Abstract
Host signaling pathways play important roles in the replication of influenza virus, but their functional effects remain to be characterized at the molecular level. Here we identify two receptor tyrosine kinase inhibitors (RTKIs) of the tyrphostin class that exhibit robust antiviral activity against influenza A virus replication in cultured cells. One of these (AG879) is a selective inhibitor of the nerve growth factor receptor and human epidermal growth factor receptor 2 (TrkA/HER2) signaling; the other, tyrphostin A9 (A9), inhibits the platelet-derived growth factor receptor (PDGFR) pathway. We find that each inhibits at least three postentry steps of the influenza virus life cycle: AG879 and A9 both strongly inhibit the synthesis of all three influenza virus RNA species, block Crm1-dependent nuclear export, and also prevent the release of viral particles through a pathway that is modulated by the lipid biosynthesis enzyme farnesyl diphosphate synthase (FPPS). Tests of short hairpin RNA (shRNA) knockdown and additional small-molecule inhibitors confirmed that interventions targeting TrkA can suppress influenza virus replication. Our study suggests that host cell receptor tyrosine kinase signaling is required for maximal influenza virus RNA synthesis, viral ribonucleoprotein (vRNP) nuclear export, and virus release and that specific RTKIs hold promise as novel anti-influenza virus therapeutics.
Influenza virus imposes substantial burdens on public health by causing annual epidemics and occasional pandemics of acute respiratory disease that may lead to potentially severe and deadly complications, such as pneumonia. Antiviral therapeutics are a critical tool in combating influenza virus infections, especially in years when the vaccine strain does not match well with the circulating virus, when vaccines are unavailable at the early pandemic stage, or when vaccines are in short supply. Development of novel anti-influenza virus drugs is urgent, as variant strains resistant to all currently available drugs have been isolated and are expected to evolve rapidly (7, 8, 26, 44, 59). Targeting host cell signaling pathways or other host factors required for influenza virus replication offers an alternative strategy for antiviral drug development. Recent proteomic screening using small interfering RNA (siRNA) libraries has identified hundreds of host factors that may promote influenza virus replication (3, 16, 22, 24, 49), but the challenge of validating, characterizing, and interdicting their respective activities through pharmacological means remains.
Influenza A virus is an enveloped, negative-strand RNA virus with a segmented RNA genome (38). Influenza virus enters cells through receptor-mediated endocytosis after binding to sialylated receptors (50). After internalization, the low-pH environment in endosomes triggers fusion of viral and endosomal membranes and facilitates the release of viral ribonucleoprotein (vRNP) complexes into the cell cytoplasm (58). The released vRNPs then enter the nucleus, where viral RNA (vRNA) replication and transcription occur (38). Newly synthesized vRNPs are exported from the nucleus via the cellular Crm1-mediated nuclear export pathway (1, 12, 28, 55). Virus budding is mediated mainly by the viral M1 protein, which interacts with viral integral membrane proteins (HA, NA, and M2) and vRNP complexes at the plasma membrane (5, 33). The final release of virions from the cell surface requires the neuraminidase activity of viral NA protein (37, 39). Despite extensive studies, many aspects of influenza virus replication are incompletely understood, including the roles of host signaling pathways and cellular factors at each step of the virus life cycle. Identification of small-molecule compounds targeting any of these processes can yield biological insights as well as potential new therapies. For example, amantadine was found to block virus uncoating (4, 29), and viruses resistant to amantadine were found to harbor mutations in the ion channel region of the M2 transmembrane domain, suggesting both that the viral M2 protein is the target of amantadine (17) and that M2 ion channel activity is essential for virus uncoating. Viral HA protein was also found to influence amantadine sensitivity, implying an interaction between HA and M2 (17).
Receptor tyrosine kinases (RTKs) are a group of growth factor receptors that, upon ligand binding, undergo autophosphorylation at Tyr residues (18, 48, 52). These phosphorylated tyrosines then recruit Src homology 2 (SH2)- and phosphotyrosine-binding (PTB) domain-containing proteins that activate or link to downstream signaling pathways, such as the Ras/ERK/MAPK, PI3K/Akt, and JAK/STAT pathways (40, 48). Together, the complex signaling network triggered by RTKs leads to regulation of cell growth, migration, metabolism, and differentiation. Due to their critical roles in the development and progression of various cancers, RTKs have recently been studied extensively as targets for anticancer therapeutics. Host signaling through RTKs and other tyrosine kinases has also been shown to play important roles in virus replication. The tyrosine kinase inhibitor genistein was found to block replication of HIV-1, herpes simplex virus type 1 (HSV-1), and arenavirus (51, 53, 61), for example, and Src family kinases are known to be important for assembly and maturation of dengue virus and West Nile virus (6, 19). The Raf/MEK/ERK (42) and PI3K/Akt (9, 10, 15) pathways downstream of RTKs play important roles in influenza virus replication. It has been shown that Raf/MEK/ERK signaling is required for the nuclear export of influenza vRNPs (42). The functional mechanism by which the PI3K pathway affects influenza virus replication is unclear, however. One recent report indicates that epidermal growth factor receptor (EGFR) signaling promotes influenza A virus uptake by cells (11).
In this study, we identify two specific RTK inhibitors (RTKIs), known as AG879 and tyrphostin A9 (A9), that have strong antiviral activity against influenza A virus, and we demonstrate that they both inhibit the Crm1-dependent nuclear export of the vRNP complex, viral RNA synthesis, and virus release. We show that diverse interventions targeting TrkA can impede influenza virus replication, thus validating this specific RTK as a candidate drug target. Our findings provide mechanistic insights into the potential roles of host RTK signaling in facilitating influenza virus replication, and they also suggest that specific RTKIs could be developed as potential anti-influenza virus therapeutics.
MATERIALS AND METHODS
Cells and viruses.
293T cells (human kidney epithelial cells) and A549 cells (human lung epithelial cells) were grown in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum. Madin-Darby canine kidney (MDCK) cells were maintained in Eagle's minimal essential medium supplemented with 5% fetal bovine serum. After infection with influenza A virus, MDCK cells were grown in L-15 medium containing 15 mM HEPES (pH 7.5), nonessential amino acids, 0.75 g of NaHCO3 per liter, and 0.125% (wt/vol) bovine serum albumin. Influenza virus strains A/WSN/33 (A/WSN) and A/PR8/34 (A/PR8) were grown in 10-day-old embryonated chicken eggs, and their titers were determined by plaque assay on MDCK cells. The WSN-LUC reporter viruses were generated as previously described (25).
Plasmids, antibodies, and inhibitors.
Plasmid expressing farnesyl diphosphate synthase (FPPS) was provided by P. Creswell (Yale University). Plasmid expressing NF-κB molecule p65 was obtained from W. Greene (UCSF). Plasmid expressing the Rev-green fluorescent protein (GFP) fusion protein was provided by A. Mergia (University of Florida). Generation of the luciferase (LUC)-encoding reporter constructs vNA-LUC and cNA-LUC was described previously (45). Anti-FPPS antibody was obtained from P. Edward (UCLA). Anti-NP monoclonal antibody was purchased from Serotec. The library of kinase inhibitors was purchased from BIOMOL and includes 80 kinase inhibitors. Tyrphostin AG879, tyrphostin A9, tyrphostin AG494, ammonium pyrrolidinedithiocarbamate (PDTC), Bay11-7082 (Bay11), ribavirin, and AG1296 were purchased from Sigma. U0126 was purchased from Promega. GW441756 was purchased from Santa Cruz. K252a, TAK-165, ZD1839, and SKI-606 were purchased from LC Lab.
MTT assay.
The MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay was used to measure the effect of selected compounds on cell viability. MDCK or A549 cells in 96-well plates were treated with sequential 10-fold dilutions of either a given compound or dimethyl sulfoxide (DMSO), in triplicates, in a total of 100 μl growth medium for 48 h. Freshly made 5 mg/ml MTT solution (20 μl) was added to each well, and the cells were incubated at 37°C for 5 h before the medium was replaced with 200 μl DMSO to dissolve the crystals. The plates were further incubated at 37°C for another 5 min to dissolve any air bubbles before the MTT signal was measured at an absorbance of 550 nm. The MTT assay was performed on each compound, and no cytotoxicity was observed for A9 at 5 μM, AG879 at 81 μM, AG494 at 27 μM, Bay11-7082 (Bay11) at 10 μM, U0126 at 50 μM, or PDTC at 100 μM. Accordingly, throughout this work, except where noted, A9 was used at 4 μM, AG879 at 10 μM, AG494 at 10 μM, Bay11 at 10 μM, U0126 at 50 μM, and PDTC at 50 μM.
Virus attachment assay.
A549 cells were preincubated with DMSO or selected compounds for 30 min and then infected with A/WSN (multiplicity of infection [MOI] of 2) at 4°C for 2 h. After cells were washed 6 times with phosphate-buffered saline (PBS), cell lysates were prepared by rapid freeze-thaw. Virus titer was determined by plaque assay.
Inhibition of nucleocytoplasmic trafficking.
A549 cells were infected with A/WSN virus at an MOI of 5 for 1 h and replaced with fresh medium containing vehicle control or respective inhibitors. Intracellular localization of influenza vRNPs in the virus-infected cells at various times postinfection was detected by immunofluorescence assay as described previously (25). To examine the effects of the inhibitors on the nucleocytoplasmic trafficking of HIV rev protein, A549 cells grown on coverslips were transfected with a plasmid expressing rev-GFP fusion protein and, 18 h later, treated with DMSO or respective inhibitors for 4 h prior to observation under a fluorescence microscope. The intensity of fluorescein isothiocyanate (FITC)-NP or rev-GFP signals in both nucleus (defined by DAPI [4′,6-diamidino-2-phenylindole] staining) and cytoplasm of individual cells was quantified using an image analysis program, and the percentage of nuclear signal (nuc%) was calculated for each cell. The mean percentage of nuclear signal was then calculated for 40 cells per drug treatment per time point, and statistical comparisons among groups were conducted using Student's t test.
Quantitative real-time RT-PCR.
The levels of influenza vRNA, cRNA, and mRNA in the virus-infected cells were quantified by real-time reverse transcription PCR (RT-PCR) as described previously (25).
Reporter-based influenza virus RNA transcription assay.
The luciferase (LUC)-based five-plasmid RNA transcription assay was conducted as described previously (25). In brief, A549 cells grown in serum-free medium were transfected with 5 plasmids, four encoding the PA, PB1, PB2, and NP proteins, respectively, and one expressing the viral RNA promoter-directed LUC gene (vNA-LUC or cNA-LUC). DMSO or inhibitors were added at 8 h posttransfection, and LUC activity was measured at 24 h posttransfection.
Virus release assay.
A549 cells were infected with influenza A virus at an MOI of 2. At 8 h postinfection (hpi), cells were washed 5 times with PBS and replaced with fresh medium containing vehicle control or respective inhibitors. At different time points (15, 30, 45, and 60 min) following addition of inhibitors, supernatants were collected and cell pellets were lysed by rapid freeze-thaw on dry ice-ethanol and a 37°C water bath. Virus titers in the supernatant (extracellular virus) and cell lysate (membrane-associated virus) were then quantified by plaque assay on MDCK cells.
shRNA knockdown.
We used two different short hairpin RNAs (shRNAs) to knock down human TrkA expression. Two different pairs of oligonucleotides, 5′ CCAGTGACCTCAACAGGAAGAttcaagagaTCTTCCTGTTGAGGTCACTGG 3′ and 5′ TCATCGAGAACCCACAATACTttcaagagaAGTATTGTGGGTTCTCGATGA 3′ (only forward sequences are shown), were each cloned into the psiRNA-hH1 vector (InvivoGen) between the Acc65I and HindIII sites downstream of the human H1 RNA promoter. The resulting plasmids, psiRNA-TrkA1 and psiRNA-TrkA3, were verified by sequence analysis. For shRNA knockdown, A549 cells were transfected twice in 48 h using Lipofectamine (Invitrogen) with either the psiRNA-hH1Luc control plasmid, which expresses luciferase-specific shRNA, or the combined psiRNA-TrkA1 and -TrkA3 plasmids and then, at 72 h posttransfection, infected with A/WSN virus at an MOI of 1. Virus titer was determined at 12 hpi.
RESULTS
Identification of two RTKIs, AG879 and A9, with anti-influenza virus activity.
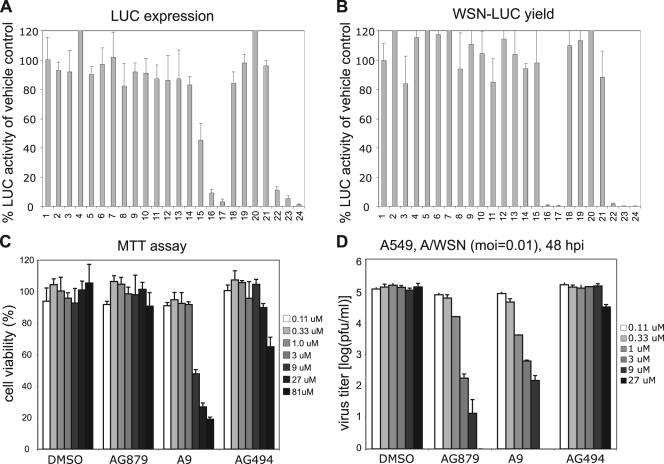
Using the WSN-LUC reporter virus system that we have previously described (25), we screened a small library of 80 kinase inhibitors for anti-influenza virus activity. MDCK cells on 96-well plates were infected with WSN-LUC virus at an MOI of 0.5 for 1 h, washed with PBS, and treated with DMSO vehicle control, respective compounds at 10 μm, or ribavirin at various concentrations as a positive control. The LUC activity in the infected MDCK cells was measured and normalized against that for the DMSO control (Fig. 1 A, lane 1). As shown in a representative screen (Fig. 1A), most of the tested compounds showed no inhibitory effects on LUC activity in the WSN-LUC-infected cells, whereas the nonspecific antiviral compound ribavirin inhibited LUC expression in a dosage-dependent manner (Fig. 1A, lanes 22 to 24). Notably, two inhibitors (Fig. 1A, lanes 16 and 17) significantly decreased LUC activity, suggesting that they each strongly suppress virus infectivity or viral gene expression. To determine whether any compound inhibited virus yield, we used the supernatants from these treated, WSN-LUC-infected MDCK cells to infect fresh cells and then assayed LUC activity to quantify infectious viruses (Fig. 1B). Most of the tested inhibitors did not greatly impair virus production, as evidenced by high levels of LUC activity. In contrast, the two inhibitors of interest (Fig. 1B, lanes 16 and 17) reduced virus yield to a degree comparable to that produced by ribavirin (Fig. 1B, lanes 22 to 24). The library of kinase inhibitors and the screening results are detailed further in the supplemental material. These two compounds, AG879 (lane 16) and tyrphostin A9 (lane 17), are tyrphostin-type receptor tyrosine kinase inhibitors (RTKIs). AG879 is known to inhibit the nerve growth factor receptor (TrkA; pp140trk) and human epidermal growth factor receptor 2 (HER2) (35), while A9 is a selective inhibitor of the receptor tyrosine kinase platelet-derived growth factor receptor (PDGFR) (27). To exclude cytotoxic effects, we evaluated cell viability under various concentrations of AG879, A9, or AG494, using the MTT assay (Fig. 1C). AG494 is a potent EGFR inhibitor in cell-free kinase assays but cannot inhibit EGFR in intact cells (36) and thus is used as a negative control. Compared to the results for the DMSO control, no cytotoxicity was observed at up to 3 μM A9, 81 μM AG879, or 27 μM AG494 (Fig. 1C). Subsequent MTT assays indicated that A9 is noncytotoxic at up to 5 μM (data not shown). To compare antiviral potencies, A549 cells were infected with A/WSN/H1N1 at an MOI of 0.01 and then treated with DMSO or the respective RTKI at various concentrations, and virus yield in the supernatants at 48 hpi was determined (Fig. 1D). Compared to results for vehicle control DMSO and negative control AG494, both AG879 and A9 showed dose-dependent inhibition of influenza virus yield in A549 cells. Note that the virus yield with AG879 at 27 μM was ∼0 PFU and is therefore undetectable in Fig. 1D and that the strong virus inhibition produced by A9 at 9 μM could partly be due to cytotoxicity at that concentration (Fig. 1C). Based on these studies of cytotoxicity and antiviral efficacy, we used A9 at 4 μM and AG879 and AG494 at 10 μM in all subsequent work. Taken together, these initial studies identified two specific RTKIs that can strongly inhibit both influenza A viral gene expression and virus yield.
FIG. 1.
Identification of RTKIs AG879 and A9 with anti-influenza virus activity. (A) Screening of a library of kinase and phosphatase inhibitors that inhibit influenza virus gene expression. MDCK cells were infected with WSN-LUC reporter viruses and treated with various compounds. The LUC activity in the infected MDCK cells was normalized by that for the DMSO control (lane 1). (B) Screening of a library of kinase and phosphatase inhibitors that inhibit influenza virus yield. The supernatants from the compound-treated, WSN-LUC-infected MDCK cells as described for panel A were used to infect fresh MDCK cells, in which LUC activity was measured and normalized to that for the DMSO control (lane 1). Lane 16, AG879; lane 17, A9; lanes 22 to 24, ribavirin at 5, 10, and 20 ng/ml, respectively. (C) Determination of cytotoxicity of the RTKIs by MTT assay. A549 cells were incubated with various concentrations of the respective RTKIs or the vehicle control DMSO for 48 h prior to the MTT assay. (D) Dose-dependent inhibition of A/WSN/H1N1 virus in A549 cells by AG879 and A9. A549 cells were infected with A/WSN virus at an MOI of 0.01 and treated with various concentrations of the respective RTKIs or the vehicle control DMSO. Virus yields at 48 hpi were quantified by plaque assay. Error bars indicate standard deviations (SD) (n = 3).
RTKIs AG879 and A9 effectively block influenza A virus replication in cell culture.
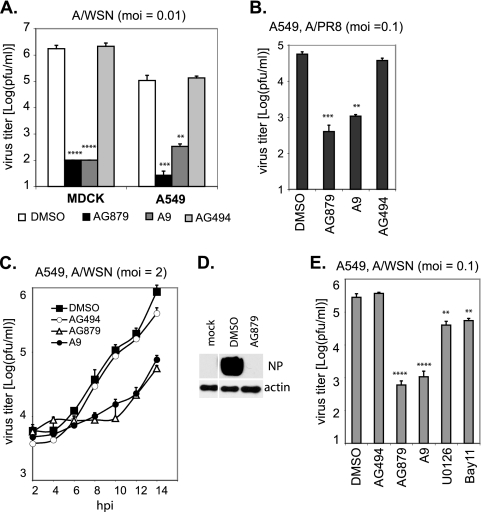
To verify their anti-influenza virus activities, we first examined whether AG879 and A9 can block replication of more than one influenza virus strain in diverse cell lines. MDCK or A549 cells were infected with A/WSN at an MOI of 0.01 in the presence of DMSO or various compounds (AG879, A9, or AG494). Virus titers in the supernatants at 48 hpi were quantified by plaque assays. Compared to results for DMSO or AG494 controls, AG879 and A9 efficiently blocked A/WSN replication >4,000-fold in MDCK cells and >200-fold in A549 cells (Fig. 2 A), suggesting that their anti-influenza virus activity is not restricted to a single cell type. We also found that, in addition to blocking laboratory-adapted A/WSN virus, AG879 and A9 lowered virus yields of a different influenza A virus strain (A/PR8) >100-fold in A549 cells 18 h after infection at an MOI of 0.1 (Fig. 2B).
FIG. 2.
RTKIs AG879 and A9 effectively block influenza A virus replication in cell culture. (A) AG879 and A9 block the multiplication of influenza A/WSN virus in both MDCK and A549 cells. Cells were infected with A/WSN at an MOI of 0.1 for 18 h in the presence of DMSO, AG879 (10 μM), tyrphostin A9 (4 μM), or AG494 (10 μM). Virus titer was determined by plaque assay. (B) AG879 and A9 inhibit influenza A/PR8 virus replication. A similar virus yield inhibition assay was performed with A549 cells infected with A/PR8 strain at an MOI of 0.1 for 18 h in the presence of DMSO or various inhibitors. (C) AG879 and A9 block single-round replication of influenza A virus. Virus titers were determined at various time points after A/WSN infection of A549 cells at a high MOI (MOI of 2) in the presence of DMSO or various inhibitors. (D) Western blot analysis of viral NP proteins at 8 hpi in cells mock infected (mock) or infected with A/WSN virus (MOI of 1) and treated with DMSO or AG879. (E) RTKIs AG879 (10 μM) and A9 (4 μM) inhibited influenza virus replication much more strongly than MEK inhibitor U0126 (50 μM) and NF-κB inhibitor Bay11-7082 (Bay11) (10 μM). Pairwise statistical comparisons to the DMSO control group were performed using Student's t test (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Error bars indicate SD (n = 3).
We further characterized the inhibitory effects of AG879 and A9 in a single-round replication assay, distinct from the multiple-round assay described above. A549 cells were infected with A/WSN at a high MOI (MOI of 2), washed three times with PBS, and then maintained in medium containing DMSO or various compounds. The virus titer in the supernatants at various time points was quantified (Fig. 2C). Newly synthesized viruses were detected as early as 8 hpi in DMSO- or A494-treated cells and increased in number exponentially at later times. In contrast, production of new viral particles (over background) in AG879- or A9-treated cells was not detectable until 12 hpi and lagged behind results for the controls by >1 log at later times. Consistent with those findings, viral NP protein was highly expressed in the DMSO-treated cells at 8 hpi but undetectable in the AG879-treated cells when analyzed by Western blotting (Fig. 2D).
RTKIs are not the only inhibitors of host signaling that can block influenza virus replication. Previous studies have identified several host signaling inhibitors with anti-influenza virus activity, including the MEK inhibitor U0126 and the NF-κB inhibitor Bay11-7082 (Bay11). To compare their antiviral potencies, we infected A549 cells with A/WSN virus at an MOI of 0.1 for 18 h in the presence of either DMSO or respective inhibitors. Each inhibitor was used at a concentration that gave the maximal antiviral effect without cytotoxicity: A9 at 4 μM, AG879 and Bay11 at 10 μM, and U0126 at 50 μM. In keeping with previous studies (25, 42), U0126 and Bay11 each decreased virus production by 1 log (Fig. 2E). AG879 and A9, however, showed much greater inhibition, reducing virus yield by ∼2 log.
AG879 and A9 inhibit the later stages of the influenza virus life cycle.
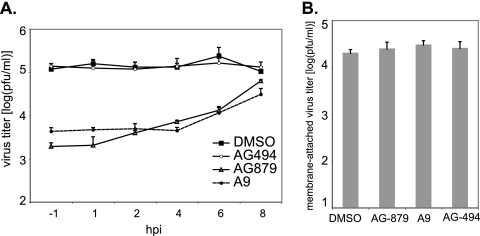
To characterize the specific step(s) of the influenza virus life cycle that is impeded by AG879 and A9, we examined the time course of their inhibitory effects. A549 cells were infected with A/WSN virus at an MOI of 1 and then, at various time points postinfection, were treated with DMSO or an inhibitor. We measured the titer of infectious viral particles released into the supernatant at 9 hpi, as a complete influenza virus life cycle (from the initial cell attachment to the final release of newly synthesized infectious viral particles) takes about 8 h (Fig. 2C). Both AG879 and A9 efficiently blocked virus production even when they were applied as late as 6 hpi (Fig. 3 A), suggesting that these compounds are able to inhibit later stages of the virus life cycle. In addition, when applied at 1, 2, or even 4 hpi, each reduced virus production comparably to the degree achieved when applied at 1 h prior to infection (−1 hpi) (Fig. 3A), indicating that neither inhibitor blocks virus entry. A virus attachment assay, performed with and without inhibitor, demonstrated that AG879 and A9 did not affect the quantity of infectious virus particles that can attach to host membranes (Fig. 3B). Furthermore, a vRNP subcellular localization assay (Fig. 4) revealed that neither AG879 nor A9 impaired the cytoplasmic accumulation or nuclear importation of vRNP complex early in infection (see below for more details). The findings together imply that AG879 and A9 block mainly later steps of the influenza virus life cycle, subsequent to vRNP nuclear importation.
FIG. 3.
AG879 and A9 inhibit the later stages of the influenza virus life cycle. (A) Time course analysis of the AG879 and A9 inhibitory effects on influenza A virus replication. A549 cells were infected with A/WSN at an MOI of 1 and, at different time points, treated with DMSO or the respective inhibitor. Virus titer at 9 hpi was determined by plaque assay. (B) Effects of AG879 and A9 on virus attachment. A549 cells were preincubated with DMSO or chemicals for 30 min and infected with A/WSN (MOI of 2) at 4°C for 2 h. After cells were washed 6 times with PBS, cell lysates were prepared by rapid freeze-thaw. Virus titer was determined by plaque assay. Results shown are the averages from at least three independent experiments. Error bars indicate SD.
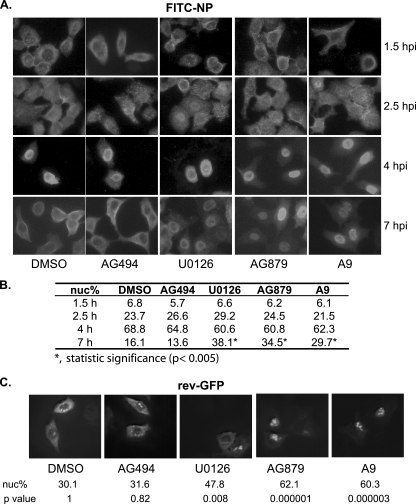
FIG. 4.
RTKIs AG879 and A9 impair the nuclear export of influenza vRNPs by inhibiting the host Crm1-dependent pathway. (A) A549 cells were infected with A/WSN virus and treated with either DMSO vehicle control or chemical inhibitors. At various time points, cells were stained with anti-NP antibody, followed by FITC-conjugated secondary antibody, and observed under a fluorescence microscope. A representative image from each condition is shown. Three independent experiments were conducted. (B) Quantification of the percentage of NP-FITC nuclear signal (nuc%). Results shown are the average from 40 cells for each drug treatment at each time point. Statistical analysis was conducted as described in Materials and Methods, and statistical significance as determined by t test is shown. (C) A549 cells were transfected with the rev-GFP fusion protein expression vector and treated with DMSO or respective inhibitors for 4 h prior to observation under a fluorescence microscope. For each treatment, the average percentage of rev-GFP nuclear signal (nuc%) and the P value of pairwise statistical comparisons to the DMSO control group are shown.
AG879 and A9 block nuclear export of influenza vRNPs by inhibiting the host Crm1-dependent pathway.
Influenza virus RNA synthesis occurs in the nucleus. Incoming vRNPs, which contain NP-encapsidated viral genomic RNAs with associated viral polymerase proteins (PA, PB1, and PB2), must be imported to the nucleus for RNA transcription or replication to occur. Newly synthesized vRNPs are exported from the nucleus via the cellular Crm1-mediated nuclear export pathway (12, 28, 55) as a (Crm1-RanGTP)-NEP-M1-vRNP complex (1) to be packaged into virions at the plasma membrane. To investigate the effects of AG879 and A9 on nucleocytoplasmic trafficking of vRNPs, we infected A549 cells with A/WSN at an MOI of 5 and then treated the cells with various inhibitors. U0126, a MEK inhibitor that has been shown to block influenza vRNP nuclear export (42), was included as a positive control. Localization of the NP protein was assessed by indirect immunofluorescence at various times postinfection. Representative images are shown in Fig. 4A. Under all conditions tested, the incoming vRNPs were detected mainly in the cytoplasm at 1.5 hpi, in both the cytoplasm and the nuclei at 2.5 hpi, and mostly within the nuclei at 4 hpi, suggesting that AG879 and A9 do not affect vRNP nuclear import early in virus infection. In contrast, at a later stage of virus replication (7 hpi), newly synthesized vRNPs were detected predominantly in the cytoplasm of cells treated with vehicle control DMSO or negative control AG494 but predominantly within the nuclei of those treated with U0126, AG879, or A9. To quantitate these findings, we calculated the mean percentage of FITC-NP signal within the nucleus (nuc%) in 40 cells for each drug treatment at each time point (Fig. 4B); statistical analysis confirmed that only samples treated with U0126, AG879, and A9 at 7 hpi were significantly different from the DMSO control (P < 0.005). These data suggest that, like the MEK inhibitor U0126, AG879 and A9 prevent the nuclear export of vRNPs late in infection.
As influenza vRNP nuclear export not only depends on the cellular Crm1 pathway but also requires the viral proteins NEP/NS2 and M1, the nuclear retention of vRNPs following AG879 or A9 treatment might be due either to direct inhibition of the cellular Crm1 nuclear export pathway or to significantly decreased levels of viral protein expression (Fig. 2D) resulting from a possible blockade of viral RNA synthesis (see below for more details). We therefore examined the effect of AG879 and A9 on the nucleocytoplasmic trafficking of the HIV Rev protein, whose nuclear export depends on the Crm1 pathway (34). As shown in Fig. 4C, the rev-GFP fusion protein showed a predominant nuclear localization in control samples that had been treated with DMSO or AG494. The MEK inhibitor U0126 moderately increased the nuclear localization of rev-GFP, while AG879 and A9 led to a predominant nuclear retention. Statistical analysis of the proportion of the rev-GFP signal found within nuclei (nuc%) confirmed that nuclear localization of rev-GFP was significantly increased by U0126, AG879, or A9 treatment compared to that for controls (P = 0.008, P = 0.000001, and P = 0.000003, respectively). These findings strongly suggest that AG879 and A9 directly inhibit the cellular Crm1 nuclear export pathway, which may largely account for their ability to cause nuclear retention of influenza vRNPs (Fig. 4A).
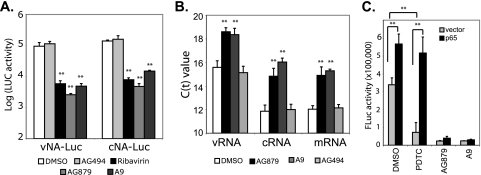
AG879 and A9 strongly inhibit influenza virus RNA synthesis.
We then evaluated whether AG879 and A9 can directly inhibit viral RNA synthesis, using a 5-plasmid assay (25) that is based on the reconstitution of all of the cis- and trans-acting elements required for influenza virus RNA replication and transcription. Compared to results for vehicle control DMSO and negative control AG494, the addition of AG879, A9, or ribavirin, a known inhibitor of influenza virus RNA synthesis, decreased the LUC activity expressed from either vRNA or cRNA templates by ∼95% (Fig. 5 A), strongly suggesting that the RTKIs inhibit viral RNA synthesis. To validate this finding in virus-infected cells, we infected A549 cells with A/WSN at an MOI of 1 and then treated cells with various inhibitors at 1 hpi. Whole-cell RNA was isolated from the infected cells at 5 hpi and quantified for vRNA, cRNA, and mRNA using an established quantitative real-time RT-PCR assay (25). The levels of all three RNA species were found to be significantly decreased in AG879- and A9-treated cells, as indicated by increased threshold cycle (CT) values (Fig. 5B).
FIG. 5.
AG879 and A9 strongly inhibit the synthesis of all three viral RNA species independently of the NF-κB pathway. (A) AG879 and A9 inhibit viral RNA transcription from the cRNA or vRNA promoter, based on the FLuc-based 5-plasmid assay. A549 cells were transfected with expression plasmids of NP, PA, PB1, and PB2, together with a vNA-LUC or cNA-LUC reporter construct. DMSO or inhibitor was added to the cells at 8 h posttransfection and FLuc activity determined 16 h later. FLuc, firefly luciferase. (B) Synthesis of all three RNA species (vRNA, cRNA, and mRNA) was decreased in the virus-infected cells treated with AG879 and A9. The viral RNA level was quantified by real-time RT-PCR and is shown as CT value. (C) Overexpression of NF-κB molecule p65 cannot reverse the inhibition of viral RNA transcription from the cRNA promoter by AG879 or A9. The 5-plasmid assay was conducted as described for panel A in cells transfected with empty vector plasmid or p65 expression vector. Compound treatment and LUC assay were similarly performed. Results shown are the averages from at least three independent experiments. Error bars indicate SD. Statistical analysis was conducted with Student's t test (**, P < 0.01).
Our previous studies have suggested that activation of NF-κB signaling promotes the efficient replication of influenza virus RNAs (25). Although NF-κB is also a downstream pathway of RTK signaling, AG879 and A9 are unlikely to block viral RNA synthesis via that pathway, as we have shown that NF-κB signaling is differentially involved in vRNA but not mRNA or cRNA synthesis (25), whereas AG879 and A9 block synthesis of all three viral RNA species (Fig. 5A and B). This is further evidenced by overexpression of NF-κB subunit p65, which we have shown can increase influenza vRNA synthesis and also rescue vRNA synthesis that has been blocked by NF-κB inhibitors (PDTC and Bay11) in the 5-plasmid assay (25) (Fig. 5C). In contrast, overexpression of p65 failed to rescue AG879- or A9-mediated inhibition of vRNA synthesis (Fig. 5C), suggesting that AG879 and A9 do not act through p65 to block vRNA synthesis. Taken together, our data suggest that AG879 and A9 effectively block synthesis of all three influenza virus RNA species independently of the NF-κB pathway.
AG879 and A9 inhibit the release of influenza virus particles.
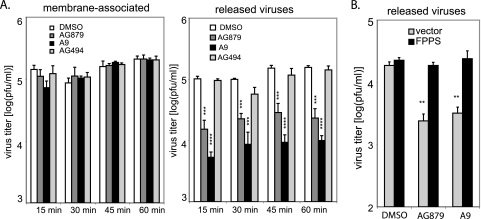
To determine whether the antiviral RTKIs affect virus assembly and release, we utilized a previously described assay (20) to quantify viruses that are either membrane associated (assembled) or released into the supernatant (budded). A549 cells were infected with A/WSN at an MOI of 2. At 8 hpi, we washed these cells extensively with PBS and then added fresh medium containing DMSO or an inhibitor. After treatment for 15, 30, 45, or 60 min, we determined the titer of infectious viruses released to the supernatants, as well as that of the membrane-associated viruses, as described in Materials and Methods. At all time points, we recovered comparable amounts of membrane-associated infectious viruses in DMSO- and inhibitor-treated samples (Fig. 6 A). In contrast, the titers of released extracellular viruses dropped significantly, by 80 to 90%, in cells treated with AG879 or A9 compared to those for controls (Fig. 6A). (Note that virus titer is shown on a log scale.) Our data suggest that the antiviral RTKIs block influenza virus release but do not impair its binding to the plasma membrane.
FIG. 6.
AG879 and A9 inhibit virus release via FPPS. (A) Determination of infectious viruses associated with membranes or released to the supernatants. A549 cells were infected with A/WSN (MOI of 2) for 8 h and treated with either DMSO or compound for 15, 30, 45, and 60 min. Titers of infectious virus in the supernatants and in the cell lysates were determined. (B) FPPS overexpression fully restored the block of virus release by AG879 and A9. A virus release assay similar to that described for panel A was conducted in cells that had been transfected with empty vector or FPPS expression vector. Results shown are the averages from at least three independent experiments. Error bars indicate SD. Statistical analysis was conducted with Student's t test (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Wang et al. (54) have shown that an interferon (IFN)-induced protein, viperin, inhibits influenza A virus release from the plasma membrane by affecting the membrane fluidity and formation of lipid rafts and that it acts through a pathway dependent upon farnesyl diphosphate synthase (FPPS), an enzyme essential for isoprenoid biosynthesis. Overexpression of FPPS was able to reverse viperin-mediated inhibition of virus production and restore normal membrane fluidity. To determine whether AG879 and A9 might also target FPPS, we examined the effect of FPPS overexpression on AG879- and A9-induced inhibition of virus release. A549 cells were transfected with either an FPPS expression vector or an empty vector control, infected with A/WSN, and then treated with DMSO or inhibitor for 30 min. As expected, AG879 and A9 significantly reduced virus release in the cells transfected with empty vector. However, overexpression of FPPS completely prevented this inhibition, restoring the levels of virus release from AG879- or A9-treated cells to those of the controls (Fig. 6B). This suggests that the two RTKIs may target FPPS to inhibit influenza virus particle release. As we did not observe any change in FPPS expression levels under the inhibitor treatment by Western blotting (data not shown), we speculate that AG879 and A9 may act by impairing FPPS function. It remains to be determined how RTK signaling regulates the activity of FPPS and how FPPS functions to facilitate influenza virus budding (5, 33).
TrkA is important for influenza virus replication.
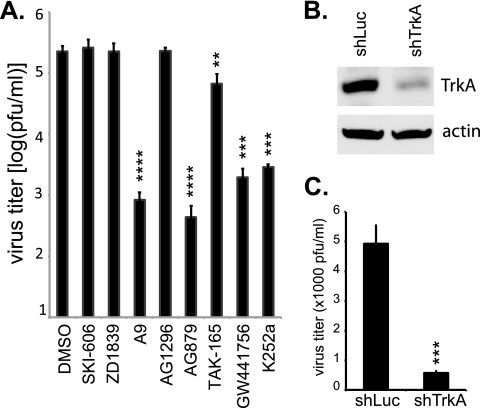
Although AG879 is a known inhibitor of TrkA and HER2 and A9 reportedly blocks PDGFR activation, we sought to determine whether their antiviral activities were actually due to inhibition of those RTKs rather than to off-target effects. To that end, we tested six additional small-molecule RTKIs, with reported specificities for the Src (SKI-606), EGFR (ZD1839), PDGFR (AG1296), HER2 (TAK-165), and TrkA (K252a and GW441756) kinases. Each compound was used at a concentration (2 to 10 μM) reported to exert RTKI activity but which we found to be noncytotoxic by the MTT assay. As depicted in Fig. 7 A, we found that the two TrkA inhibitors GW441756 and K252a each blocked influenza virus replication to a degree comparable to that of AG879 (i.e., ∼2 log) at the concentrations tested. TAK-165, a HER2 inhibitor, had only modest efficacy, reducing virus production by 70%. The other compounds showed no appreciable antiviral activity. Thus, three different TrkA inhibitors (AG879, GW441756, and K252a) suppressed influenza A virus replication in this assay, whereas compounds targeting other host RTKs did not. Additional studies are needed to verify the antiviral target(s) of A9, however, as another reported PDGFR inhibitor (AG1296) did not block virus production in our assay.
FIG. 7.
TrkA signaling is important for influenza virus replication. (A) TrkA inhibitors significantly reduce influenza virus production in vitro. A549 cells were infected with A/WSN at an MOI of 0.1, in the presence of either vehicle control (DMSO alone) or various inhibitors: SKI-606 (10 μM), ZD1839 (10 μM), A9 (4 μM), AG1296 (10 μM), AG879 (10 μM), TAK-165 (10 μM), GW441756 (10 μM), or K252a (2 μM). Virus production at 18 hpi was quantified by plaque assay. (B) TrkA-specific shRNAs decreased the TrkA expression level. A549 cells were transfected with the shRNA-expressing plasmid specific for either LUC or TrkA and then infected with influenza A virus. TrkA protein expression was detected 48 h after transfection by Western blot analysis using anti-TrkA antibody. (C) TrkA-specific shRNAs significantly decreased virus production. A549 cells transfected 48 h previously with either LUC- or TrkA-specific shRNA plasmid were infected with A/WSN at an MOI of 1. Virus production was quantified at 12 hpi. Results shown are the averages from at least three independent experiments. Error bars indicate SD. Statistical analysis was conducted with Student's t test (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
We then used a pair of shRNA-expressing plasmids to specifically knock down TrkA expression in target cells prior to influenza virus infection. The TrkA-specific shRNA vectors significantly reduced TrkA protein expression in A549 cells, as determined by Western blotting, whereas a control LUC-specific shRNA had no effect (Fig. 7B). Upon infection with influenza A virus at an MOI of 1, cells transfected with the TrkA-specific shRNA vectors showed significantly reduced virus replication, yielding titers ∼1 log lower than those of control cells (Fig. 7C). Taken together, our findings using three TrkA-specific small-molecule inhibitors as well as shRNA knockdown strongly imply that signals emanating from TrkA are needed for maximal influenza A virus replication.
DISCUSSION
Host cell RTKs are growth factor receptors that regulate a variety of cellular activities related to growth, metabolism, and differentiation. In this study, we have shown that two small-molecule RTKIs, AG879 and A9, can each potently block influenza virus replication at multiple steps of the virus life cycle, impairing vRNP nuclear export, RNA synthesis, and virus release. AG879 and A9 are tyrphostin-class compounds that selectively antagonize the TrkA/HER2 and PDGFR pathways, respectively. By testing additional pharmacologic inhibitors and specific shRNA knockdown, we were able to verify the importance of at least one of these target RTKs (TrkA) in influenza A virus replication. These findings serve to extend prior studies that have also suggested important roles for RTK signaling in the influenza virus life cycle. Several independent genome-wide screens conducted to search for host factors involved in influenza virus replication have previously implicated particular RTKs and many of their downstream targets (3, 16, 22, 24, 49, 56). In those studies, siRNA knockdown of at least 5 known RTKs, transforming growth factor receptor (TGFR) (49), fibroblast growth factor 1 (FGFR-1) through FGFR-4 (24), NTRK2/TrkB (24), EphB6 (22), and EphB2 (24), resulted in reduced influenza virus replication, supporting the functional role of RTK signaling in influenza virus replication. Another recent report (11) suggests, moreover, that EGFR signaling is important to promote influenza A virus uptake by infected cells. Thus, mounting evidence indicates that diverse pathways of RTK signaling may be required at multiple discrete steps of the influenza virus life cycle and so may present novel targets for antiviral drug development. To the extent that any given drug might target multiple host components and multiple steps of virus replication, drug-resistant viral variants are less likely to occur. It is worth noting that, given their low therapeutic indexes in vitro, AG879 and A9 are unlikely to be useful as antiviral agents themselves, but nonetheless they point the way to developing or discovering better antiviral drugs.
That RTK signaling is involved in the nuclear export of influenza vRNPs is not surprising, as one RTK signaling pathway, the Raf/MEK/ERK pathway, has previously been reported to be important for this process (42). Our data further reveal, however, that host RTK signaling is involved in directly regulating the host Crm1-dependent nuclear export pathway. Crm1 (also called exportin1, or Xpo1) is a major nuclear export receptor for proteins and for many RNAs (21); it forms trimeric transport complexes with RanGTP and export cargo molecules, a process promoted by the Ran-binding protein RanBP3. RTK signaling might be proposed to regulate Crm1 nuclear export through various mechanisms. Yoon et al. have shown that growth factor-mediated modulation of nuclear export occurs through phosphorylation of RanBP3 by RSK and Akt, which are the respective downstream targets of the Ras/ERK/RSK and PI3K/Akt pathways (60). We have not yet evaluated whether AG879 or A9 can block the phosphorylation of RanBP3 or affect other components of the Crm1 nuclear export complex. In addition, interaction of Crm1 with cargo proteins can be regulated by cargo phosphorylation (21). Several viral protein components of influenza vRNPs are known to be phosphoproteins, including PA (47), NP (23, 43), M1 (13, 14, 23), and NEP/NS2 (46), and hyperphosphorylation of a mutant M1 protein has been shown to cause its aberrant nuclear retention (57). Conflicting results concerning the relationship between NP phosphorylation and nuclear export have been reported: one study reported that phosphorylated NP accumulated in the nucleus and cytoplasm with similar kinetics (41), suggesting that phosphorylation did not affect NP nucleocytoplasmic trafficking, whereas another study found that vRNPs isolated from the nucleus contained much more phosphorylated NP than those from the cytoplasm, consistent with differential nuclear export (2). Whether phosphorylation of vRNP components regulates nuclear export and whether AG879 and A9 cause nuclear retention of vRNPs by specifically blocking that process require further investigation.
A variety of host signaling pathways and other host factors have been implicated in regulating influenza virus RNA synthesis, but their underlying mechanisms are largely unknown. Several cellular factors that stimulate influenza virus RNA synthesis have been identified, including Hsp90 (31), the splicing-related factor AP56/BAT1 (30), and the chaperone Tat-SF1 (32). Recent proteomic screens using siRNA libraries have identified hundreds of candidate host factors that affect influenza virus replication (3, 16, 22, 24, 49), but exactly which host factors are functionally required for viral RNA synthesis and how they function await further research. We have previously shown that NF-κB signaling can differentially regulate influenza virus RNA synthesis by promoting vRNA but not mRNA or cRNA synthesis (25). Here we present evidence that host RTK signaling, by contrast, is important for the synthesis of all three influenza virus RNA species, through mechanisms that remain to be characterized.
Influenza virus particles are assembled and bud at the plasma membrane at sites that are enriched in cholesterol and glycosphingolipids, forming lipid raft microdomains (5, 33). The eventual release of virus from the plasma membrane requires closure of the bud and separation of the virus particle from the host membrane, processes that are influenced by viral components as well as host factors. It has been shown that inhibitors of certain G proteins and protein kinases can inhibit influenza virus budding (20), suggesting an important role for host signaling in this process. An enzyme essential for isoprenoid biosynthesis, FPPS, appears to be critically involved in influenza virus budding, possibly owing to its role in the formation of lipid rafts (54). At least two different classes of influenza virus inhibitors, the IFN response protein viperin (54) and the antiviral RTKIs AG879 and A9 (Fig. 6B), block influenza virus release via FPPS. Moreover, siRNA-mediated knockdown of FPPS significantly reduces influenza virus replication (54), confirming that FPPS is a potential target for developing anti-influenza virus drugs. Further studies will cast light on the functional mechanisms by which FPPS, as well as other host factors, affects influenza virus budding and release.
Supplementary Material
Acknowledgments
We thank Y. Kawaoka (University of Wisconsin—Madison) for the influenza virus protein plasmids, P. Creswell (Yale University) for the FPPS expression plasmid, W. Greene (UCSF) for the plasmid expressing NF-κB molecule p65, A. Mergia (University of Florida) for the rev-GFP expression plasmid, and P. Edward (UCLA) for the anti-FPPS antibody.
This work was supported by NIH grants AI067704 to Tristram G. Parslow and AI083409 to Yuying Liang.
Footnotes
Published ahead of print on 5 January 2011.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Akarsu, H., et al. 2003. Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2). EMBO J. 22:4646-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almond, J. W., and V. Felsenreich. 1982. Phosphorylation of the nucleoprotein of an avian influenza virus. J. Gen. Virol. 60:295-305. [DOI] [PubMed] [Google Scholar]
- 3.Brass, A. L., et al. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukrinskaya, A. G., N. K. Vorkunova, G. V. Kornilayeva, R. A. Narmanbetova, and G. K. Vorkunova. 1982. Influenza virus uncoating in infected cells and effect of rimantadine. J. Gen. Virol. 60:49-59. [DOI] [PubMed] [Google Scholar]
- 5.Chen, B. J., and R. A. Lamb. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu, J. J., and P. L. Yang. 2007. c-Src protein kinase inhibitors block assembly and maturation of dengue virus. Proc. Natl. Acad. Sci. U. S. A. 104:3520-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, N. J., and K. Subbarao. 1999. Influenza. Lancet 354:1277-1282. [DOI] [PubMed] [Google Scholar]
- 8.de Jong, M. D., et al. 2005. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N. Engl. J. Med. 353:2667-2672. [DOI] [PubMed] [Google Scholar]
- 9.Ehrhardt, C., et al. 2006. Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defence. Cell. Microbiol. 8:1336-1348. [DOI] [PubMed] [Google Scholar]
- 10.Ehrhardt, C., et al. 2007. Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 81:3058-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eierhoff, T., E. R. Hrincius, U. Rescher, S. Ludwig, and C. Ehrhardt. 2010. The epidermal growth factor receptor (EGFR) promotes uptake of influenza A viruses (IAV) into host cells. PLoS Pathog. 6:e1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elton, D., et al. 2001. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 75:408-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregoriades, A., T. Christie, and K. Markarian. 1984. The membrane (M1) protein of influenza virus occurs in two forms and is a phosphoprotein. J. Virol. 49:229-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregoriades, A., G. G. Guzman, and E. Paoletti. 1990. The phosphorylation of the integral membrane (M1) protein of influenza virus. Virus Res. 16:27-41. [DOI] [PubMed] [Google Scholar]
- 15.Hale, B. G., D. Jackson, Y. H. Chen, R. A. Lamb, and R. E. Randall. 2006. Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol-3-kinase signaling. Proc. Natl. Acad. Sci. U. S. A. 103:14194-14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hao, L., et al. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hay, A. J., A. J. Wolstenholme, J. J. Skehel, and M. H. Smith. 1985. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 4:3021-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heldin, C. H. 1995. Dimerization of cell surface receptors in signal transduction. Cell 80:213-223. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch, A. J., et al. 2005. The Src family kinase c-Yes is required for maturation of West Nile virus particles. J. Virol. 79:11943-11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hui, E. K., and D. P. Nayak. 2002. Role of G protein and protein kinase signalling in influenza virus budding in MDCK cells. J. Gen. Virol. 83:3055-3066. [DOI] [PubMed] [Google Scholar]
- 21.Hutten, S., and R. H. Kehlenbach. 2007. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 17:193-201. [DOI] [PubMed] [Google Scholar]
- 22.Karlas, A., et al. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818-822. [DOI] [PubMed] [Google Scholar]
- 23.Kistner, O., K. Muller, and C. Scholtissek. 1989. Differential phosphorylation of the nucleoprotein of influenza A viruses. J. Gen. Virol. 70:2421-2431. [DOI] [PubMed] [Google Scholar]
- 24.Konig, R., et al. 2010. Human host factors required for influenza virus replication. Nature 463:813-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar, N., Z. T. Xin, Y. Liang, H. Ly, and Y. Liang. 2008. NF-kappaB signaling differentially regulates influenza virus RNA synthesis. J. Virol. 82:9880-9889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le, Q. M., et al. 2005. Avian flu: isolation of drug-resistant H5N1 virus. Nature 437:1108. [DOI] [PubMed] [Google Scholar]
- 27.Levitzki, A., and C. Gilon. 1991. Tyrphostins as molecular tools and potential antiproliferative drugs. Trends Pharmacol. Sci. 12:171-174. [DOI] [PubMed] [Google Scholar]
- 28.Ma, K., A. M. Roy, and G. R. Whittaker. 2001. Nuclear export of influenza virus ribonucleoproteins: identification of an export intermediate at the nuclear periphery. Virology 282:215-220. [DOI] [PubMed] [Google Scholar]
- 29.Martin, K., and A. Helenius. 1991. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67:117-130. [DOI] [PubMed] [Google Scholar]
- 30.Momose, F., et al. 2001. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J. Virol. 75:1899-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Momose, F., et al. 2002. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 277:45306-45314. [DOI] [PubMed] [Google Scholar]
- 32.Naito, T., et al. 2007. An influenza virus replicon system in yeast identified Tat-SF1 as a stimulatory host factor for viral RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 104:18235-18240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nayak, D. P., E. K. Hui, and S. Barman. 2004. Assembly and budding of influenza virus. Virus Res. 106:147-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neville, M., F. Stutz, L. Lee, L. I. Davis, and M. Rosbash. 1997. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 7:767-775. [DOI] [PubMed] [Google Scholar]
- 35.Ohmichi, M., et al. 1993. The tyrosine kinase inhibitor tyrphostin blocks the cellular actions of nerve growth factor. Biochemistry 32:4650-4658. [DOI] [PubMed] [Google Scholar]
- 36.Osherov, N., A. Gazit, C. Gilon, and A. Levitzki. 1993. Selective inhibition of the epidermal growth factor and HER2/neu receptors by tyrphostins. J. Biol. Chem. 268:11134-11142. [PubMed] [Google Scholar]
- 37.Palese, P., and R. W. Compans. 1976. Inhibition of influenza virus replication in tissue culture by 2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA): mechanism of action. J. Gen. Virol. 33:159-163. [DOI] [PubMed] [Google Scholar]
- 38.Palese, P., and M. L. Shaw. 2007. Orthomyxoviridae: the viruses and their replication, p. 1647-1689. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 39.Palese, P., K. Tobita, M. Ueda, and R. W. Compans. 1974. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 61:397-410. [DOI] [PubMed] [Google Scholar]
- 40.Pawson, T. 1995. Protein modules and signalling networks. Nature 373:573-580. [DOI] [PubMed] [Google Scholar]
- 41.Petri, T., and N. J. Dimmock. 1981. Phosphorylation of influenza virus nucleoprotein in vivo. J. Gen. Virol. 57:185-190. [DOI] [PubMed] [Google Scholar]
- 42.Pleschka, S., et al. 2001. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat. Cell. Biol. 3:301-305. [DOI] [PubMed] [Google Scholar]
- 43.Privalsky, M. L., and E. E. Penhoet. 1978. Influenza virus proteins: identity, synthesis, and modification analyzed by two-dimensional gel electrophoresis. Proc. Natl. Acad. Sci. U. S. A. 75:3625-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puthavathana, P., et al. 2005. Molecular characterization of the complete genome of human influenza H5N1 virus isolates from Thailand. J. Gen. Virol. 86:423-433. [DOI] [PubMed] [Google Scholar]
- 45.Regan, J. F., Y. Liang, and T. G. Parslow. 2006. Defective assembly of influenza A virus due to a mutation in the polymerase subunit PA. J. Virol. 80:252-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson, J. C., and R. K. Akkina. 1991. NS2 protein of influenza virus is found in purified virus and phosphorylated in infected cells. Arch. Virol. 116:69-80. [DOI] [PubMed] [Google Scholar]
- 47.Sanz-Ezquerro, J. J., et al. 1998. The PA influenza virus polymerase subunit is a phosphorylated protein. J. Gen. Virol. 79:471-478. [DOI] [PubMed] [Google Scholar]
- 48.Schlessinger, J. 2000. Cell signaling by receptor tyrosine kinases. Cell 103:211-225. [DOI] [PubMed] [Google Scholar]
- 49.Shapira, S. D., et al. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 51.Stantchev, T. S., I. Markovic, W. G. Telford, K. A. Clouse, and C. C. Broder. 2007. The tyrosine kinase inhibitor genistein blocks HIV-1 infection in primary human macrophages. Virus Res. 123:178-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ullrich, A., and J. Schlessinger. 1990. Signal transduction by receptors with tyrosine kinase activity. Cell 61:203-212. [DOI] [PubMed] [Google Scholar]
- 53.Vela, E. M., G. C. Bowick, N. K. Herzog, and J. F. Aronson. 2008. Genistein treatment of cells inhibits arenavirus infection. Antiviral Res. 77:153-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, X., E. R. Hinson, and P. Cresswell. 2007. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2:96-105. [DOI] [PubMed] [Google Scholar]
- 55.Watanabe, K., et al. 2001. Inhibition of nuclear export of ribonucleoprotein complexes of influenza virus by leptomycin B. Virus Res. 77:31-42. [DOI] [PubMed] [Google Scholar]
- 56.Watanabe, T., S. Watanabe, and Y. Kawaoka. 2010. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 7:427-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whittaker, G., I. Kemler, and A. Helenius. 1995. Hyperphosphorylation of mutant influenza virus matrix protein, M1, causes its retention in the nucleus. J. Virol. 69:439-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whittaker, G. R., and P. Digard. 2006. Entry and intracellular transport of influenza virus, p. 37-64. In Y. Kawaoka (ed.), Influenza virology—current topics. Caister Academic Press, Norwich, United Kingdom.
- 59.Winquist, A. G., K. Fukuda, C. B. Bridges, and N. Cox. 1999. Neuraminidase inhibitors for treatment of influenza A and B infections. MMWR Surveill. Summ. 48:1-9. [Google Scholar]
- 60.Yoon, S. O., et al. 2008. Ran-binding protein 3 phosphorylation links the Ras and PI3-kinase pathways to nucleocytoplasmic transport. Mol. Cell 29:362-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yura, Y., H. Yoshida, and M. Sato. 1993. Inhibition of herpes simplex virus replication by genistein, an inhibitor of protein-tyrosine kinase. Arch. Virol. 132:451-461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.