Abstract
Human norovirus (HuNoV) is a major causative agent of food-borne gastroenteritis worldwide. Currently, there are no vaccines or effective therapeutic interventions for this virus. Development of an attenuated vaccine for HuNoV has been hampered by the inability to grow the virus in cell culture. Thus, a vector-based vaccine may be ideal. In this study, we constructed a recombinant vesicular stomatitis virus (rVSV-VP1) expressing VP1, the major capsid protein of HuNoV. Expression of the capsid protein by VSV resulted in the formation of HuNoV virus-like particles (VLPs) that are morphologically and antigenically similar to native virions. Recombinant rVSV-VP1 was attenuated in cultured mammalian cells as well as in mice. Mice inoculated with a single dose of rVSV-VP1 through intranasal and oral routes stimulated a significantly stronger humoral and cellular immune response than baculovirus-expressed VLP vaccination. Moreover, we demonstrated that mice inoculated with rVSV-VP1 triggered a comparable level of fecal and vaginal IgA antibody. Taken together, the VSV recombinant system not only provides a new approach to generate HuNoV VLPs in vitro but also a new avenue for the development of vectored vaccines against norovirus and other noncultivable viruses.
Human norovirus (HuNoV), formerly called Norwalk-like virus, was initially isolated in the outbreak of gastroenteritis in an elementary school in Norwalk, OH, in 1968 (35). HuNoV, a member of the Caliciviridae family, is a major causative agent of food-borne gastroenteritis in both developed and developing countries. It has been estimated that over 90% of outbreaks of acute nonbacterial gastroenteritis are caused by noroviruses (14, 15, 37, 45). HuNoV is transmitted primarily through the fecal-oral route, either by direct person-to-person contact or by fecally contaminated food or water. HuNoV is highly contagious, and only a few virus particles are thought to be sufficient to cause an infection (12, 14, 37). Outbreaks frequently occur in restaurants, hotels, day care centers, schools, nursing homes, cruise ships, swimming pools, hospitals, and military installations (14, 15, 27, 37, 45). Despite the significant economic impact and high morbidity caused by HuNoV, no vaccines or antiviral drugs are currently available for this virus. This is due in major part to the lack of a cell culture system and an animal model for HuNoV (13, 15). For these reasons, HuNoV and other caliciviruses have been classified as NIAID category B priority biodefense pathogens.
HuNoV is a nonenveloped, positive-sense RNA virus. The genome of HuNoV contains 7.3 to 7.7 kb and encodes three open reading frames (ORFs) (29). ORF1 encodes a polyprotein that is cleaved to produce six nonstructural proteins, including the RNA-dependent RNA polymerase (RdRp) (5, 29). ORF2 encodes the major capsid protein (VP1) that contains the antigenic and receptor binding sites (5, 10, 29, 30, 57, 58) and plays an essential role in viral attachment and entry (57, 58, 62). ORF3 encodes a minor capsid protein (VP2) that may play a role in stabilizing virus particles (5). It is known that the expression of VP1 alone in cell culture yields self-assembled virus-like particles (VLPs) that are structurally and antigenically similar to native virions (10, 30, 47). Consequently, most HuNoV vaccine studies have focused on VLPs. To date, HuNoV VLPs have been expressed in Escherichia coli (58), Pichia pastoris (63), insect cells (1, 4, 25, 30), mammalian cell lines (59), and plants (such as tobacco and potatoes) (64). Immunization of mice with VLPs orally or intranasally induced variable humoral, mucosal, and cellular immunities (1, 15, 21, 36, 63, 64). It was reported that human volunteers who received 250 to 2,000 μg of VLPs developed significant increases in IgA anti-VLP antibody-secreting cells, and 30 to 40% of volunteers developed mucosal anti-VLP IgA (55). Although these studies are very promising, there are several limitations to the development of in vitro-expressed VLPs into a vaccine candidate. Preparation of VLPs in vitro is time-consuming and expensive. Immunization usually requires a high dose of VLPs (usually more than 100 μg) and multiple booster immunizations (15, 53). The efficacy of VLP-based vaccines relies on the addition of mucosal adjuvants such as cholera toxin (CT) and E. coli toxin (LT), which may have side effects, such as neurotoxicity and induction of immune tolerance (11, 46). Also, the duration of the antigen stimulation may be limited because VLPs are actually proteins, a nonreplicating immunogen.
Generally, a live attenuated virus vaccine stimulates strong systemic immunity and provides durable protection because replication in vivo results in high level intracellular synthesis of the full complement of viral antigens over a prolonged period. However, such a vaccine is not realistic for viruses that cannot be grown in cell culture. Given this limitation, the virus cannot be attenuated, and even if an attenuated strain were available, it could not be mass produced. In this situation, a vectored vaccine may be ideal to overcome this obstacle. Vesicular stomatitis virus (VSV) has been shown to be an excellent vector to deliver foreign antigens (reviewed in references 41 and 50). VSV is a nonsegmented negative-sense (NNS) RNA virus that belongs to the virus family Rhabdoviridae. Recombinant VSV can be recovered entirely from an infectious cDNA clone by a reverse genetics system (38, 61). With this technique, an exogenous gene can be inserted into the VSV genome, and recombinant VSV expressing this antigen can be recovered. The exogenous antigen is expressed continuously in vivo once the recombinant viruses are inoculated into animals, and thus it triggers specific immune responses. To date, VSV has been successfully examined as a vaccine candidate for a number of pathogens, including human immunodeficiency virus (HIV) (23, 31, 51, 56), severe acute respiratory syndrome (SARS) virus (17, 34), hepatitis C virus (HCV) (8, 16), influenza virus (49, 50), papillomavirus (48), human respiratory syncytial virus (33), poxvirus (7), arenavirus (19), Ebola virus, and Marburg virus (20, 32). These studies have shown that VSV-based vaccines trigger strong immunity in animal models even after a single immunizing dose. Particularly, the VSV-based HIV vaccine has been applied for clinical study through the HIV Vaccine Design and Development Teams (HVDDT) program at NIAID in partnership with Wyeth Pharmaceuticals (54). However, the exploration of VSV as a vector to deliver vaccines against noncultivable viruses has not been reported.
In this study, we successfully recovered a recombinant VSV expressing HuNoV capsid protein (rVSV-VP1) which was attenuated in cell culture. Infection of mammalian cells with recombinant rVSV-VP1 resulted in high-level production of HuNoV VLPs. Importantly, we further demonstrated that rVSV-VP1 was attenuated in mice and elicited a high level of HuNoV-specific humoral, cellular, and mucosal immune responses in a mouse model. Thus, the VSV recombinant system not only provided a new approach to generate HuNoV VLPs but also resulted in a novel live vectored vaccine candidate for HuNoV, and perhaps for other noncultivable food-borne viruses as well.
MATERIALS AND METHODS
Plasmid construction.
Plasmids encoding VSV N (pN), P (pP), and L (pL) genes, and an infectious cDNA clone of the viral genome, pVSV1(+), were generous gifts from Gail Wertz (61). Plasmid pVSV1(+) GxxL, which contains SmaI and XhoI at the G and L gene junction, was kindly provided by Sean Whelan. The capsid VP1 gene of HuNoV genogroup II.4 strain HS66 (kindly provided by Linda Saif) was amplified by high-fidelity PCR. The resulting DNA fragment was digested with SmaI and XhoI and cloned into pVSV(+)GxxL at the same sites. The resulting plasmid was designated pVSV1(+)-VP1, in which the HuNoV VP1 gene was inserted into the G and L gene junction. The firefly luciferase gene was amplified from the pGL2 luciferase reporter vector (Promega, Madison, WI) by PCR and cloned into pVSV1(+) at the gene junction between the leader and N, resulting in construction of pVSV1(+)-Luc. All the inserted genes contain VSV gene start and gene end sequences. The HuNoV VP1 gene was also cloned into a pFastBac-Dual expression vector (Invitrogen, Carlsbad, CA) at SmaI and XhoI sites under the control of the p10 promoter, which resulted in of pFastBac-Dual-VP1. All constructs were confirmed by sequencing.
Recovery and purification of recombinant VSV.
Recovery of recombinant VSV from the infectious clone was carried out as described previously (61). Briefly, recombinant VSV was recovered by cotransfection of pVSV1(+)-VP1, pN, pP, and pL into BSRT7 cells infected with a recombinant vaccinia virus (vTF7-3) expressing T7 RNA polymerase. At 96 h posttransfection, cell culture fluids were collected and filtered through a 0.2-μm filter, and the recombinant virus was further amplified in BSRT7 cells. Subsequently, the viruses were plaque purified as described previously (39, 40). Individual plaques were isolated, and seed stocks were amplified in BSRT7 cells. The viral titer was determined by a plaque assay performed in Vero cells.
Single-cycle growth curves.
Confluent BSRT7 cells were infected with individual viruses at a multiplicity of infection (MOI) of 10. After 1 h of absorption, the inoculum was removed, the cells were washed twice with Dulbecco's modified Eagle's medium (DMEM), fresh DMEM (supplemented with 2% fetal bovine serum) was added, and the infected cells were incubated at 37°C. Aliquots of the cell culture fluid were removed at the indicated intervals, and virus titers were determined by plaque assay in Vero cells.
Analysis of protein synthesis.
Confluent BSRT7 cells were infected with either rVSV or rVSV-VP1 as described above. After 3 h postinfection, cells were washed with methionine- and cysteine-free (M−C−) medium and incubated with fresh M−C− medium supplemented with actinomycin D (15 μg/ml). After 1 h of incubation, the medium was replaced with M−C− medium supplemented with EasyTag 35S-Express (4 μCi/ml; Perkin-Elmer, Wellesley, MA). After 4 h of incubation, cytoplasmic extracts were prepared and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (39, 40). Labeled proteins were detected either by autoradiography or by using a phosphorimager.
RT-PCR.
Viral RNA was extracted from either rVSV or rVSV-VP1 by using an RNeasy minikit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Two primers (5′-CGAGTTGGTATTTATCTTTGC-3′ and 5′-GTACGTCATGCGCTCATCG-3′) were designed to target the VSV G gene at position 4524 and the L gene at position 4831 (numbering refers to the complete VSV Indiana genome sequence), respectively. Reverse transcription-PCR (RT-PCR) was performed using a One Step RT-PCR kit (Qiagen). The amplified products were analyzed on 1% agarose gel electrophoresis.
Western blotting.
BSRT7 cells were infected with either rVSV or VSV-VP1 as described above. At the indicated times postinfection, cell culture medium was harvested and clarified at 3,000 rpm for 15 min and further concentrated at 30,000 rpm for 1.5 h. In the meantime, cells were lysed in lysis buffer containing 5% β-mercaptoethanol, 0.01% NP-40, and 2% SDS. Proteins were separated by 12% SDS-PAGE and transferred to a Hybond enhanced chemiluminescence nitrocellulose membrane (Amersham) in a Mini Trans-Blot electrophoretic transfer cell (Bio-Rad). The blot was probed with guinea pig anti-HuNoV VP1 antiserum (a generous gift from Xi Jiang) at a dilution of 1:5,000, followed by horseradish peroxidase-conjugated goat anti-guinea pig IgG secondary antibody (Santa Cruz) at a dilution of 1:20,000. The blot was developed with SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) and exposed to Kodak BioMax MR film.
Production and purification of VLPs by a baculovirus expression system.
The baculovirus expression plasmid encoding the HuNoV VP1 gene (pFastBac-VP1) was transformed into DH10Bac. Baculovirus expressing the VP1 protein was generated by transfection of bacmids into Spodoptera frugiperda (Sf9) cells with a Cell-fectin transfection kit (Invitrogen), according to the instructions of the manufacturer. Purification of VLPs from insect cells was as described previously with some minor modifications (1, 30, 53). Sf9 cells were infected with baculovirus at an MOI of 10, and the infected Sf9 cells and cell culture supernatants were harvested at 6 days postinoculation. The VLPs were purified from cell culture supernatants and cell lysates by ultracentrifugation through a 40% (wt/vol) sucrose cushion, followed by CsCl isopycnic gradient (0.39 g/cm3) ultracentrifugation. Purified VLPs were analyzed by SDS-PAGE, Western blotting, and electron microscopy (EM). The protein concentrations of the VLPs were measured by using the Bradford reagent (Sigma Chemical Co., St. Louis, MO).
Production and purification of VLPs by a VSV vector.
Recombinant rVSV-VP1 was inoculated into 10 confluent T150 flasks of BSRT7 cells at an MOI of 0.01 in a volume of 2 ml of DMEM. At 1 h postabsorption, 15 ml of DMEM (supplemented with 2% fetal bovine serum) was added to the cultures, and infected cells were incubated at 37°C for 24 to 48 h. Cell culture fluids were harvested when extensive cytopathic effect (CPE) was observed. Cell culture fluids were clarified by centrifugation at 3,000 rpm for 30 min. Virus was concentrated through a 40% (wt/vol) sucrose cushion by centrifugation at 30,000 rpm for 2 h at 4°C in a Ty 50.2 rotor (Beckman). The pellet was resuspended in TNC buffer (0.05 M Tris-HCl, 0.15 M NaCl, 15 mM CaCl2 [pH 6.5]) and further purified through a CsCl isopycnic gradient by centrifugation at 35,000 rpm for 18 h at 4°C in an SW55 rotor (Beckman). The final pellet was resuspended in 0.3 ml of TNC buffer. Purified VLPs were analyzed by SDS-PAGE, Western blotting, and EM. The protein concentrations of the VLPs were measured by using the Bradford reagent (Sigma Chemical Co., St. Louis, MO).
Transmission electron microscopy.
Negative-staining electron microscopy of purified VLPs was performed as described previously (35). Briefly, 20 μl of VLP suspension was fixed in copper grids (Electron Microscopy Sciences, Inc.) and negatively stained with 1% ammonium molybdate. Virus particles were visualized by using a FEI Tecnai G2 Spirit transmission electron microscope (TEM) at 80 kV at the Microscopy and Imaging Facility at The Ohio State University. Images were captured on a MegaView III side-mounted charge-coupled-device camera (Soft Imaging System, Lakewood, CO), and figures were processed using Adobe Photoshop software (Adobe Systems, San Jose, CA).
Animal experiment.
Twenty-five 4-week-old specific-pathogen-free female BALB/c mice (Charles River laboratories, Wilmington, MA) were randomly divided into five groups (five mice per group). Mice in groups 1 to 3 were inoculated with 106 PFU of rVSV, rVSV expressing luciferase (rVSV-Luc), or rVSV-VP1, respectively. Mice in group 4 were inoculated with 100 μg of VLPs (purified from the baculovirus expression system). Mice in group 5 were inoculated with 200 μl of DMEM and served as unimmunized controls. All mice were inoculated through the combination of intranasal and oral routes. Half of the antigens were inoculated intranasally, and the other half was administrated orally. After inoculation, the animals were evaluated on a daily basis for mortality, weight loss, and the presence of any symptoms of VSV infection. Blood samples were collected from each mouse weekly by facial vein bleed, and serums were isolated for IgG antibody detection. Fecal and vaginal homogenate samples were isolated weekly for the detection of norovirus specific IgA. At 5 weeks postinoculation, all mice were sacrificed. The spleens were isolated from each mouse, and mononuclear cell (MNC) suspensions were prepared for a T cell proliferation assay.
T cell proliferation assay.
Ninety-six-well plates were coated with 50 μl of highly purified HuNoV VLPs (10 μg/ml) in 200 mM NaCO3 buffer (pH 9.6) at 4°C overnight. After homogenization, spleen cells were washed twice with phosphate-buffered saline (PBS) and plated in triplicate at 5 ×105 cells/well in a 96-well-plate in RPMI 1640 medium with 2% naive mouse serum. After 48 h of incubation at 37°C, 0.5 μCi of [3H]thymidine was added to each well, and 16 h later cells were harvested onto glass filters and counted with a Betaplate counter (Wallac, Turku, Finland). The stimulation index (SI) was calculated as the mean of the following ratio: proliferation of HuNoV VLP-stimulated cells/proliferation of cells in medium (in cpm).
Serum IgG ELISA.
Ninety-six-well plates were coated with 50 μl of highly purified HuNoV VLPs (7.5 μg/ml) in 50 mM NaCO3 buffer (pH 9.6) at 4°C overnight. Individual serum samples were tested for HuNoV-specific IgG on VLP-coated plates. Briefly, serum samples were 2-fold-serially diluted and added to VLP-coated wells. After incubation at room temperature for 1 h, the plates were washed five times with PBS-Tween (0.05%), followed by incubation with 50 μl of goat anti-mouse IgG horseradish peroxidase-conjugated secondary antibodies (Sigma) at a dilution of 1:80,000 for 1 h. Plates were washed and developed with 75 μl of 3,3′,5,5′-tetramethylbenzidine (TMB), and the optical density (OD) at 450 nm was determined using an enzyme-linked immunosorbent assay (ELISA) plate reader. End point titer values were determined as the reciprocal of the highest dilution that had an absorbance value greater than background level (DMEM control).
Fecal IgA ELISA.
For each stool sample, HuNoV-specific and total fecal IgA were determined as described previously (21). Fecal pellets were diluted 1:2 (wt/vol) in PBS containing 0.1% Tween and a Complete EDTA-free proteinase inhibitor cocktail tablet (Roche). Samples were vortexed twice for 30 s and clarified twice by centrifugation at 10,000 × g for 10 min. Ninety-six-well plates were coated with 50 μl of highly purified HuNoV VLPs (1 μg/ml) in 50 mM NaCO3 buffer (pH 9.6) at 4°C overnight for detection of HuNoV-specific IgA, while total fecal IgA was determined by capturing all fecal extract IgA molecules with 1 μg/ml sheep anti-mouse IgA (Sigma). To block nonspecific protein binding, the plates were incubated for 4 h at 4°C with 10% (wt/vol) dry milk in PBS (10% BLOTTO). The level of IgA was calculated from a standard curve that was determined by the absorbance values of the mouse IgA standard (Sigma). The HuNoV-specific IgA level was expressed in nanograms per milliliter, and each corresponding total IgA level was expressed in micrograms per milliliter.
Vaginal HuNoV-specific IgA ELISA.
Ninety-six-well plates were coated with HuNoV VLPs in selected columns as described above. After an overnight blocking at 4°C with 5% BLOTTO, 75 μl of an undiluted vaginal sample per well or a 1:5 dilution of the sample was added, the sample was serially diluted 2-fold down the plate and incubated for 2 h at 37°C. The remaining protocol was identical as described above for the HuNoV-specific fecal IgA ELISA or the serum IgG ELISA.
Quantitative and statistical analyses.
Quantitative analysis was performed by either densitometric scanning of autoradiographs or by using a Typhoon PhosphorImager (GE Healthcare) and ImageQuant TL software (GE Healthcare, Piscataway, NJ). Each experiment was performed three to six times. Statistical analysis was performed by a paired Student's t test. A P value of <0.05 was considered statistically significant.
RESULTS
Recovery of recombinant VSV expressing HuNoV capsid protein.
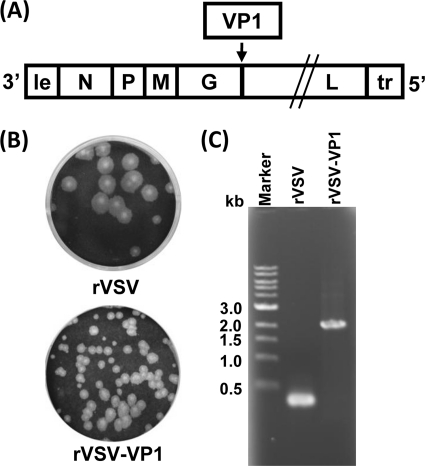
The feasibility for the delivery of VLPs using VSV as a vector is poorly understood, although many antigens have been successfully expressed by VSV. Since the abundance of gene expression decreases with distance from the 3′ end to the 5′end of the VSV genome (28), we attempted to recover recombinant VSV harboring the VP1 gene at the 3′-proximal end of the genome. Unfortunately, after multiple attempts, we failed to recover recombinant viruses with the VP1 gene inserted in any of the following gene junctions: leader-N, N-P, P-M, or M-G. However, recombinant VSV expressing HuNoV VP1 was successfully recovered when the VP1 gene was inserted at the gene junction G-L in the VSV genome (Fig. 1 A). Recombinant rVSV-VP1 formed much smaller plaques than rVSV (Fig. 1B). After 24 h of incubation, rVSV formed plaques that were 4.3 ± 0.8 mm (mean ± standard deviation) in diameter. However, the average plaque size for rVSV-VP1 was 1.7 ± 0.6 mm even after 48 h of incubation, suggesting that rVSV-VP1 may have a defect in viral growth. To confirm that the recovered virus indeed contained the VP1 gene, viral genomic RNA was extracted followed by RT-PCR using two primers annealing to the VSV G and L genes, respectively. As shown in Fig. 1C, a 2.0-kb cDNA band containing the VP1 gene was amplified from genomic RNA extracted from rVSV-VP1, while a 300-bp cDNA was amplified from rVSV. The cDNA was purified and sequenced, confirming that VP1 of HuNoV was indeed inserted into the VSV genome. We also sequenced the entire rVSV-VP1 genome to confirm that no additional mutation was introduced. By using a similar strategy, we also recovered rVSV expressing firefly luciferase (rVSV-Luc), in which luciferase was inserted between the leader and N gene junction (data not shown).
FIG. 1.
Recovery of recombinant VSV expressing HuNoV VP1 (rVSV-VP1). (A) Insertion of VP1 into the VSV genome at the gene junction between G and L. Le, VSV leader sequence; N, nucleocapsid gene; P, phosphoprotein gene; M, matrix protein gene; G, glycoprotein gene; L, large polymerase gene; Tr, VSV trailer sequence. (B) The plaque morphology of recombinant viruses compared to rVSV. Plaques of rVSV-VP1 developed after 48 h of incubation compared to rVSV, which developed plaques after 24 h of incubation. (C) Amplification of the VP1 gene from recombinant rVSV-VP1 by RT-PCR. Genomic RNA was extracted from each of the viruses, and the VP1 gene was amplified by RT-PCR using two primers annealing to the G and L genes.
The replication of rVSV-VP1 is delayed in cell culture.
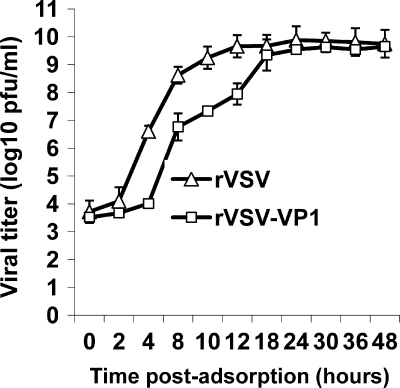
To further characterize recombinant rVSV-VP1, we monitored the kinetics of release of infectious virus by using a single-step growth assay in BSRT7 cells. Briefly, BSRT7 cells were infected with each of the recombinant viruses at an MOI of 10, and viral replication was determined at time points from 0 to 48 h postinfection. As shown in Fig. 2, rVSV-VP1 had a significant delay in viral replication compared to that of rVSV. Wild-type rVSV reached a peak titer (4.6 × 109 PFU/ml) at 12 h postinfection. However, rVSV-VP1 reached a peak titer of 4.0 × 109 PFU/ml at approximately 30 h postinfection. At an MOI of 10, recombinant rVSV exhibited significant CPE by 6 h postinfection (data not shown), and cells were completely killed by 14 h postinfection. However, rVSV-VP1 first showed mild CPE after 12 h postinfection, most cells were still healthy at 24 h, and most cells were killed by 36 h postinfection. These results suggested that rVSV-VP1 had delayed replication and was attenuated in cell culture.
FIG. 2.
Single-step growth curve of recombinant VSV in BSRT7 cells. Confluent BSRT7 cells were infected with individual viruses at an MOI of 10. After 1 h of incubation, the inoculum was removed, the cells were washed with DMEM, and fresh medium (containing 2% fetal bovine serum) was added, followed by incubation at 37°C. Samples of supernatant were harvested at the indicated intervals over a 48-h time period, and the virus titer was determined by plaque assay. Titers are expressed as the mean ± the standard deviation of three independent single-step growth experiments.
High-level expression of HuNoV VP1 protein by the VSV vector.
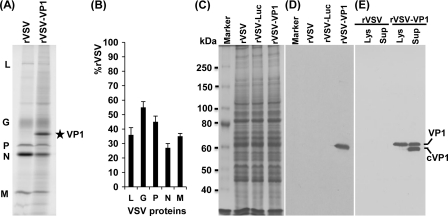
To examine the expression of HuNoV VP1 by VSV, we first determined protein synthesis in virus-infected cells by metabolic labeling as described in Materials and Methods. Briefly, BSRT7 cells were infected with either rVSV or rVSV-VP1 and, at 3 h postinfection, the cells were incubated with [35S]methionine-cysteine for 4 h. After incubation, cytoplasmic extracts were prepared, and total protein was analyzed by SDS-PAGE. As shown in Fig. 3 A, rVSV synthesized five viral proteins, L, G, P, N, and M. In rVSV-VP1-infected cells, an additional protein band with a molecular mass of approximately 58 kDa was detected. This protein is the correct size for the HuNoV capsid protein VP1. The abundance of this protein increased when cells were infected with a higher MOI of rVSV-VP1 (data not shown). Significantly less VSV proteins were synthesized from rVSV-VP1-infected cells than from rVSV-infected cells (Fig. 3A, compare lanes 1 and 2). Quantitative analysis of three independent experiments showed that there is approximately 25 to 50% as much of the VSV proteins synthesized by rVSV-VP1 relative to the wild type (Fig. 3B). The combination of decreased viral plaque size, delayed single step viral replication, and reduced protein synthesis suggests that rVSV-VP1 is attenuated in cell culture. To further characterize the expression of the VP1 protein, we performed a Western blot analysis using a polyclonal antibody against the VP1 protein. Briefly, BSRT7 cells were infected with rVSV, rVSV-VP1, or rVSV-Luc at an MOI of 10, and cell lysates were harvested at 8 h postinfection. The cell lysates were analyzed by SDS-PAGE, followed by Western blotting. As shown in Fig. 3D, a 58-kDa protein band was visualized in rVSV-VP1, but not in rVSV or rVSV-Luc lysates. For comparison, cell culture medium was harvested at 54 h postinfection. After 30,000 rpm ultracentrifugation, the pellets were analyzed by Western blotting. Interestingly, two protein bands with molecular masses of 58 and 52 kDa were detected from the supernatant by Western blotting (Fig. 3E). If the 58-kDa protein is the native full-length VP1 protein, the 52-kDa protein must be a cleaved form of VP1 (cVP1) protein. However, in the cell lysate, the majority of VP1 remained uncleaved. This was consistent with the earlier observation that HuNoV VP1 can be cleaved when expressed in mammalian and insect cells (4, 25). Taken together, these results demonstrated that (i) expression of VP1 by VSV resulted in two forms of VP1 protein and (ii) the expressed VP1 protein was antigenic and reacted with anti-HuNoV VP1 antibody.
FIG. 3.
Expression of HuNoV VP1 by the VSV vector. (A) Viral protein synthesis in BSRT7 cells. BSRT7 cells were infected with rVSV or rVSV-VP1 at an MOI of 10. After 3 h postinfection, proteins were metabolically labeled by incorporation of [35S]methionine-cysteine in the presence of actinomycin D. After 4 h of incubation, cytoplasmic extracts were harvested, and proteins were analyzed by SDS-PAGE and detected by using a phosphorimager. The identities of the proteins are shown on the left. (B) Quantitative analysis of VSV structural proteins between rVSV and rVSV-VP1. Data were generated in three independent experiments. For each protein the mean ± the standard deviation is shown as a percentage of that observed for rVSV. (C) SDS-PAGE analysis of total cell lysate from virus-infected cells. A total of 2 × 105 of BSRT7 cells in 60-mm dishes were infected with rVSV, rVSV-VP1, or rVSV-Luc at an MOI of 10, cells were lysed in 500 μl of lysis buffer at 8 h postinfection, and 10 μl of lysate was analyzed by SDS-PAGE. (D) Analysis of VP1 expression in cell lysate by Western blotting. Identical samples from those shown in panel C were blotted with guinea pig anti-HuNoV VP1 antiserum. (E) Analysis of VP1 protein in cell culture medium by Western blotting. BSRT7 cells were infected with rVSV or rVSV-VP1, and cell culture medium was harvested at 54 h postinfection. After ultracentrifugation at 29,000 rpm, the pellets were subjected to Western blotting. Lys, cell lysate; Sup, cell culture supernatant; cVP1, cleaved VP1 protein.
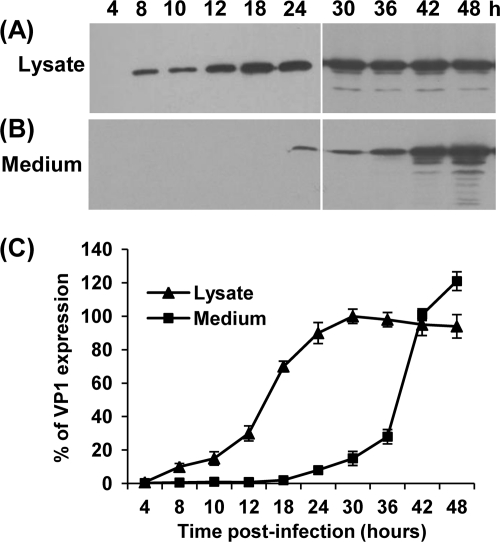
We also monitored the kinetics of VP1 expression in BSRT7 cells. Briefly, BSRT7 cells were infected with rVSV-VP1 at an MOI of 10, and cell culture medium and cell lysates were harvested separately at the indicated times. The expression of VP1 was determined by Western blotting. In the cell lysate, VP1 expression was detected at 4 h postinfection, gradually increased, and reached a peak at 30 h postinfection (Fig. 4 A and C). VP1 protein was secreted into cell culture medium but was not detectable until 24 h postinfection (Fig. 4B and C). A high level of VP1 protein was released to the supernatant after 24 to 48 h postinfection. Thus, VP1 protein was not only expressed in the cytoplasm but also released into cell culture medium.
FIG. 4.
Kinetics of VP1 expression by the VSV vector. (A) Dynamics of VP1 expression in cell lysate by Western blotting. BSRT7 cells were infected with rVSV or rVSV-VP1 at an MOI of 10. Cytoplasmic extracts were harvested at the indicated time points. Equal amounts of total cytoplasmic lysate were analyzed by SDS-PAGE, followed by Western blot analysis using guinea pig anti-HuNoV VP1 antiserum. (B) Analysis of the dynamics of VP1 expression in cell culture medium by Western blotting. Cell culture supernatants were harvested at the indicated time points. VP1 proteins were pelleted from cell culture supernatants through ultracentrifugation at 29,000 rpm and resuspended in 200 μl of NTE (0.1 M NaCl, 1 mM EDTA, 0.01 M Tris [pH 7.4]). Equal amounts of suspensions were subjected to Western blotting. (C) Quantitative analysis of VP1 expression in cytoplasmic lysate and cell culture supernatants. Three independent experiments were used to generate the quantitative analysis shown. Data are expressed as the means ± the standard deviations.
Characterization of HuNoV VLPs expressed by the VSV vector.
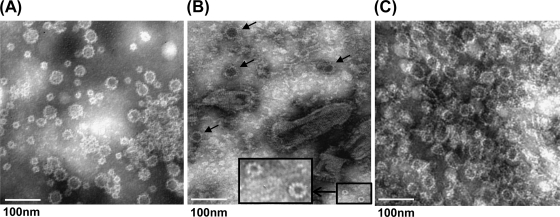
To determine whether expression of VP1 from VSV leads to the assembly of VLPs, BSRT7 cells were infected with rVSV-VP1 and the cell culture medium was harvested at 48 h postinfection. The expressed VP1 protein was purified as described in Materials and Methods. Crude cell culture medium (unpurified) and purified VP1 proteins were subjected to negative-stain EM. HuNoV VLPs purified from insect cells by baculovirus were used as a control. As shown in Fig. 5 A, particles of two sizes (35 to 38 and 18 to 20 nm) were observed in baculovirus-expressed VLPs. In unpurified cell culture medium, two types of virus particles, VSV and HuNoV VLPs, were found (Fig. 5B). VSV is a bullet-shaped particle around 120 nm in length and 70 nm in diameter, while HuNoV VLPs are small round structured particles, 38 nm in diameter. After CsCl isopycnic gradient purification, a large number of HuNoV VLPs were obtained. The size of a majority of the VLPs expressed by VSV had a diameter of approximately 38 nm, although 20 nm-particles were also found (Fig. 5B and C). Therefore, these results confirm that expression of VP1 protein by the VSV vector resulted in the assembly of VLPs that are structurally similar to native virions.
FIG. 5.
Electron microscope analysis of purified VLPs. Twenty microliters of VLP suspension was fixed in copper grids and negatively stained with 1% ammonium molybdate. Virus-like particles were visualized by using a FEI Tecnai G2 Spirit transmission electron microscope. (A) VLPs purified from insect cells by baculovirus. (B) Cell culture supernatant. BSRT7 cells were infected with rVSV-VP1, and cell culture supernatants were harvested at 48 h postinfection. The larger VLPs, 38 nm in diameter, are indicated by arrows. The smaller VLPs, 19 nm in diameter, are shown inside the box (with a magnified image provided). (C) VLPs purified from BSRT7 cells infected by rVSV-VP1.
Recombinant rVSV-VP1 is attenuated in a mouse model.
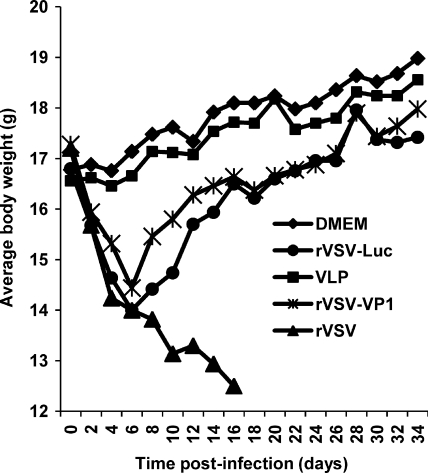
It has been well documented that wild-type rVSV Indiana strain is highly virulent for mice (2, 41). To test the safety of rVSV-VP1, mice were inoculated with 106 PFU of either rVSV or rVSV-VP1 through a combination of intranasal and oral routes. After inoculation, animals were evaluated daily for weight loss and the presence of any clinical symptoms. Consistent with previous reports (2), mice infected with rVSV had severe weight loss (Fig. 6) and exhibited typical clinical signs, including ataxia, hyperexcitability, tremors, circling, and paralysis. At 10 days postinoculation, two of the five mice were dead in the rVSV group. At day 16, we euthanized the remaining three mice in the rVSV group, since they were extremely sick. Mice inoculated with rVSV-VP1 also showed a significant weight loss (P < 0.05) within the first week postinoculation but started to gain weight after 10 days postinoculation (Fig. 6). After 3 weeks postinoculation, there was no significant difference in weight compared to DMEM controls (P > 0.05). Mice inoculated with rVSV-Luc exhibited similar dynamics of weight changes. However, mice inoculated with rVSV-Luc had significantly more weight loss (P < 0.05) than those in rVSV-VP1 group at days 8 to 12 postinoculation. After 12 days postinoculation, mice in the rVSV-VP1 group gained slightly more weight than those in the rVSV-Luc group, although there was no significant difference between these two groups (P > 0.05). In addition, mice inoculated with rVSV-VP1 and rVSV-Luc showed no significant clinical signs, although some mice in the rVSV-Luc group had a ruffled coat at days 6 to 10 postinoculation. Mice inoculated with DMEM did not have any weight loss or clinical signs. This experiment suggests that both rVSV-VP1 and rVSV-Luc were attenuated in mice. Overall, it appears that rVSV-VP1 was more attenuated in mice than rVSV-Luc.
FIG. 6.
Dynamics of mouse body weight after inoculation with recombinant viruses. Five BALB/c mice in each group were inoculated with 106 PFU of rVSV, 106 PFU of rVSV-VP1, or 100 μg of VLPs (purified from insect cells by baculovirus) through a combination of the intranasal and oral routes. The body weight for each mouse was evaluated every other day. The average body weights of five mice are shown. At day 10, two out of five mice were dead in the rVSV group. The remaining three mice were euthanized at day 16 because of severe illnesses.
Intranasal and oral administration of rVSV-VP1 induces a strong serum IgG immune response in mice.
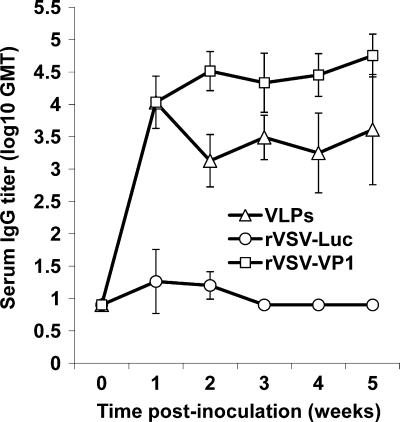
To evaluate whether rVSV-VP1 induces antibodies against HuNoV, blood samples were isolated from each mouse and the serum IgG antibody response was determined by ELISA as described in Materials and Methods. The geometric mean titers (GMT) were calculated for each group of mice and compared. Prior to inoculation, all mice were negative (titer, <10) for HuNoV-specific IgG (data not shown). As shown in Fig. 7, mice inoculated with rVSV-VP1 triggered a much higher serum IgG response than the mice that received baculovirus-produced VLPs during the 5-week experimental period (P < 0.05). At 1 week postinoculation, all mice inoculated with rVSV-VP1 had a high level of serum IgG with a GMT of 10,809. The IgG antibody gradually increased at week 2 postinoculation with a GMT of 32,768 and remained at a high level through week 5. Mice inoculated with 100 μg of baculovirus-produced VLPs had a similar level of HuNoV-specific IgG antibodies at week 1 postinoculation. However, the IgG antibody in the VLP group had decreased by week 2 postinoculation. Moreover, from weeks 2 to 5, the GMT in rVSV-VP1 group was significantly higher than that of the VLP group (P < 0.05). As controls, mice inoculated with rVSV-Luc and DMEM lacked HuNoV-specific serum IgG antibody responses during the experimental period. Thus, this experiment demonstrates that (i) a single-dose inoculation of mice with recombinant rVSV-VP1 stimulated a high level of serum IgG antibody response and (ii) the IgG antibody response induced by rVSV-VP1 was significantly stronger than that of the baculovirus-produced VLP-based vaccine candidate.
FIG. 7.
Serum IgG immune responses to HuNoV vaccine. Groups of five BALB/c mice were inoculated with either 106 PFU of rVSV-VP1 or 100 μg of VLPs through a combination of intranasal and oral routes. Serum samples were collected weekly and analyzed by ELISA for HuNoV-specific serum IgG antibody. Data are expressed as the GMT of five mice. Error bars at each time point represent the standard deviation between mice.
Immunization of mice with rVSV-VP1 induces a strong cellular immune response in mice.
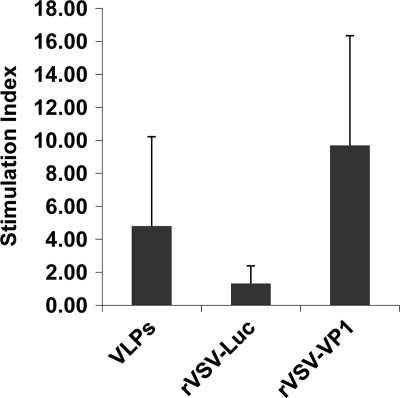
To determine the cellular immune response of the VSV-based HuNoV vaccine, spleens were isolated from each mouse at week 5 postinoculation, and the cellular immune responses were measured by a T cell proliferation assay. As shown in Fig. 8, mice inoculated with rVSV-VP1 stimulated much higher HuNoV-specific T cell proliferation than that of traditional VLP-based vaccines (P < 0.05). All mice in the rVSV-VP1 group had strong HuNoV-specific T cell responses, with an average stimulation index of 9.7. However, only three out of the five mice in the VLP group had a T cell immune response, with an average stimulation index of 4.8. Mice inoculated with rVSV-Luc and DMEM had no HuNoV-specific T cell immune responses. Therefore, these data demonstrate that rVSV-VP1 stimulates a significantly stronger T cell immune response than the VLP-based vaccine candidate.
FIG. 8.
T cell proliferative responses to HuNoV vaccine. Spleen cells were harvested from all mice in each group at week 5 postinoculation and stimulated with HuNoV VLPs. T cell proliferation was measured by [3H]thymidine incorporation. The SI was calculated as the mean of the ratio of the proliferation of HuNoV VLP-stimulated cells versus proliferation of cells in medium (in cpm). Data are expressed as the means of five mice ± the standard deviations.
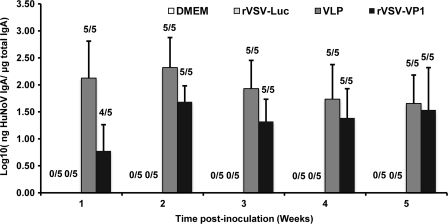
Immunization of mice with rVSV-VP1 induces a mucosal immune response in mice.
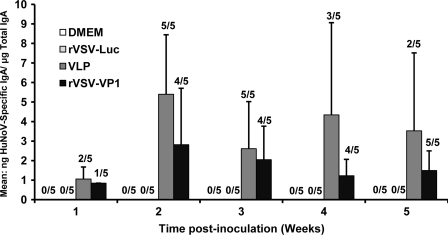
Norovirus causes gastroenteritis; it is likely that mucosal antibodies play an important role in protection from infection. To measure the mucosal immune response, HuNoV-specific and total IgA in fecal and vaginal extracts were assayed by ELISA. The level of IgA response was expressed as the ratio between HuNoV-specific IgA and total IgA. Prior to antigen inoculation, there was no HuNoV-specific IgA in either fecal or vaginal samples in any mice. Figure 9 shows the fecal IgA antibody response from weeks 1 to 5. In week 1, only one and two out of five mice had an IgA response in the rVSV-VP1 group and VLP groups, respectively. At week 2, all mice in the VLP group developed HuNoV-specific IgA, while four out of five mice in the rVSV-VP1 group exhibited an IgA response. At week 3, the IgA antibody started to decrease in both the rVSV-VP1 and the VLP group. There was no significant difference in IgA response between these two groups at weeks 2 and 3 (P > 0.05). At weeks 4 and 5, the VLP group had a higher IgA antibody response than that of the rVSV-VP1 group. However, the number of mice that had detectable IgA in the VLP group (two and three mice) was less than that of the rVSV-VP1 group (four and five mice). Overall, these results demonstrated that rVSV-VP1 was able to trigger a HuNoV-specific mucosal IgA immune response in the intestine. The recombinant rVSV-VP1 group showed a comparable level of IgA at weeks 1 to 3 but had a lower IgA level at weeks 4 and 5 compared to the VLP group. None of the mice in the rVSV-Luc and DMEM groups showed HuNoV-specific IgA antibody over the entire experimental period.
FIG. 9.
Fecal IgA responses to HuNoV vaccine. Fecal samples were collected from all mice in each group weekly. Samples were diluted in PBS, vortexed, and clarified by centrifugation, and HuNoV-specific and total IgA antibodies were determined by ELISA. The ratio between HuNoV-specific IgA and total IgA was calculated for each mouse. Data are expressed as the average titer of IgA-positive mice ± the standard deviation.
Using an identical approach, the vaginal IgA antibody responses were also determined. Interestingly, the ratio between HuNoV-specific IgA and total IgA in vaginal samples was much higher than that of fecal samples. Thus, the level of IgA response was expressed as the log10 of the ratio between HuNoV-specific IgA and total IgA. As shown in Fig. 10, both the rVSV-VP1 and VLP groups triggered a high level of vaginal IgA antibody. At week 1 postinoculation, all mice in the VLP group had vaginal IgA antibody. However, four out of five mice in the rVSV-VP1 group had an IgA response. In the following 4 weeks, all mice developed an IgA response in both the rVSV-VP1 and VLP groups. In weeks 1 and 2, the average IgA titer in the VLP group was higher than in the rVSV-VP1 group (P < 0.05). In weeks 3 to 5, however, there was no significant difference in vaginal IgA response between these two groups (P > 0.05). Mice inoculated with rVSV-Luc and DMEM did not have any HuNoV-specific IgA antibody in vaginal samples during the 5-week experimental period. Overall, these results demonstrated that mice inoculated with rVSV-VP1 and VLPs stimulated a comparable vaginal IgA antibody response. We concluded that the VSV-based HuNoV vaccine is capable of inducing a high level of mucosal immunity in mice, in both the intestinal and vaginal extracts.
FIG. 10.
Vaginal IgA responses to HuNoV vaccine. Vaginal samples were collected weekly from each mouse, and HuNoV-specific and total IgA antibodies were determined by ELISA. The level of vaginal IgA is reported as the log10 of the ratio between HuNoV-specific IgA and total IgA. Data are expressed as the GMT of IgA-positive mice ± the standard deviation.
DISCUSSION
In this study, we recovered a recombinant rVSV expressing the HuNoV VP1 protein. We demonstrated that the VP1 protein was not only highly expressed by the VSV vector but also self-assembled into VLPs that were morphologically and antigenically similar to the native virions. Recombinant rVSV-VP1 was attenuated in cell culture as well as in a mouse model. We further demonstrated that mice inoculated with the VSV-based HuNoV vaccine responded with a high level of HuNoV-specific humoral, cellular, and mucosal immunities. These results showed that VSV is an excellent vector to deliver HuNoV VLPs and that it is a novel vaccine candidate against HuNoV. To our knowledge, this is the first demonstration using VSV as the vector to deliver VLPs of noncultivable viruses in vitro as well as in vivo.
A new approach to generate HuNoV VLPs using VSV as the vector.
VSV is one of the most attractive viral vectors for vaccines, oncolytic therapy, and gene delivery (reviewed in references 41 and 60). Since the establishment of the reverse genetics system for VSV in 1995, hundreds of exogenous genes have been expressed by using VSV as a vector. However, the feasibility of using VSV as the vector to express and deliver VLPs has not been well studied. To date, there has only been one report which demonstrated use of VSV to generate VLPs. Specifically, it demonstrated that the expression of the HCV core, E1, and E2 proteins by VSV assembled to form HCV-like particles in BHK-21 cells which were similar to the ultrastructural properties of HCV virions (16). However, Blanchard et al. in 2003 argued that these particles may be the endogenous viruses of BHK-21 cells, such as intracisternal R-type particles, but not the complete budded HCV-like particles (6). Later, it was shown that expression of HCV E1 and E2 by propagating and nonpropagating (G-protein-deleted) VSV vectors resulted in correctly folded E1/E2 heterodimers (44). However, detailed characterization of these HCV-like particles was lacking in their studies.
In contrast to HCV, expression of HuNoV VP1 alone led to the formation of VLPs (30). We initially inserted the VP1 gene into the 3′-proximal end of the VSV genome. However, recombinant VSV expressing VP1 was recovered only when the VP1 gene was inserted between the gene junction of G and L, which is located at the 5′-proximal end of the VSV genome. It was likely that the high expression level of VP1 at the 3′-proximal end of the VSV genome inhibited the recovery of the virus. Indeed, recovery of rVSV was inhibited when a plasmid encoding the VP1 gene was cotransfected with a VSV infectious clone (data not shown). We found that recombinant rVSV-VP1 showed diminished plaque sizes in Vero cells and delayed replication in BSRT7 cells. In addition, there was significantly less VSV protein synthesized in rVSV-VP1-infected cells than in cells infected with rVSV (P < 0.05). However, a high level of VP1 protein was found in cell lysates at early time points in rVSV-VP1-infected cells. At later times, a high level of VP1 was also found in cell culture supernatants. The expressed VP1 was antigenic, as shown by Western blotting with a polyclonal antibody against VP1. While the expression of VP1 occurs in the cytoplasm, it is likely that some VP1 protein is secreted into the medium across the cell membrane, since most cells were healthy at 24 h postinfection. At late times postinfection, the release of VP1 protein may due to the loss of cell membrane integrity and cell lysis. EM analysis confirmed that the expressed VP1 protein assembled into VLPs. These results demonstrated that VSV-expressed VLPs are structurally and antigenically similar to native virions. In our study, we used BSRT7 cells, since VSV grew to a higher titer in this cell line. The delayed replication of rVSV-VP1 and the formation of VLPs were also observed in other cell lines, such as BHK-21 (data not shown). In addition, there are a number of advantages of using VSV as the vector to express HuNoV VLPs. First, VSV grows to a high titer in a wide range of mammalian cells. It is easy to grow VSV and thus facilitate the large-scale production of VLPs. Second, it is a time-saving approach. It only took 2 days to generate VLPs using VSV as the vector. However, it took 6 days when a baculovirus system was used. Third, it is a highly productive system. A large number of VLPs can be found by EM analysis using cell culture supernatants (without purification). Therefore, a VSV vector is a novel and efficient approach to generate HuNoV VLPs.
A new live vaccine candidate against noncultivable viruses.
Noroviruses are responsible for more than 90% of nonbacterial gastroenteritis worldwide and cause up to 200,000 deaths in children of less than 5 years of age in developing countries (14, 15, 27, 37). Many attempts have been made to develop an effective vaccine against this biodefense agent. To date, most studies have focused on the VLPs purified from the baculovirus expression system. It has been shown that HuNoV VLP vaccination induces humoral and cellular immune responses in both humans and mice (1, 15, 21, 36, 63, 64). Since HoNoV is noncultivable, live vectored vaccines may provide a novel and effective vaccine strategy. Two live viral vectors, Venezuelan equine encephalitis virus (VEE) and adenovirus, have been shown in studies to deliver HuNoV VLPs in vivo. It was shown that the VEE replicon expressing HuNoV VLPs induced HuNoV-specific systemic, mucosal, and heterotypic immunity in mice (3, 26). By using cultivable murine norovirus (MNV) as a model, it was shown that a VEE-adjuvanted vaccine induced homotypic and heterotypic humoral and cellular immunity and protected mice from MNV challenge (42, 43). Most recently, it was reported that a recombinant adenovirus expressing HuNoV capsid protein stimulated a specific immune response in mice (22). These studies demonstrated the feasibility of using vectored vaccine against HuNoV. However, there are some potential disadvantages to use of VEE and adenovirus as vectors. Although the VEE replicon is a single-cycle replicating vector, the biosafety of VEE has been questioned, since VEE is a biodefense pathogen, and the use of functional VEE genes is restricted. For adenovirus, in vivo delivery of the vectored vaccine may be hampered by the host immune response, since a large portion of the global population has preexisting immunities against the adenovirus vector. In fact, recent clinical trials suggest that adenovirus has performed poorly as a vaccine vector (52).
In this study, we developed a VSV-based vaccine candidate against HuNoV. We first demonstrated that recombinant rVSV-VP1 was attenuated in mice. Since VSV transcription results in a gradient of mRNA and protein expression, it would be expected that an exogenous gene inserted at 3′-proximal end of the VSV genome will have more impact on virus attenuation than a gene inserted at the 5′-proximal end. However, it appears that rVSV-VP1 was more attenuated in mice than rVSV-Luc despite the fact that the luciferase and VP1 genes were inserted at the leader-N and G-L gene junctions, respectively. This suggests that the insertion of the VP1 gene had more impact on VSV virulence than that of the luciferase gene. Excitingly, this VSV-based HuNoV vaccine stimulated approximately 10 times more serum IgG than the traditional VLP-based vaccine. High levels of serum antibody lasted at least 5 weeks postinoculation. However, antibody induced by traditional VLP-based vaccines began to decline at only 2 weeks postinoculation. The VSV-based HuNoV vaccine stimulated a strong T cell proliferation with an average stimulation index of 9.7, which was more than two times higher than that of the VLP groups. Since HuNoV causes acute gastroenteritis, it is likely that mucosal immunity plays an important role in protecting humans from disease. Thus, we used a combination of intranasal and oral routes for vaccination. Consistent with earlier observations (1, 21), we also found that not all of the mice developed IgA responses in fecal extracts despite the high dose of VLPs used. There were five mice that had a fecal IgA response at weeks 2 and 3, but only two to three mice had a detectable fecal IgA response at weeks 4 and 5. However, four to five mice from the rVSV-VP1 group had a fecal IgA response from weeks 2 to 5. Mice inoculated with rVSV-VP1 and VLPs stimulated a comparable fecal IgA antibody response at weeks 1 to 3, although the rVSV-VP1 group had a lower level of fecal IgA antibody at weeks 4 and 5. Interestingly, all mice in both the VLP and rVSV-VP1 groups had vaginal IgA antibody. Moreover, the rVSV-VP1 group had an equivalent level of vaginal IgA response as with VLP vaccination at weeks 4 and 5. Taken together, these results suggest that the VSV-based HuNoV vaccine induced a significantly stronger humoral and cellular immunity than the traditional VLP-based vaccine. In addition, rVSV-VP1 was able to trigger a comparable level of mucosal immunity. Thus, our data demonstrated that mice inoculated with a single dose of rVSV expressing HuNoV VLPs triggered a high level of humoral, cellular, and mucosal immunity. This is likely related to the extremely high level of intracellular synthesis of VLPs in infected cells once inoculated into mice. Furthermore, VLPs may be continuously expressed in vivo by the VSV vector, which in turn stimulated long-lasting immune responses. In contrast, conventional purified VLPs are nonreplicating antigens, and thus the duration of the immune response may be limited. A VSV-based vaccine offers a number of other distinctive advantages, including genetic stability, expression of multiple antigens, simplicity of production, multiple routes of administration, and ease of manipulation. Unlike adenovirus vector, human infection with VSV is very rare (18, 24), and the general population is free of preexisting immunity against VSV (41). Therefore, these advantages will facilitate the clinical trials of a VSV-vectored vaccine in the future.
Unfortunately, there is no robust small animal model for HuNoV infection. In this study, we did not show any protection data, since the mouse is not susceptible to HuNoV infection. Recently, it was shown that gnotobiotic pigs infected with HuNoV genogroup II.4 strain HS66 developed diarrhea, viral shedding, and histopathological lesions in their intestines (9). Subsequently, vaccination of gnotobiotic pigs with baculovirus-expressed VLPs generated systemic immune responses and provided protection against viral shedding and diarrhea (53). It will be of great interest to determine whether a VSV-based HuNoV vaccine can protect such animal models from virulent challenge.
In summary, our study highlights a major gap in our understanding of whether VSV can be used as a vector to deliver VLP in vitro as well as in vivo. Our study has three important applications for the development of (i) a highly productive bioreactor to facilitate large-scale purification of HuNoV VLPs using VSV as a vector, (ii) a high titer of HuNoV-specific antibody for virus detection, disease diagnosis, and therapy, and (iii) a VSV-based vaccine as a novel vaccine candidate against HuNoV as well as other noncultivable viruses.
Acknowledgments
This study was supported by a grant from the OSU Public Health Preparedness for Infectious Diseases program to J.L.
We thank Gail Wertz for her generous gift of the VSV infectious clone and Sean Whelan for providing the pVSV(+)GxxL plasmid. We thank Linda Saif for the VP1 gene of human norovirus genogroup II.4 strain HS66 and Xi Jiang for human norovirus VP1 antibody. We acknowledge Stefan Niewiesk for his help on the T cell proliferation assay and his helpful discussion on this study. We are grateful to Mark Peeples, Ronald Iorio, and members of the Li laboratory for critical reviews of the manuscript.
All animal studies in this paper were approved by the Institutional Animal Care and Use Committee (protocol no. 2009A0160) at The Ohio State University.
Footnotes
Published ahead of print on 12 January 2011.
REFERENCES
- 1.Ball, J. M., M. E. Hardy, R. L. Atmar, M. E. Conner, and M. K. Estes. 1998. Oral immunization with recombinant Norwalk virus-like particles induces a systemic and mucosal immune response in mice. J. Virol. 72:1345-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball, L. A., C. R. Pringle, B. Flanagan, V. P. Perepelitsa, and G. W. Wertz. 1999. Phenotypic consequences of rearranging the P, M, and G genes of vesicular stomatitis virus. J. Virol. 73:4705-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baric, R. S., et al. 2002. Expression and self-assembly of Norwalk virus capsid protein from Venezuelan equine encephalitis virus replicons. J. Virol. 76:3023-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertolotti-Ciarlet, A., L. J. White, R. Chen, B. V. Venkataram Prasad, and M. K. Estes. 2002. Structural requirements for the assembly of Norwalk virus-like particles. J. Virol. 76:4044-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertolotti-Ciarlet, A., S. E. Crawford, A. M. Hutson, and M. K. Estes. 2003. The 3′ end of Norwalk virus mRNA contains determinants that regulate the expression and stability of the viral capsid protein VP1: a novel function for the VP2 protein. J. Virol. 77:11603-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchard, E., D. Brand, and P. Roingeard. 2003. Endogenous virus and hepatitis C virus-like particle budding in BHK-21 cells. J. Virol. 77:3888-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braxton, C. L., S. H. Puckett, S. B. Mizel, and D. S. Lyles. 2010. Protection against lethal vaccinia virus challenge by using an attenuated matrix protein mutant vesicular stomatitis virus vaccine vector expressing poxvirus antigens. J. Virol. 84:3552-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buonocore, L., K. J. Blight, C. M. Rice, and J. K. Rose. 2002. Characterization of vesicular stomatitis virus recombinants that express and incorporate high levels of hepatitis C virus glycoproteins. J. Virol. 76:6865-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheetham, S., et al. 2006. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J. Virol. 80:10372-10381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, R., J. D. Neill, M. K. Estes, and B. V. Prasad. 2006. X-ray structure of a native calicivirus: structural insights into antigenic diversity and host specificity. Proc. Natl. Acad. Sci. U. S. A. 103:8048-8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clements, J. D., N. M. Hartzog, and F. L. Lyon. 1988. Adjuvant activity of Escherichia coli heat-labile enterotoxin and effect on the induction of oral tolerance in mice to unrelated protein antigens. Vaccine 6:269-277. [DOI] [PubMed] [Google Scholar]
- 12.Donaldson, E. F., L. C. Lindesmith, A. D. Lobue, and R. S. Baric. 2008. Norovirus pathogenesis: mechanisms of persistence and immune evasion in human populations. Immunol. Rev. 225:190-211. [DOI] [PubMed] [Google Scholar]
- 13.Duizer, E., et al. 2004. Laboratory efforts to cultivate noroviruses. J. Gen. Virol. 85:79-87. [DOI] [PubMed] [Google Scholar]
- 14.Estes, M. K., B. V. Prasad, and R. L. Atmar. 2006. Noroviruses everywhere: has something changed? Curr. Opin. Infect. Dis. 19:467-474. [DOI] [PubMed] [Google Scholar]
- 15.Estes, M. K., et al. 2000. Norwalk virus vaccines: challenges and progress. J. Infect. Dis. 181:S367-S373. [DOI] [PubMed] [Google Scholar]
- 16.Ezelle, H. J., D. Markovic, and G. N. Barber. 2002. Generation of hepatitis C virus-like particles by use of a recombinant vesicular stomatitis virus vector. J. Virol. 76:12325-12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faber, M., et al. 2005. A single immunization with a rhabdovirus-based vector expressing severe acute respiratory syndrome coronavirus (SARS-CoV) S protein results in the production of high levels of SARS-CoV-neutralizing antibodies. J. Gen. Virol. 86:1435-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fellowes, O. N., G. T. Dimopoullos, and J. J. Callis. 1955. Isolation of vesicular stomatitis virus from an infected laboratory worker. Am. J. Vet. Res. 16:623-626. [PubMed] [Google Scholar]
- 19.Garbutt, M., et al. 2004. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J. Virol. 78:5458-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geisbert, T. W., et al. 2008. Vesicular stomatitis virus-based vaccines protect nonhuman primates against aerosol challenge with Ebola and Marburg viruses. Vaccine 26:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerrero, R. A., et al. 2001. Recombinant Norwalk virus-like particles administered intranasally to mice induce systemic and mucosal (fecal and vaginal) immune responses. J. Virol. 75:9713-9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo, L., et al. 2009. A recombinant adenovirus prime-virus-like particle boost regimen elicits effective and specific immunities against norovirus in mice. Vaccine 27:5233-5238. [DOI] [PubMed] [Google Scholar]
- 23.Haglund, K., et al. 2002. High-level primary CD8+ T-cell response to human immunodeficiency virus type 1 Gag and Env generated by vaccination with recombinant vesicular stomatitis viruses. J. Virol. 76:2730-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanson, R. P., A. F. Rasmussen, Jr., C. A. Brandly, and J. W. Brown. 1950. Human infection with the virus of vesicular stomatitis. J. Lab. Clin. Med. 36:754-758. [PubMed] [Google Scholar]
- 25.Hardy, M. E., L. J. White, J. M. Ball, and M. K. Estes. 1995. Specific proteolytic cleavage of recombinant Norwalk virus capsid protein. J. Virol. 69:1693-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrington, P. R., et al. 2002. Systemic, mucosal, and heterotypic immune induction in mice inoculated with Venezuelan equine encephalitis replicons expressing Norwalk virus-like particles. J. Virol. 76:730-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris, J. P., W. J. Edmunds, R. Pebody, D. W. Brown, and B. A. Lopman. 2008. Deaths from norovirus among the elderly, England and Wales. Emerg. Infect. Dis. 14:1546-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iverson, L. E., and J. K. Rose. 1981. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell 23:477-484. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, X., D. Y. Graham, K. Wang, and M. K. Estes. 1990. Norwalk virus genome cloning and characterization. Science 250:1580-1583. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, X., M. Wang, D. Y. Graham, and M. K. Estes. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, J. E., M. J. Schnell, L. Buonocore, and J. K. Rose. 1997. Specific targeting to CD4+ cells of recombinant vesicular stomatitis viruses encoding human immunodeficiency virus envelope proteins. J. Virol. 71:5060-5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, S. M., et al. 2005. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat. Med. 11:786-790. [DOI] [PubMed] [Google Scholar]
- 33.Kahn, J. S., A. Roberts, C. Weibel, L. Buonocore, and J. K. Rose. 2001. Replication-competent or attenuated, nonpropagating vesicular stomatitis viruses expressing respiratory syncytial virus (RSV) antigens protect mice against RSV challenge. J. Virol. 75:11079-11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapadia, S. U., et al. 2005. Long-term protection from SARS coronavirus infection conferred by a single immunization with an attenuated VSV-based vaccine. Virology 340:174-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kapikian, A. Z., et al. 1972. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J. Virol. 10:1075-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitamoto, N., et al. 2002. Cross-reactivity among several recombinant calicivirus virus-like particles (VLPs) with monoclonal antibodies obtained from mice immunized orally with one type of VLP. J. Clin. Microbiol. 40:2459-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koopmans, M. 2008. Progress in understanding norovirus epidemiology. Curr. Opin. Infect. Dis. 21:544-552. [DOI] [PubMed] [Google Scholar]
- 38.Lawson, N. D., E. A. Stillman, M. A. Whitt, and J. K. Rose. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. U. S. A. 92:4477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, J., E. C. Fontaine-Rodriguez, and S. P. Whelan. 2005. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J. Virol. 79:13373-13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, J., J. T. Wang, and S. P. Whelan. 2006. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl. Acad. Sci. U. S. A. 103:8493-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lichty, B. D., A. T. Power, D. F. Stojdl, and J. C. Bell. 2004. Vesicular stomatitis virus: re-inventing the bullet. Trends Mol. Med. 10:210-216. [DOI] [PubMed] [Google Scholar]
- 42.LoBue, A. D., J. M. Thompson, L. Lindesmith, R. E. Johnston, and R. S. Baric. 2009. Alphavirus-adjuvanted norovirus-like particle vaccines: heterologous, humoral, and mucosal immune responses protect against murine norovirus challenge. J. Virol. 83:3212-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LoBue, A. D., L. C. Lindesmith, and R. S. Baric. 2010. Identification of cross-reactive norovirus CD4+ T cell epitopes. J. Virol. 84:8530-8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Majid, A. M., H. Ezelle, S. Shah, and G. N. Barber. 2006. Evaluating replication-defective vesicular stomatitis virus as a vaccine vehicle. J. Virol. 80:6993-7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mead, P. S., et al. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petrovsky, N., and J. C. Aguilar. 2004. Vaccine adjuvants: current state and future trends. Immunol. Cell Biol. 82:488-496. [DOI] [PubMed] [Google Scholar]
- 47.Prasad, B. V., et al. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 48.Reuter, J. D., et al. 2002. Intranasal vaccination with a recombinant vesicular stomatitis virus expressing cottontail rabbit papillomavirus L1 protein provides complete protection against papillomavirus-induced disease. J. Virol. 76:8900-8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts, A., et al. 1998. Vaccination with a recombinant vesicular stomatitis virus expressing an influenza virus hemagglutinin provides complete protection from influenza virus challenge. J. Virol. 72:4704-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts, A., L. Buonocore, R. Price, J. Forman, and J. K. Rose. 1999. Attenuated vesicular stomatitis viruses as vaccine vectors. J. Virol. 73:3723-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rose, N. F., et al. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106:539-549. [DOI] [PubMed] [Google Scholar]
- 52.Sekaly, R. P. 2008. The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development? J. Exp. Med. 205:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Souza, M., V. Costantini, M. S. Azevedo, and L. J. Saif. 2007. A human norovirus-like particle vaccine adjuvanted with ISCOM or mLT induces cytokine and antibody responses and protection to the homologous GII.4 human norovirus in a gnotobiotic pig disease model. Vaccine 25:8448-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spearman, P. 2003. HIV vaccine development: lessons from the past and promise for the future. Curr. HIV Res. 1:101-120. [DOI] [PubMed] [Google Scholar]
- 55.Tacket, C. O., M. B. Sztein, G. A. Losonsky, S. S. Wasserman, and M. K. Estes. 2003. Humoral, mucosal, and cellular immune responses to oral Norwalk virus-like particles in volunteers. Clin. Immunol. 108:241-247. [DOI] [PubMed] [Google Scholar]
- 56.Tan, G. S., et al. 2005. Strong cellular and humoral anti-HIV Env immune responses induced by a heterologous rhabdoviral prime-boost approach. Virology 331:82-93. [DOI] [PubMed] [Google Scholar]
- 57.Tan, M., et al. 2009. Conservation of carbohydrate binding interfaces: evidence of human HBGA selection in norovirus evolution. PLoS One 4:e5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan, M., W. Zhong, D. Song, S. Thornton, and X. Jiang. 2004. E. coli-expressed recombinant norovirus capsid proteins maintain authentic antigenicity and receptor binding capability. J. Med. Virol. 74:641-649. [DOI] [PubMed] [Google Scholar]
- 59.Taube, S., A. Kurth, and E. Schreier. 2005. Generation of recombinant norovirus-like particles (VLP) in the human endothelial kidney cell line 293T. Arch. Virol. 150:1425-1431. [DOI] [PubMed] [Google Scholar]
- 60.Whelan, S. P., J. N. Barr, and G. W. Wertz. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:61-119. [DOI] [PubMed] [Google Scholar]
- 61.Whelan, S. P., L. A. Ball, J. N. Barr, and G. T. Wertz. 1995. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc. Natl. Acad. Sci. U. S. A. 92:8388-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White, L. J., et al. 1996. Attachment and entry of recombinant Norwalk virus capsids to cultured human and animal cell lines. J. Virol. 70:6589-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia, M., T. Farkas, and X. Jiang. 2007. Norovirus capsid protein expressed in yeast forms virus-like particles and stimulates systemic and mucosal immunity in mice following an oral administration of raw yeast extracts. J. Med. Virol. 79:74-83. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, X., N. A. Buehner, A. M. Hutson, M. K. Estes, and H. S. Mason. 2006. Tomato is a highly effective vehicle for expression and oral immunization with Norwalk virus capsid protein. Plant Biotechnol. J. 4:419-432. [DOI] [PubMed] [Google Scholar]