Abstract
Rice dwarf virus (RDV), with 12 double-stranded RNA (dsRNA) genome segments (S1 to S12), replicates in and is transmitted by vector insects. The RDV-plant host-vector insect system allows us to examine the evolution, adaptation, and population genetics of a plant virus. We compared the effects of long-term maintenance of RDV on population structures in its two hosts. The maintenance of RDV in rice plants for several years resulted in gradual accumulation of nonsense mutations in S2 and S10, absence of expression of the encoded proteins, and complete loss of transmissibility. RDV maintained in cultured insect cells for 6 years retained an intact protein-encoding genome. Thus, the structural P2 protein encoded by S2 and the nonstructural Pns10 protein encoded by S10 of RDV are subject to different selective pressures in the two hosts, and mutations accumulating in the host plant are detrimental in vector insects. However, one round of propagation in insect cells or individuals purged the populations of RDV that had accumulated deleterious mutations in host plants, with exclusive survival of fully competent RDV. Our results suggest that during the course of evolution, an ancestral form of RDV, of insect virus origin, might have acquired the ability to replicate in a host plant, given its reproducible mutations in the host plant that abolish vector transmissibility and viability in nature.
Most plant viruses are transmitted by insects in various ways that are classified into nonpersistent, semipersistent, and persistent transmissions, depending primarily on the times of acquisition, latency, and retention (5). A variety of interactions among plants, viruses, and insects are involved in viral propagation, and the prerequisite intimate relationships impose selective pressure, shape viral populations, and influence viral evolution. Plant viruses maintained exclusively in host plants without transmission by insect vectors accumulate mutations that are detrimental to vector transmissibility, regardless of the mode of transmission, and emerge as transmission-defective (TD) strains (3). Representative examples of this phenomenon are provided by potyviruses (nonpersistent) (18), cucumovirus (nonpersistent) (10), and tospoviruses (propagative) (24), in which mutations occur mostly in genes for structural proteins and/or transmission helper components (nonstructural proteins) assumed to interact with insect partners. Plant viruses transmitted in a propagative manner, a persistent mode of transmission that involves replication in vectors, allow us to examine the genetic effects of the maintenance of such viruses in either host.
Rice dwarf virus (RDV) is a member of the family Reoviridae, one of the largest virus families, with a wide range of hosts from fungi to plants, insects, nonhuman vertebrates, and humans (1). RDV has a double-stranded RNA (dsRNA) genome consisting of 12 segments (S1 to S12). It induces dwarf symptoms with leaf specks in monocotyledonous plant hosts and can replicate in vector insects, such as leafhoppers. Seven segments, S1, S2, S3, S5, S7, S8, and S9, encode structural proteins P1, P2, P3, P5, P7, P8, and P9, respectively, which form double-layered virions, while the remaining segments code for nonstructural proteins Pns4, Pns6, Pns10, Pns11, and Pns12 (16, 26). The RDV-plant host-vector insect system is an amenable and suitable system for exploring virus evolution, adaptation, and population structures, because of the availability of established cultured cell lines of the vector insect and the ease of long-term maintenance of RDV-infected rice plants. Similar to another phytoreovirus, Wound tumor virus (15), RDV gradually lost its transmissibility by insect vectors, becoming a TD strain, when RDV-infected plants were propagated vegetatively for more than 4 years in a greenhouse (7). Possibly relevant mutations have been mapped to a nonsense substitution in S2, which encodes an outer capsid protein required for insect transmission (17, 19, 25, 27).
The closer association of RDV with insect vectors than with plant hosts was postulated previously on the basis of the asymptomatic infection of vector insects with RDV and the high frequency (100%) of vertical transmission in insect vectors (4). However, the possible involvement of plant host factors could not be discounted, because such insects were reared on rice plants. To gain nonbiased insights into possible effects of hosts on virus population structures, we used cultured insect vector cells, with which plant host factors can never interact during viral replication, in the present study. By analyzing the expression of the 12 virus-encoded proteins in RDV-infected plants maintained vegetatively in the absence of the vector insects and in RDV-infected vector cell monolayers (VCM) maintained in the absence of the plant host, we examined the effect of each viral host on the integrity of the viral genome. We also studied the effects of vector insects on the selection of competent viruses from mixtures of transmission-competent (TC) and TD viruses from rice plants.
MATERIALS AND METHODS
Viruses, plants, and cells.
The O strain of RDV (9), which we used as a TC isolate, was maintained in a greenhouse (27 ± 3°C) by inoculation of healthy rice seedlings by the viruliferous insect Nephotettix cincticeps (leafhopper) at least once a year. TD isolates of RDV were obtained by vegetative propagations of RDV-infected plants as described by Kimura (7). Three months after inoculation, tillers were separated from vegetatively propagated plants and propagated separately. After the first winter season, one plant was selected from a set of vegetatively propagated infected plants and used for vegetative propagation the following year. The same cycle of selection and vegetative propagation was repeated for 6 years. After inoculation of VCM (8) with strain O (VCM0), cells were cultured to confluence, and about half of such cells were used for subsequent weekly passaging. RDV-infected cells were maintained for 6 years (VCM6).
Transmission assay.
Second- to third-instar nymphs of N. cincticeps were allowed to feed on RDV-infected rice plants for 2 days. The insects were subsequently reared for 18 days on healthy rice seedlings with occasional replacement by fresh seedlings. The insects were then allowed 2 days of inoculation access feeding on healthy rice seedlings. The inoculated plants were grown in a greenhouse (27 ± 3°C) for 40 days for development of symptoms.
Immunoblotting, RT-PCR, and sequence analysis.
The expression of proteins of each strain was examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with protein-specific antibodies as reported previously (20, 21, 22). For reverse transcription (RT)-PCR, RNA was extracted from RDV-infected rice plants and VCM by using an RNeasy Plus Mini Kit (Qiagen), following the manufacturer's protocol. Finally, the RNA was eluted in 60-μl diethyl pyrocarbonate (DEPC)-treated water (Sigma). Then, RT was carried out with a Qiagen Omniscript Kit (Qiagen GmbH, Hilden, Germany), following the manufacturer's protocol, to prepare cDNA, using random hexamers as primers. The primers used for PCR were as follows: 5′-GGCGATGGCTTATCCTAACG and 5′-CCATCAACAGAGCAGAATCC for S2 and 5′-GGTAAACTTGCGCCTTTCTG and 5′-ATCAGAATCCCTG for S10. The program for PCR of S2 was as follows: denaturation for 2 min at 94°C and 30 cycles of denaturation for 15 s at 94°C, annealing for 20 s at 52°C, and extension for 60 s at 68°C. The program for PCR of S10 was the same as that for S2, except the annealing temperature was 55°C. The PCR products were inserted into pCR-Blunt II-Topo (Invitrogen). The primers used for sequence analysis were universal M13 primers.
Immunofluorescence staining.
VCM were inoculated with RDV at a multiplicity of infection (MOI) of 0.0002. Two hours later, virus-neutralizing antibodies were added to the medium. Ten days later, the cells were fixed, stained with viral-particle-specific IgG conjugated to Alexa Fluor 488 (Invitrogen) and Pns10-specific IgG conjugated to rhodamine (Invitrogen), and examined by confocal fluorescence microscopy (LSM 510; Carl Zeiss, New York, NY) using a 10× lens, and images were obtained with LSM 510 image browser software as described previously (22). Coverslips with mock-infected cells were included as controls in each experiment and were processed in the same way as coverslips with infected cells.
RESULTS
Changes in population structures of RDV maintained in different hosts.
The 12 RDV proteins encoded by S1 through S12 were analyzed by immunoblotting. Maintenance of RDV for 6 years in infected plants resulted in the disappearance of detectable P2 and Pns10 (Table 1, TD6). In contrast, all protein products were detected when the virus was alternated between plants and insects (Table 1, strain O). Moreover, all 12 proteins were detected in RDV-infected VCM that had been maintained for 6 years with weekly transfer (Table 1, VCM6), with slightly reduced accumulation of P2, P9, and Pns11 compared to levels in VCM newly inoculated with strain O (Table 1, VCM0). The P2 outer capsid protein functions in virus adsorption to and penetration into vector insect cells (16, 17, 19, 27). Pns10 forms tubules approximately 85 nm in diameter that mediate transportation of viral particles to neighboring healthy vector insect cells (6, 20). Thus, all 12 viral proteins might be required for RDV propagation in insects, while the P2 and Pns10 proteins might be dispensable in plants. Sequence analysis of cDNA of TD6-S2 revealed the disappearance of nucleotide G at position 433 from the wild-type sequence and a resultant termination codon at nucleotides 460 to 462. We showed earlier that a mutation at position 47 of S2 in 12-year old TD RDV resulted in a termination codon and absence of functional P2 (25). Thus, mutations appear to occur randomly, and position 47 is not a mutational hot spot. In TD6-S10, we detected an A-to-U point mutation at position 120, which generated a UAA termination codon. Thus, loss of P2 and Pns10 was due to mutations in the corresponding open reading frames (ORFs).
TABLE 1.
Immunodetection of proteins encoded by 12 genome segments of Rice dwarf virus in different hosts
| Strain or isolate | Protein dilutiona |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | P2 | P3 | Pns4 | P5 | Pns6 | P7 | P8 | P9 | Pns10 | Pns11 | Pns12 | |
| Ob | 1:5,120 | 1:10,240 | 1:40,960 | 1:5,120 | 1:320 | 1:20,480 | 1:10,240 | 1:40,960 | 1:10,240 | 1:20,480 | 1:5,120 | 1:320 |
| TD6c | 1:2,560 | >1:10 | 1:40,960 | 1:1,280 | 1:320 | 1:5,120 | 1:5,120 | 1:40,960 | 1:640 | >1:10 | 1:5,120 | 1:320 |
| VCM0d | 1:5,120 | 1:2,560 | 1:40,960 | 1:10,240 | 1:320 | 1:10,240 | 1:40,960 | 1:40,960 | 1:2,560 | 1:10,240 | 1:10,240 | 1:1,280 |
| VCM6e | 1:1,280 | 1:640 | 1:40,960 | 1:5,120 | 1:320 | 1:2,560 | 1:5,120 | 1:40,960 | 1:160 | 1:1,280 | 1:320 | 1:1,280 |
The values are the highest dilutions of the original materials that were detected by immunoblotting.
O, RDV which was alternatively maintained between plants and insects.
TD6, RDV which was maintained in rice plant for 6 years.
VCM0, RDV in VCM newly inoculated with strain O.
VCM6, RDV in VCM maintained for 6 years with weekly transfer.
Genetic distances between strains O, TD6, VCM0, and VCM6 using the unweighted pair group method with arithmetic mean (UPGMA) (3) ranged from 0.0000 to 0.0083 in segment S2 and from 0.0000 to 0.0031 in segment S10, values very close to each other compared to those for RDV strains O and China, with 0.0346 for S2 and 0.0267 for S10 (see Fig. S1 and S2 in the supplemental material).
Dysfunctional mutations gradually accumulated when RDV was maintained exclusively in host plants.
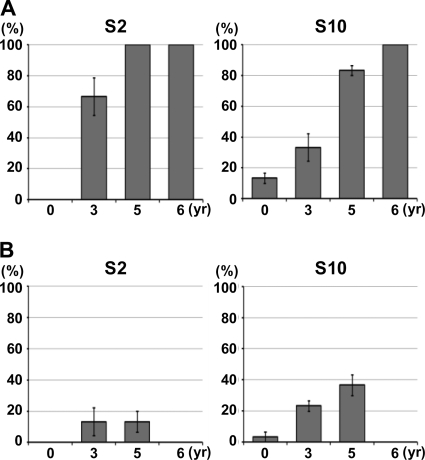
Postulating that insect cells might select RDV populations that express all the viral proteins necessary for the replication cycle in the vector insect, we first examined the changes in population structures of RDV S2 and S10 in rice plants. The analyzed regions were 876 bp starting from the first AUG of the open reading frame for S2 and 1,321 bp covering the whole region of the segment for S10 (see Fig. S3A and C in the supplemental material). Ten clones were analyzed for each sample, and all assays were repeated three times using different sets of samples. As shown in Fig. 1A, the percentages of cDNA clones with deleterious mutations in both S2 and S10 increased with increasing duration of vegetative propagation. “Deleterious mutations” are defined as nonsense, deletion, and/or insertion mutations that would lead to production of only short, truncated forms of viral proteins. The results with a set of assays are illustrated in Fig. S3 in the supplemental material. With respect to S2, deleterious mutations were found in 6 out of 10 clones derived from RDV maintained for 3 years in rice plants but not in those from the RDV O strain (Fig. 1; see Fig. S3 in the supplemental material). All 10 clones contained deleterious mutations, either nonsense mutations or single-nucleotide deletions, resulting from maintenance for 5 or 6 years in plants (Fig. 1; see Fig. S3B in the supplemental material). Similarly, RDV accumulated nonsense mutations, in addition to missense and silent mutations, on S10 after being maintained for 5 or 6 years in rice (see Fig. S3D in the supplemental material). It is interesting that deleterious mutations were found at different positions and that different types of mutated S2 and S10 became dominant in three samples of RDV that had been maintained in rice plants for 3, 5, and 6 years. These results suggest dynamic population structures of RDV in host plants. Similar results were obtained by using different samples. Detailed sequence information on the analyzed cDNA clones is available upon request.
FIG. 1.
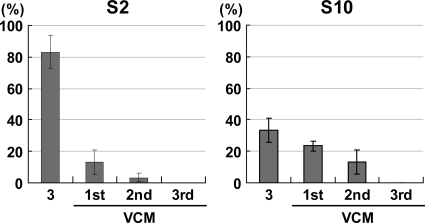
Frequency of deleterious mutations in S2 and S10 of RDV strain O during long-term vegetative propagation (A) and exclusion of RDV with such mutants by a single passage through VCM (B). (A) RDV genomic RNA was prepared from infected rice plants in which RDV had been maintained by vegetative propagation for 0, 3, 5, and 6 years. cDNA clones of S2 and S10, respectively, were sequenced, and the ratios of cDNA clones with deleterious mutations to wild-type clones are shown as percentages. (B) Genomic RNA was recovered from virus populations that had been passaged once through VCM that had been inoculated with the populations of RDV used for the analysis shown in panel A. The bars represent the percentages of cDNA clones with deleterious mutations. Ten clones were sequenced in each assay, all assays were performed three times, and the values are expressed as means ± standard deviations (SD). RDV maintained for 6 years in rice plants was unable to infect VCM and was not included in this analysis.
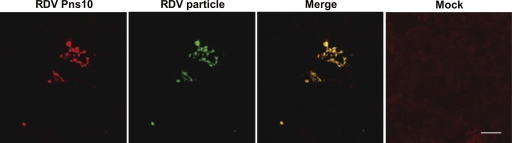
Approximately 10% of S10 cDNA clones of strain O that had replicated in plants for less than 1 year (90 days after inoculation) harbored mutations (Fig. 1A, 0 year). As noted above, Pns10, encoded by S10, is involved in cell-to-cell movement in VCM (6, 20). Thus, some populations of strain O might not be able to move to neighboring cells in insect vector cells, a hypothesis verified by the following observation. We inoculated VCM with RDV and analyzed RDV distribution 10 days after inoculation following the previously reported method (20). As shown in Fig. 2, a small percentage of viruses, probably corresponding to mutated cDNA clones (10%) (Fig. 1A), were confined to a single cell and seemed unable to move to neighboring cells, in contrast to the clusters of approximately 10 infected cells seen in most cases, as reported previously (20).
FIG. 2.
Focal spread of RDV strain O in the presence of virus-neutralizing antibodies. VCM were inoculated for 2 h with RDV strain O at an MOI of 0.0002 and cultured for 10 days in the presence of virus-neutralizing antibodies, which prevent infection by non-cell-associated virus particles. Viruses were labeled with Pns10-specific IgG conjugated with rhodamine (red; RDV Pns10) and viral-particle-specific IgG conjugated to Alexa 488 (green; RDV particle). Immunostaining was visualized by confocal fluorescence microscopy. “Merge” is a merged view of “RDV Pns10” and “RDV particle.” “Mock” is a view of cells inoculated with healthy plant materials. Scale bar, 30 μm.
Vector insect cells eliminated RDV populations with dysfunctional genomes.
We next examined the effects of insect vector cells as hosts on the selection of wild-type RDV and the exclusion of mutants during viral proliferation. As shown in Fig. 1B, the relative percentages of deleteriously mutated cDNA clones in the population decreased greatly after one passage in VCM compared to those in corresponding populations of RDV that had been maintained for 3 and 5 years in plants exclusively (Fig. 1A). RDV TD6, maintained for 6 years in rice plants, no longer infected VCM. Although we failed to detect cDNA clones of intact S2 in the RDV population maintained in plants for 5 years, a very minor population of RDV was believed to retain functional S2 and infectivity for VCM (Fig. 1B). Such populations were probably purified by passage through the VCM. Thus, VCM apparently served as a filter for the selection of functional genomes. Dysfunctional mutated S2 and S10, detected after one passage in VCM, suggesting the presence of mixed populations of functional and dysfunctional segments that perhaps resulted from transcapsidation (11), were excluded after a further two passages through VCM (Fig. 3). S2 and S10, with silent and missense mutations that would be still able to direct the synthesis of full-length protein products, were not filtered out by passage though VCM (see Fig. S4 to S6 in the supplemental material). This may suggest that mutations occurred randomly across the genome segments but that those tolerable for virus viability were detected.
FIG. 3.
Elimination of deleterious mutations of RDV after three passages through VCM. Cells in VCM were inoculated with crude sap of rice leaves infected with RDV strain O that had been maintained for 3 years in rice plants (3) at an MOI of 1 in the first passage. Macerates of the infected cells were further inoculated into healthy VCM as a second passage. The procedure was repeated up to the third passage. Sequence analysis was performed 10 days after inoculation for each passage. The bars represent the percentages of cDNA clones with deleterious mutations. Ten cDNAs were sequenced in each assay, all assays were performed three times, and the values are expressed as means ± SD.
Viruses with functional genomes were purified after transmission by vector insects.
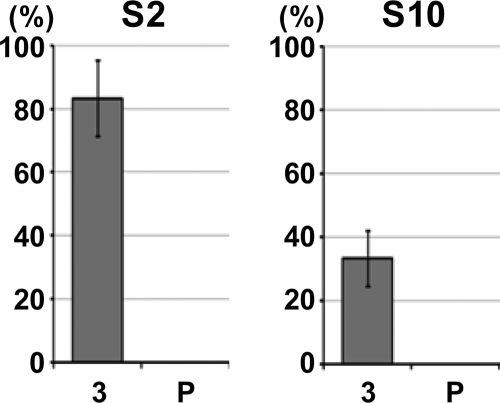
Given the ability of VCM to select functional RDV genomes, we postulated that the vector insect, N. cincticeps, might play the same role as a filter during a natural transmission process, so that RDV with translatable segments would be selected and RDV with untranslatable segments would be rejected. To test our hypothesis, we used strain O of RDV maintained for 3 years in rice plants (Fig. 1A) and allowed insects to acquire the viruses and transmit them to healthy rice plants by the method reported previously (4). As shown in Fig. 4, no deleteriously mutated cDNA was detected after transmission in either genomic segment, although a few silent and missense mutations appeared randomly in translatable genomes (see Fig. S7 in the supplemental material). Thus, insects played an important role as an alternate host in selection of RDV populations with functional genome segments under natural conditions. RDV is maintained in nature via its ability to infect and multiply in vector insects. The fact that all the cDNAs of S10 that we isolated had wild-type sequences, in slight contrast to the data in Fig. 1, might be attributable to the difference in the numbers of days after inoculation: 40 days in this experiment versus 90 days in Fig. 1.
FIG. 4.
Complete elimination of deleterious mutations in S2 and S10 after a single passage through vector insects. RDV strain O-infected rice plants, maintained for 3 years (Fig. 1A; designated 3 here) were exposed to vector insects, which then transmitted RDV to healthy rice seedlings (P). The bars represent the percentages of deleterious mutations in 10 cDNAs sequenced in each assay. Each assay was performed three times, and the values are expressed as means ± SD.
DISCUSSION
Populations of RDV have different structures when the viruses are maintained in cultured insect cells and in its plant host. The integrity of all 12 functional proteins of RDV was maintained for 6 years in vector insect cells, while during prolonged maintenance of RDV in rice plants, random point mutations accumulated in S2 and S10 and the encoded proteins were no longer expressed (Table 1 and Fig. 1). It is evident that accumulation of deleterious mutations during replication cycles is much greater in rice than in VCM. The number of replication rounds over 6 years was estimated to be 2312 × 0.1 (≫1031) for VCM6 and 206 × 0.3, equal to 19.2 × 106 (<108), for TD6. The factors considered in the estimation included the number of passages (312 weekly transfers in 6 years), the ratio of insect cells passaged (half), and the reduced virus content of VCM6 (10%) relative to VCM0 (data not shown) for VCM, and the number of passages (six annual transfers), the rate of increased plant mass (approximately 20 times in a year), and the reduced content of TD6 virus (30%) relative to strain O (data not shown) for rice. The estimated number of RDV replication cycles was at least 1023-fold greater in VCM than in rice. Therefore, we concluded that the accumulation pace of deleterious mutations in plants is much greater than in VCM on a replication round basis.
RDV with deleterious mutant S2 and S10 was unable to infect insect vector cells (25) and thus could not be transmitted to rice plants (7). Furthermore, passage through insect vector cells or intact insects efficiently selected for RDV that was able to replicate in both hosts from among mixed populations of RDV generated in plants (Fig. 1 and 4). These observations provide insight into the functional roles of RDV S2 and S10 and RDV evolution.
Segments S2 and S10 are clearly dispensable in the plant host, while all 12 viral proteins appear necessary for replication in vector insects. S2 encodes outer capsid protein P2, which is involved in vector transmission, probably via a role in adsorption and entry (25). Pns10, together with actin-based filopodia, forms tubular inclusion bodies that encase viral particles in infected cells and extend toward neighboring cells to enhance intercellular viral propagation (20, 23). A defect in this machinery might be associated with the inability of mutant RDV to replicate infectiously, as shown in Fig. 1. The dispensability of Pns10 in the plant host is of interest, given its anti-RNA-silencing activities (2), which should enhance RDV replication. The observation that RDV with S10 nonsense mutations is favored in the plant host remains to be explored.
The insect origin of plant reoviruses, including RDV, has been postulated on the basis of their closer association with insects than with the host plant (13). While RDV was maintained for 6 years, possibly by vertical transmission, in insects under laboratory conditions (4), no seed-mediated transmission occurs in plants. Thus, RDV can survive for generations in insects alone, while plants are “dead-end” hosts. Moreover, while RDV causes severe symptoms in plant hosts, RDV and other plant-infecting reoviruses induce asymptomatic infection in leafhoppers, a well-established phenomenon for a better-adapted virus. In addition, the experimental data gained here showing (i) that RDV maintained exclusively in rice plants gradually accumulated nonsense mutations in S2 and S10 and that lack of the encoded proteins resulted in complete loss of transmissibility, (ii) that RDV retained an intact functional protein-coding genome when maintained exclusively in cultured insect cells, and (iii) that vector insects can serve as a filter for selection of RDV with an intact functional genome and only such RDV can infect insect cells and thus be transmitted to the host plant strongly support the hypothesis that the ancestor of RDV might have originated in the vector insect. The ancestral insect virus might have been transmitted vertically at a high frequency through insects and/or horizontally via plants as a transitory reservoir, and it might have evolved into RDV after acquiring the ability to replicate in plants. It was reported previously that Nilaparvata lugens reovirus (NLRV) (12, 14), which replicates in planthoppers but not in rice plants, can be transmitted laterally between insects reared on rice. Thus, it might not be surprising that such insect viruses might evolve into plant reoviruses by gaining molecular tools that allow propagation in rice plants. Acquisition of the ability to replicate in plant hosts might have enhanced ecological fitness potential by enabling the virus to spread horizontally among insects and plants, as well as between them. It is unlikely that RDV was of plant virus origin, given that mutations detrimental to replication in insects accumulate relatively rapidly in plant hosts.
Supplementary Material
Acknowledgments
This study was supported by funds from the Ministry of Agriculture, People's Republic of China (grant nyhyzx07-051), and by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) to T.O.
Footnotes
Published ahead of print on 29 December 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Boccardo, G., and R. G. Milne. 1984. Plant reovirus group, p. 1-7. In A. F. Morant and B. D. Harrison (ed.), CM/AAB descriptions of plant viruses, no. 294. Commonwealth Mycological Institute and Association of Applied Biologists. Gresham Press, Old Woking, England.
- 2.Cao, X., et al. 2005. Identification of an RNA silencing suppressor from a plant double-stranded RNA virus. J. Virol. 79:13018-13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elena, S. F., P. Agudelo-Romero, and J. Lalic. 2009. The evolution of viruses in multi-host fitness landscapes. Open Virol. J. 3:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honda, K., et al. 2007. Retention of Rice dwarf virus by descendants of pairs of viruliferous vector insects after rearing for 6 years. Phytopathology 97:712-716. [DOI] [PubMed] [Google Scholar]
- 5.Hull, R. 2002. Matthews' plant virology, 4th ed. Academic Press, San Diego, CA.
- 6.Katayama, S., T. Wei, T. Omura, J. Takagi, and K. Iwasaki. 2007. Three-dimensional architecture of virus-packed tubule. J. Electron Microsc. (Tokyo) 56:77-81. [DOI] [PubMed] [Google Scholar]
- 7.Kimura, I. 1976. Loss of vector-transmissibility in an isolate of rice dwarf virus. Phytopathol. Soc. Jpn. 42:322-324. [Google Scholar]
- 8.Kimura, I., and T. Omura. 1988. Leafhopper cell cultures as a means for phytoreovirus research. Adv. Dis. Vector Res. 5:111-135. [Google Scholar]
- 9.Kimura, I., Y. Minobe, and T. Omura. 1987. Changes in a nucleic acid and a protein component of Rice dwarf virus particle associated with an increase in symptom severity. J. Gen. Virol. 68:3211-3215. [Google Scholar]
- 10.Liu, S., X. He, G. Park, C. Josefsson, and K. L. Perry. 2002. A conserved capsid protein surface domain of Cucumber mosaic virus is essential for efficient aphid vector transmission. J. Virol. 76:9756-9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyazaki, N., et al. 2005. Transcapsidation and the conserved interactions of two major structural proteins of a pair of phytoreoviruses confirm the mechanism of assembly of the outer capsid layer. J. Mol. Biol. 345:229-237. [DOI] [PubMed] [Google Scholar]
- 12.Nakashima, N., and H. Noda. 1995. Nonpathogenic Nilaparvata lugens reovirus is transmitted to the brown planthopper through rice plant. Virology 207:303-307. [DOI] [PubMed] [Google Scholar]
- 13.Nault, L. R., and E. D. Ammar. 1989. Leafhopper and planthopper transmission of plant viruses. Annu. Rev. Entomol. 34:503-529. [Google Scholar]
- 14.Noda, H. 1995. Nonpathogenic reoviruses of leafhoppers and planthoppers. Semin. Virol. 6:109-116. [Google Scholar]
- 15.Nuss, D. L. 1984. Molecular biology of wound tumor virus. Adv. Virus Res. 29:57-93. [DOI] [PubMed] [Google Scholar]
- 16.Omura, T., and J. Yan. 1999. Role of outer capsid proteins in transmission of Phytoreovirus by insect vectors. Adv. Virus Res. 54:15-43. [DOI] [PubMed] [Google Scholar]
- 17.Omura, T., et al. 1998. The P2 protein of rice dwarf phytoreovirus is required for adsorption of the virus to cells of the insect vector. J. Virol. 72:9370-9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sako, N. 1980. Loss of aphid transmissibility of turnip mosaic virus. Phytopathology 70:647-649. [Google Scholar]
- 19.Tomaru, M., et al. 1997. The loss of outer capsid protein P2 results in nontransmissibility by the insect vector of rice dwarf phytoreovirus. J. Virol. 71:8019-8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei, T., et al. 2006. The spread of Rice dwarf virus among cells of its insect vector exploits virus-induced tubular structures. J. Virol. 80:8593-8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei, T., et al. 2006. Pns4 of rice dwarf virus is a phosphoprotein, is localized around the viroplasm matrix, and forms minitubules. Arch. Virol. 151:1701-1712. [DOI] [PubMed] [Google Scholar]
- 22.Wei, T., et al. 2006. Pns12 protein of Rice dwarf virus is essential for formation of viroplasms and nucleation of viral-assembly complexes. J. Gen. Virol. 87:429-438. [DOI] [PubMed] [Google Scholar]
- 23.Wei, T., T. Shimizu, and T. Omura. 2008. Endomembranes and myosin mediate assembly into tubules of Pns10 of Rice dwarf virus and intercellular spreading of the virus in cultured insect vector cells. Virology 372:349-356. [DOI] [PubMed] [Google Scholar]
- 24.Whitfield, A. E., D. E. Ullman, and T. L. German. 2005. Tospovirus-thrips interactions. Annu. Rev. Phytopathol. 43:459-489. [DOI] [PubMed] [Google Scholar]
- 25.Yan, J., et al. 1996. P2 protein encoded by genome segment S2 of rice dwarf phytoreovirus is essential for virus infection. Virology 224:539-541. [DOI] [PubMed] [Google Scholar]
- 26.Zhong, B., et al. 2003. A minor outer capsid protein, P9, of Rice dwarf virus. Arch. Virol. 148:2275-2280. [DOI] [PubMed] [Google Scholar]
- 27.Zhou, F., et al. 2007. The P2 capsid protein of the nonenveloped rice dwarf phytoreovirus induces membrane fusion in insect host cells. Proc. Natl. Acad. Sci. U. S. A. 104:19547-19552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.