, including breast tumors, uterine cervical adenocarcinomas, carcinoma metastases to the brain, and ovarian tumors. Although the main candidate so far has been STK11/LKB1 (0.3 Mb away from PLK5), recent evidence suggests additional targets in this region (25).
The roles of PLK1 in cell division and its overexpression in human tumors have suggested that inhibition of PLK1 activity may have beneficial effects in cancer. Indeed, current therapeutic strategies are directed to identify not only small molecular ATP competitors for PLK1 but also drugs that interact with the PBD (39, 42). However, both PLK2 and PLK3 have been suggested to have tumor suppressor functions, and PLK2 may be inactivated by promoter hypermethylation in B-cell malignancies or liver tumors (38, 52). On the other hand, Plk3-null mice displayed an increase in weight and developed tumors in multiple organs with age (63). Thus, Plk family members may behave both as oncogenes and as tumor suppressors (30). As suggested by our results, the new member of the Plk family, PLK5, may also function as a tumor suppressor in the brain. In fact, PLK5 has a relevant antiproliferative effect, and it is quite potent in the induction of apoptosis. Thus, reexpression of PLK5 by gene therapy strategies or by using epigenetic drugs may have beneficial effects on these aggressive tumors. The detailed analysis of the PBD of PLK5 and its comparison to other Plk family members may provide a molecular basis for new compounds targeting PBD partners for cancer intervention.
Supplementary Material
Acknowledgments
We thank Susana Temiño for technical assistance, Marta Cañamero for help in histology, and Jorge Martinalbo and Erich Nigg for preliminary assays on the overexpression of Plk5 constructs.
This work was funded by grants from the Association for International Cancer Research (AICR number 08-0188), Foundation Ramon Areces, Ministerio de Ciencia e Innovación (MICINN; SAF2009-07973), and NHGRI HG004164-01. The Cell Division and Cancer Group of the CNIO is supported by the OncoCycle Programme (S-BIO-0283-2006) from the Comunidad de Madrid, the OncoBIO Consolider-Ingenio 2010 Programme (CSD2007-00017) from the MICINN, Madrid, Spain, and the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement MitoSys (no. 241548).
Footnotes
Published ahead of print on 18 January 2011.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alberts, G. F., and J. A. Winkles. 2004. Murine FGF-inducible kinase is rapidly degraded via the nuclear ubiquitin-proteosome system when overexpressed in NIH 3T3 cells. Cell Cycle 3:678-684. [PubMed] [Google Scholar]
- 2.Andrysik, Z., et al. 2010. The novel mouse polo-like kinase 5 responds to DNA damage and localizes in the nucleolus. Nucleic Acids Res. 38:2931-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ang, X. L., D. P. Seeburg, M. Sheng, and J. W. Harper. 2008. Regulation of postsynaptic RapGAP SPAR by polo-like kinase 2 and the SCFbeta-TRCP ubiquitin ligase in hippocampal neurons. J. Biol. Chem. 283:29424-29432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archambault, V., and D. M. Glover. 2009. Polo-like kinases: conservation and divergence in their functions and regulation. Nat. Rev. Mol. Cell Biol. 10:265-275. [DOI] [PubMed] [Google Scholar]
- 5.Bahassi el, M., et al. 2002. Mammalian polo-like kinase 3 (Plk3) is a multifunctional protein involved in stress response pathways. Oncogene 21:6633-6640. [DOI] [PubMed] [Google Scholar]
- 6.Bahassi el, M., R. F. Hennigan, D. L. Myer, and P. J. Stambrook. 2004. Cdc25C phosphorylation on serine 191 by Plk3 promotes its nuclear translocation. Oncogene 23:2658-2663. [DOI] [PubMed] [Google Scholar]
- 7.Banker, G. 1998. Culturing nerve cells, 2nd ed. MIT Press, Cambridge, MA.
- 8.Barr, F. A., H. H. Sillje, and E. A. Nigg. 2004. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5:429-440. [DOI] [PubMed] [Google Scholar]
- 9.Bettencourt-Dias, M., et al. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15:2199-2207. [DOI] [PubMed] [Google Scholar]
- 10.Bradham, C. A., et al. 2006. The sea urchin kinome: a first look. Dev. Biol. 300:180-193. [DOI] [PubMed] [Google Scholar]
- 11.Bueno, M. J., et al. 2008. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell 13:496-506. [DOI] [PubMed] [Google Scholar]
- 12.Caenepeel, S., G. Charydczak, S. Sudarsanam, T. Hunter, and G. Manning. 2004. The mouse kinome: discovery and comparative genomics of all mouse protein kinases. Proc. Natl. Acad. Sci. U. S. A. 101:11707-11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cizmecioglu, O., S. Warnke, M. Arnold, S. Duensing, and I. Hoffmann. 2008. Plk2 regulated centriole duplication is dependent on its localization to the centrioles and a functional polo-box domain. Cell Cycle 7:3548-3555. [DOI] [PubMed] [Google Scholar]
- 14.Conn, C. W., R. F. Hennigan, W. Dai, Y. Sanchez, and P. J. Stambrook. 2000. Incomplete cytokinesis and induction of apoptosis by overexpression of the mammalian polo-like kinase, Plk3. Cancer Res. 60:6826-6831. [PubMed] [Google Scholar]
- 15.de Carcer, G., I. Perez de Castro, and M. Malumbres. 2007. Targeting cell cycle kinases for cancer therapy. Curr. Med. Chem. 14:969-985. [DOI] [PubMed] [Google Scholar]
- 16.Elia, A. E., et al. 2003. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the polo-box domain. Cell 115:83-95. [DOI] [PubMed] [Google Scholar]
- 17.Fink, J., et al. 2007. Cell type-dependent effects of polo-like kinase 1 inhibition compared with targeted polo box interference in cancer cell lines. Mol. Cancer Ther. 6:3189-3197. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Alvarez, B., G. de Carcer, S. Ibanez, E. Bragado-Nilsson, and G. Montoya. 2007. Molecular and structural basis of polo-like kinase 1 substrate recognition: implications in centrosomal localization. Proc. Natl. Acad. Sci. U. S. A. 104:3107-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greene, L. A., J. M. Aletta, A. Rukenstein, and S. H. Green. 1987. PC12 pheochromocytoma cells: culture, nerve growth factor treatment, and experimental exploitation. Methods Enzymol. 147:207-216. [DOI] [PubMed] [Google Scholar]
- 20.Guerrero, I., H. Wong, A. Pellicer, and D. E. Burstein. 1986. Activated N-ras gene induces neuronal differentiation of PC12 rat pheochromocytoma cells. J. Cell Physiol. 129:71-76. [DOI] [PubMed] [Google Scholar]
- 21.Habedanck, R., Y. D. Stierhof, C. J. Wilkinson, and E. A. Nigg. 2005. The polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7:1140-1146. [DOI] [PubMed] [Google Scholar]
- 22.Hanisch, A., A. Wehner, E. A. Nigg, and H. H. Sillje. 2006. Different Plk1 functions show distinct dependencies on polo-box domain-mediated targeting. Mol. Biol. Cell 17:448-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inglis, K. J., et al. 2009. Polo-like kinase 2 (PLK2) phosphorylates alpha-synuclein at serine 129 in central nervous system. J. Biol. Chem. 284:2598-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, N., X. Wang, M. Jhanwar-Uniyal, Z. Darzynkiewicz, and W. Dai. 2006. Polo box domain of Plk3 functions as a centrosome localization signal, overexpression of which causes mitotic arrest, cytokinesis defects, and apoptosis. J. Biol. Chem. 281:10577-10582. [DOI] [PubMed] [Google Scholar]
- 25.Kato, N., M. Romero, L. Catasus, and J. Prat. 2004. The STK11/LKB1 Peutz-Jegher gene is not involved in the pathogenesis of sporadic sex cord-stromal tumors, although loss of heterozygosity at 19p13.3 indicates other gene alteration in these tumors. Hum. Pathol. 35:1101-1104. [DOI] [PubMed] [Google Scholar]
- 26.Kauselmann, G., et al. 1999. The polo-like protein kinases Fnk and Snk associate with a Ca(2+)- and integrin-binding protein and are regulated dynamically with synaptic plasticity. EMBO J. 18:5528-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleylein-Sohn, J., et al. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell 13:190-202. [DOI] [PubMed] [Google Scholar]
- 28.Ko, M. A., et al. 2005. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat. Genet. 37:883-888. [DOI] [PubMed] [Google Scholar]
- 29.Lee, K. S., T. Z. Grenfell, F. R. Yarm, and R. L. Erikson. 1998. Mutation of the polo-box disrupts localization and mitotic functions of the mammalian polo kinase Plk. Proc. Natl. Acad. Sci. U. S. A. 95:9301-9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lens, S. M., E. E. Voest, and R. H. Medema. 2010. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat. Rev. Cancer 10:825-841. [DOI] [PubMed] [Google Scholar]
- 31.Li, L. C., and R. Dahiya. 2002. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18:1427-1431. [DOI] [PubMed] [Google Scholar]
- 32.Llamazares, S., et al. 1991. Polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev. 5:2153-2165. [DOI] [PubMed] [Google Scholar]
- 33.Lopez-Sanchez, I., M. Sanz-Garcia, and P. A. Lazo. 2009. Plk3 interacts with and specifically phosphorylates VRK1 in Ser342, a downstream target in a pathway that induces Golgi fragmentation. Mol. Cell. Biol. 29:1189-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malumbres, M., and M. Barbacid. 2007. Cell cycle kinases in cancer. Curr. Opin. Genet. Dev. 17:60-65. [DOI] [PubMed] [Google Scholar]
- 35.Manning, G., G. D. Plowman, T. Hunter, and S. Sudarsanam. 2002. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 27:514-520. [DOI] [PubMed] [Google Scholar]
- 36.Manning, G., D. B. Whyte, R. Martinez, T. Hunter, and S. Sudarsanam. 2002. The protein kinase complement of the human genome. Science 298:1912-1934. [DOI] [PubMed] [Google Scholar]
- 37.Matthew, E. M., et al. 2007. Replication stress, defective S-phase checkpoint and increased death in Plk2-deficient human cancer cells. Cell Cycle 6:2571-2578. [DOI] [PubMed] [Google Scholar]
- 38.Pellegrino, R., et al. 2010. Oncogenic and tumor suppressive roles of polo-like kinases in human hepatocellular carcinoma. Hepatology 51:857-868. [DOI] [PubMed] [Google Scholar]
- 39.Perez de Castro, I., G. de Carcer, G. Montoya, and M. Malumbres. 2008. Emerging cancer therapeutic opportunities by inhibiting mitotic kinases. Curr. Opin. Pharmacol. 8:375-383. [DOI] [PubMed] [Google Scholar]
- 40.Petronczki, M., P. Lenart, and J. M. Peters. 2008. Polo on the rise—from mitotic entry to cytokinesis with Plk1. Dev. Cell 14:646-659. [DOI] [PubMed] [Google Scholar]
- 41.Quereda, V., J. Martinalbo, P. Dubus, A. Carnero, and M. Malumbres. 2007. Genetic cooperation between p21Cip1 and INK4 inhibitors in cellular senescence and tumor suppression. Oncogene 26:7665-7674. [DOI] [PubMed] [Google Scholar]
- 42.Reindl, W., J. Yuan, A. Kramer, K. Strebhardt, and T. Berg. 2008. Inhibition of polo-like kinase 1 by blocking polo-box domain-dependent protein-protein interactions. Chem. Biol. 15:459-466. [DOI] [PubMed] [Google Scholar]
- 43.Rodrigues-Martins, A., M. Riparbelli, G. Callaini, D. M. Glover, and M. Bettencourt-Dias. 2007. Revisiting the role of the mother centriole in centriole biogenesis. Science 316:1046-1050. [DOI] [PubMed] [Google Scholar]
- 44.Ruan, Q., et al. 2004. Polo-like kinase 3 is Golgi localized and involved in regulating Golgi fragmentation during the cell cycle. Exp. Cell Res. 294:51-59. [DOI] [PubMed] [Google Scholar]
- 45.Scheeff, E. D., J. Eswaran, G. Bunkoczi, S. Knapp, and G. Manning. 2009. Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure 17:128-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seeburg, D. P., M. Feliu-Mojer, J. Gaiottino, D. T. Pak, and M. Sheng. 2008. Critical role of CDK5 and polo-like kinase 2 in homeostatic synaptic plasticity during elevated activity. Neuron 58:571-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seeburg, D. P., D. Pak, and M. Sheng. 2005. Polo-like kinases in the nervous system. Oncogene 24:292-298. [DOI] [PubMed] [Google Scholar]
- 48.Seeburg, D. P., and M. Sheng. 2008. Activity-induced polo-like kinase 2 is required for homeostatic plasticity of hippocampal neurons during epileptiform activity. J. Neurosci. 28:6583-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sotillo, R., et al. 2001. Wide spectrum of tumors in knock-in mice carrying a Cdk4 protein insensitive to INK4 inhibitors. EMBO J. 20:6637-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strebhardt, K., and A. Ullrich. 2006. Targeting polo-like kinase 1 for cancer therapy. Nat. Rev. Cancer 6:321-330. [DOI] [PubMed] [Google Scholar]
- 51.Sunkel, C. E., and D. M. Glover. 1988. Polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J. Cell Sci. 89(Pt. 1):25-38. [DOI] [PubMed] [Google Scholar]
- 52.Syed, N., et al. 2006. Transcriptional silencing of polo-like kinase 2 (SNK/PLK2) is a frequent event in B-cell malignancies. Blood 107:250-256. [DOI] [PubMed] [Google Scholar]
- 53.Takaki, T., K. Trenz, V. Costanzo, and M. Petronczki. 2008. Polo-like kinase 1 reaches beyond mitosis—cytokinesis, DNA damage response, and development. Curr. Opin. Cell Biol. 20:650-660. [DOI] [PubMed] [Google Scholar]
- 54.van de Weerdt, B. C., et al. 2008. Polo-box domains confer target specificity to the Polo-like kinase family. Biochim. Biophys. Acta 1783:1015-1022. [DOI] [PubMed] [Google Scholar]
- 55.van de Weerdt, B. C., and R. H. Medema. 2006. Polo-like kinases: a team in control of the division. Cell Cycle 5:853-864. [DOI] [PubMed] [Google Scholar]
- 56.Wang, L., J. Gao, W. Dai, and L. Lu. 2008. Activation of polo-like kinase 3 by hypoxic stresses. J. Biol. Chem. 283:25928-25935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, Q., et al. 2002. Cell cycle arrest and apoptosis induced by human polo-like kinase 3 is mediated through perturbation of microtubule integrity. Mol. Cell. Biol. 22:3450-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warnke, S., et al. 2004. Polo-like kinase-2 is required for centriole duplication in mammalian cells. Curr. Biol. 14:1200-1207. [DOI] [PubMed] [Google Scholar]
- 59.Winkles, J. A., and G. F. Alberts. 2005. Differential regulation of polo-like kinase 1, 2, 3, and 4 gene expression in mammalian cells and tissues. Oncogene 24:260-266. [DOI] [PubMed] [Google Scholar]
- 60.Xie, S., et al. 2004. MEK1-induced Golgi dynamics during cell cycle progression is partly mediated by polo-like kinase-3. Oncogene 23:3822-3829. [DOI] [PubMed] [Google Scholar]
- 61.Xie, S., et al. 2001. Reactive oxygen species-induced phosphorylation of p53 on serine 20 is mediated in part by polo-like kinase-3. J. Biol. Chem. 276:36194-36199. [DOI] [PubMed] [Google Scholar]
- 62.Xie, S., et al. 2001. Plk3 functionally links DNA damage to cell cycle arrest and apoptosis at least in part via the p53 pathway. J. Biol. Chem. 276:43305-43312. [DOI] [PubMed] [Google Scholar]
- 63.Yang, Y., et al. 2008. Polo-like kinase 3 functions as a tumor suppressor and is a negative regulator of hypoxia-inducible factor-1 alpha under hypoxic conditions. Cancer Res. 68:4077-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zimmerman, W. C., and R. L. Erikson. 2007. Polo-like kinase 3 is required for entry into S phase. Proc. Natl. Acad. Sci. U. S. A. 104:1847-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.