Abstract
Although transcription from unintegrated human immunodeficiency virus type 1 (HIV-1) DNA can occur inside infected cells, yielding all classes of viral mRNA transcripts, the translation of viral proteins is very limited. One of the proteins made is Nef, but it is unclear whether Nef produced in this way is able to play a role in immune evasion as occurs with integrated virus. We therefore asked whether transcription from preintegrated HIV-1 cDNAs could result in Nef-mediated modulation of cell surface major histocompatibility complex class I (MHC-I) expression. We infected a Rev-CEM green fluorescent protein (GFP) reporter cell line with virus and blocked integration though use of either an inactive integrase or the integrase inhibitor raltegravir. Infected cells were assayed by flow cytometry for cell surface expression of the HLA-A, HLA-B, and HLA-C allotypes (HLA-ABC), HLA-A31, and HLA-E. Viral RNA and DNA products were assayed via quantitative PCR (qPCR). The prevention of integration had no effect, relative to productively infected cells, on levels of expression of multiply spliced viral mRNA transcripts and Nef protein. Downregulation of HLA-ABC and HLA-A31 also occurred at levels similar to those seen in cells in which integration had occurred. Parallel experiments assaying cell surface HLA-ABC expression in infected activated primary CD4+ T cells produced a similar pattern of results. Hence, the capacity of HIV-1 to modulate MHC-I is not linked to its ability to integrate. Thus, Nef-mediated evasion of host immune responsiveness might be attributable, in part at least, to transcription from unintegrated viral DNA.
Retroviruses are defined by the integration of their reverse-transcribed genome into host cell chromatin. This process enables transcription and translation of viral genes by host cells, ultimately resulting in new viral progeny. However, human immunodeficiency virus type 1 (HIV-1) gene transcription and translation can also occur prior to, or even in the absence of, viral integration (8, 53, 57), since unintegrated, reverse-transcribed viral cDNAs can also serve as a template for transcription (22). Three species of unintegrated HIV cDNAs are found in natural infections; these are linear reverse-transcribed cDNA, which is the template for integration, and 1-long terminal repeat (LTR) and 2-LTR circular forms, which are the products of autointegration or nonhomologous recombination and nonhomologous end-joining events of linear cDNAs, respectively (15, 26, 37). The circular cDNAs were long considered to be “dead end” products, which cannot serve as templates for integration, though it is now understood that unintegrated cDNA can be complemented by superinfecting virus to yield productive infection (17, 39, 53).
Transcription of preintegrated HIV-1 cDNA can yield all classes of viral RNA transcripts (25, 37, 52); however, only the accessory and regulatory proteins Nef (18, 58), Tat (2, 14, 46), and Rev (22, 29) are translated in readily detectable amounts, and the full extent of the function of these proteins needs to be further characterized. Differences in transcription between integrated and unintegrated HIV-1 may be due to the fact that unintegrated HIV-1 cDNA is organized into chromatin structures, with histone modifications typical of silenced chromatin (23). Additionally, the low levels of Rev synthesized prior to integration may also limit the translation of unspliced viral RNA transcripts and ultimate expression of late gene products (58).
Studies of integrase-defective HIV-1 mutants that bear mutations in the catalytic D(64)D(116)E(152) triad of integrase have been particularly useful in the study of preintegration transcription (18, 36). Indeed, patterns of transcription and translation arising from unintegrated DNA following infection with D116N mutated HIV-1 are identical to those observed from preintegrated viral DNA and in infections of T-cell lines, activated CD4+ T cells, resting T cells, and macrophages (25, 56, 58). Transcription and translation from unintegrated cDNA following use of integrase strand transfer inhibitors (INSTIs) are also indistinguishable from those seen with preintegrated virus or integrase-defective virus (21, 58).
2-LTR circles were previously proposed as a likely transcriptional template, as their levels were found to be elevated when viral integration was inhibited (14, 21). Moreover, a novel viral transcript spanning the LTR-LTR junction was detected, demonstrating that 2-LTR circles can act as a transcriptional template (7). In contrast, a recent study calculated that there were insufficient levels of 2-LTR circles to account for the numbers of cells bearing transcriptionally active preintegrated virus (22, 54, 55).
Translation of nef, tat, and rev from preintegrated templates has been linked to a number of cellular effects which aid viral infection. For example, preintegration translation of tat and nef has been shown to increase the activation state of resting T cells, making them more amenable to productive infection (58). Preintegration translation of nef has been linked to reduced cell surface expression of CD4 in primary T cells and T cell lines (18, 36), and we have recently demonstrated that the CXCR4 and CCR5 coreceptors are also affected in this manner (45). In a study of macrophages, transcription of preintegrated HIV-1 cDNA was also linked to altered patterns of cytokine expression (25).
CD8+ cytotoxic T-lymphocytes (CTLs) play a central role in the adaptive immune response to control HIV-1, as they recognize viral antigens presented through major histocompatibility complex class I (MHC-I) on infected cells and can limit infection either by direct lysis (5) or through release of inhibitory factors, such as RANTES, MIP-1 alpha, or MIP-1 beta (9). Therefore, modulation of cell surface expression of MHC-I is a common viral immune evasion strategy and avoids the presentation of viral antigens to CTLs, thereby preventing lysis of the infected cell (20). In the case of HIV-1, the virus-encoded protein Nef performs this function (11, 44). Nef downregulates MHC-I by forming a complex with the cytoplasmic tail of MHC-I and the clathrin adaptor AP-1 in the trans-Golgi network (TGN). This allows it to divert normal migration of newly synthesized MHC-I to the cell surface and instead targets it for endosomal degradation (28, 41, 47, 51). However, there is also some evidence that Nef may mediate accelerated endocytosis of MHC-I from the plasma membrane (28), and it has been suggested that cell type differences might also be important (24).
HIV-1 Nef can mediate downregulation of the MHC-I/human leukocyte antigen (HLA) HLA-A and HLA-B allotypes, which are recognized by CTLs. In contrast, HIV-1 does not downregulate HLA-C and HLA-E, which is advantageous since a reduction in cell surface expression of these allotypes would lead to natural killer (NK) cell-mediated lysis, as NK cells respond to reduced MHC-I levels (10). We therefore hypothesized that the preintegration translation of nef has the capacity to modulate cell surface MHC-I expression.
Here we show that preintegration transcription and translation of nef can modulate cell surface MHC-I in the same manner as when integration occurs. This suggests that transcription from preintegrated viral DNA can influence viral immune evasion even prior to viral integration into the host genome.
MATERIALS AND METHODS
Plasmids and cloning.
The HIV-1 molecular clone pNL4-3 was altered through site-directed mutagenesis (Stratagene) to introduce termination codons in the first and third amino acids of the env gene (construct termed pNL4-3-ΔEnv). Further modifications by mutagenesis included the substitution D116N in the integrase coding sequence of the pol gene (construct termed pNL4-3-D116N-ΔEnv) and the introduction of termination codons into the first and third codons of the nef gene (constructs termed pNL4-3-ΔEnv-ΔNef and pNL4-3-D116N-ΔEnv-ΔNef). A pNL4-3-ΔNef construct with an intact env was also prepared in a similar manner.
Virus production.
For infections of T-cell lines, pseudovirus was produced by cotransfection of 7 × 106 293T cells with 4 μg pVPack-VSV-G (Stratagene), a vesicular stomatitis virus G protein (VSV-G) envelope-encoding construct, in combination with 12 μg of a pNL4-3 derivative (pNL4-3-ΔEnv, pNL4-3-D116N-ΔEnv, pNL4-3-ΔEnv-ΔNef, or pNL4-3-D116N-ΔEnv-ΔNef) via Lipofectamine (Invitrogen). For infections of primary CD4+ T cells, virus was prepared by the same method using either pNL4-3 or pNL4-3-ΔNef.
All transfection supernatants were harvested at 72 h posttransfection (p.i.), clarified by centrifugation for 5 min at 470 × g, and passed through a 0.45-μm filter. Virus was treated with 50 U/ml benzonase (Sigma) at 37°C for 20 min to digest contaminating plasmid DNA (40) and then stored at −80°C until use.
Cell culture and viral infections.
Rev-CEM cells were chosen for our infections and were derived by transduction of the CEM-SS cell line (54). CEM-SS cells are a clone of virus “syncytium-sensitive” cells derived from CEM cells, also known as the CEM-CCRF cell line (33). The CEM cell line was itself derived from a patient with acute leukemia (16); as the HLA serological profile of the original CEM line is known, so too is that of the Rev-CEM cell line (IMGT/HLA database [www.ebi.ac.uk]), which expresses the HLA-A31 haplotype, recognized by CTLs.
Rev-CEM cells (54), obtained through the NIH AIDS Research and Reference Reagent Program (courtesy of Yuntao Wu and Jon Marsh), were maintained in RPMI 1640 medium (Invitrogen), and 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen), each supplemented with 10% fetal bovine serum, 1% l-glutamine, and 1% penicillin-streptomycin.
Rev-CEM cells (2.5 × 105) were infected with 250 ng p24 of virus in 24-well plates by spinoculation at 1,200 × g at 37°C for 2 h, followed by 1 h at 37°C, after which medium was replaced, resulting in a multiplicity of infection (MOI) of 0.1 for the wild-type (wt) virus as determined by green fluorescent protein (GFP) expression. Cells were infected with wt or Δnef virus, which was integrase competent or contained a defective D116N mutated integrase, respectively. In some cases, media were pretreated with 1 μM raltegravir (a gift of Merck Canada, Inc.) for 1 h prior to infection; after spinoculation, raltegravir-containing media were again used at a concentration of 1 μM.
For infections of primary CD4+ T cells, fresh peripheral blood mononuclear cells (PBMCs) were isolated from whole blood of individual donors by using the Ficoll-Hypaque method. PBMCs were cultured in RPMI 1640 and stimulated with 10 μg/ml phytohemagglutinin A (PHA) and 20 U/ml human interleukin-2 (IL-2) for 72 h. CD4+ T cells were then purified by negative selection using an Invitrogen CD3+ CD4+ Untouched kit.
Activated CD4+ T cells were pooled (2 × 105) and infected with 80 ng p24 of either NL4-3 or NL4-3-ΔNef virus via spinoculation at 1,500 × g at 37°C for 2 h in the presence of 8 μg/ml Polybrene, followed by incubation for 1 h at 37°C. Thereafter, cells were cultured with RPMI 1640 supplemented with 20 U/ml human IL-2. Some infections were incubated with 2 μM raltegravir for 1 h prior to infection, during infection, and after infection.
Total viral cDNA qPCR.
For both integrated viral DNA and total viral cDNA quantitative PCR (qPCR) assays, cellular DNA was extracted with a DNeasy blood and tissue kit (Qiagen). At 72 h p.i., PCR was performed with Platinum qPCR SuperMix-UDG (Invitrogen) on a Corbett Rotor-Gene 6000 thermocycler. Total viral cDNA was amplified and quantified as described previously (4).
Integrated DNA qPCR.
A previously described Alu-HIV PCR analysis was used (60), with the following modifications (12). The first-round reaction was performed on undiluted samples (100 ng template) and 1:10 dilutions of each sample (10 ng template diluted with uninfected DNA; 100 ng DNA total) in the presence of 2 mM MgCl2 and 200 μM deoxynucleoside triphosphates (dNTPs). A portion (9 μl) of the resulting first-round product was used as the template for the second-round nested reaction in the presence of 5 mM MgCl2 (final concentration including MgCl2 carryover from the first round) and 200 μM dNTPs, using the “wild-type” probe only. Second-round cycling conditions were 50°C for 2 min, 95°C for 1 min, and 45 cycles of 95°C for 15 s and 60°C for 30 s. Dually labeled probes were obtained from Biosearch Technologies (Novato, CA). To generate a standard curve for relative quantification of integrated DNA, Alu-gag PCR was first performed on a dilution series of DNA from infected Rev-CEM cells (diluted with DNA from uninfected cells).
Multiply spliced viral RNA transcript qRT-PCR.
Total cellular RNA was extracted from infected cells 72 h p.i. with an RNeasy kit (Qiagen). Quantitative reverse transcription-PCR (qRT-PCR) was performed using a one-step Superscript III Platinum Taq kit (Invitrogen) with primers MSJ3F, 5′-CAGACTCATCAAGCTTCTCTATCAA-3′ (nucleotides [nt] 6016 to 6041 of NL4-3), and MSJ3R, 5′-CTATTCCTTCGGGCCTGTC-3′ (nt 8368 to 8390 of NL4-3), in conjunction with the dually labeled probe 5′-FAM-AACCCACCTCCCAATCCCGAGG-BHQ-1-3′ (where FAM is 6-carboxyfluorescein and BHQ-1 is black hole quencher 1) (nt 8396 to 8415 of NL4-3) (Biosearch Technologies). A PCR amplicon was produced with the primers MSJ3F and MSJ3R but incorporated a 5′ overhang containing the T7 promoter on the forward primer. An in vitro RNA standard was transcribed from the PCR amplicon using a MegaScript kit (Ambion), and serial dilutions of the standard were prepared in uninfected-cell total RNA extract. The amount of RNA amplified per reaction condition was 250 ng, with 150 nm probe, 0.2 μM primers, and 2.0 mM MgSO4. Cycling conditions were 50°C for 15 min, 95°C for 8 min, and then cycling of 95°C for 15 s and 60°C for 30 s. Cellular GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA was additionally amplified as a loading control using the same reaction and cycling conditions, with the primers GAPDHF, 5′-AGGTCGGAGTCAACGGATTTGG-3′, and GAPDHR, 5′-GATGGCAACAATATCCACTTTACCA-3′, in conjunction with the GAPDH probe, 5′-FAM-TCTTATTGGGCGCCTGGTCAC-BHQ-1-3′.
Western blot analysis.
Cells were collected at 72 h p.i. and pelleted by low-speed centrifugation at 470 × g. The pellet was resuspended in RIPA buffer (0.15 M NaCl, 20 mM Tris, pH 7.4, 2 mM EDTA, 1% Triton X-100, and 1% deoxycholate). Cell lysates were normalized by the Bradford assay (Pierce) to 1 mg/ml total protein and resolved in a 12% SDS-polyacrylamide gel. The blot was incubated for 60 min with 1:4,000 polyclonal anti-HIV-1 Nef antibody obtained from the NIH AIDS Research and Reference Reagent Program (catalog number 2949) and anti-rabbit IgG-alkaline phosphotase conjugate (secondary antibody) (1:10,000). The chemiluminescent reagent West-Pico (Pierce) was used to develop the blots.
Cell surface HLA-ABC, HLA-A31, and HLA-E staining.
Cells that had been infected with virus or pseudovirus were stained at 72 h p.i. in phosphate-buffered saline (PBS) containing 3% fetal bovine serum and 0.1% sodium azide (FACs buffer) for 30 min at 4°C with one of the following mouse monoclonal antibodies (MAbs): phycoerythrin (PE)-conjugated anti-human HLA-A, HLA-B, and HLA-C allotype (HLA-ABC) MAb (clone DX17; BD Biosciences), unconjugated anti-human HLA-A31/30 (clone 4i103; Abcam), or PE-conjugated anti-human HLA-E (clone 3D12; eBioscience). Cells stained with unconjugated antibodies were washed twice in FACs buffer and then resuspended in FACs buffer containing PE-Cy5-conjugated monoclonal rat anti-mouse IgM (clone II/41; eBioscience). Cells were then fixed in a final concentration of 1% paraformaldehyde and resuspended in FACs buffer. A total of 10,000 to 20,000 cell events were assayed on a FACSCalibur instrument (BD PharMingen); analysis was performed with BD CellQuest Pro 4.0.2 (BD PharMingen) and FCS Express 3 (DeNovo) software. Levels of receptors were quantified relative to those found after infection by Δnef virus. These studies were controlled by subtracting background isotype fluorescence or secondary antibody fluorescence values from antibody receptor fluorescence measurements.
Statistical analysis.
All statistical analyses were performed with GraphPad Prism 4.0 software. To test for statistically significant differences between pairs, unpaired two-tailed t tests were performed, with confidence intervals set at 95%. To test for significant differences between treatment groups, one-way analysis of variance (ANOVA) was performed, with confidence intervals set at 95%.
RESULTS
Nef is expressed in the absence of integration.
We first sought to confirm that Nef is expressed from unintegrated DNA. Rev-CEM cells were infected with VSV-G pseudotyped NL4-3 virus bearing either wt integrase or the integrase-defective mutation D116N, in conjunction with either wt nef or a Δnef mutation. For some infections with wt integrase coding sequences, 1 μM integrase inhibitor raltegravir was added.
Analysis of late reverse transcripts (total viral cDNA) by qPCR revealed that there were no significant differences between the capacities of any of the infecting viruses to reverse transcribe the genome, regardless of ability to integrate (P = 0.38) (Fig. 1 a), in accord with previous findings (4, 21).
FIG. 1.
Transcription from nonintegrated HIV-1 cDNA templates results in expression of Nef. Late reverse transcription DNA products (total viral cDNA) (a), integrated viral DNA (b), and multiply spliced viral RNA (c) were analyzed by qPCR 72 h p.i. The presence of Nef was assayed by Western blot analysis (d). Error bars show standard deviations (SD). Results for panels a to c are derived from 3 independent experiments, and panel d shows representative data. RAL, raltegravir.
Integration was then assayed with an Alu-HIV qPCR for integrated provirus. This assay confirmed that virus bearing the integrase mutation D116N did not integrate, as previously described (14). Nor was integration detectable with wt-integrase-bearing virus in the presence of 1 μM raltegravir. The presence or absence of an intact Nef coding sequence had no influence on the extent of integration observed (Fig. 1b).
We next used qRT-PCR to assay the capacity of our wt or nef-deleted integrating and nonintegrating viruses to produce multiply spliced viral RNA transcripts. The design of our primers should allow amplification of all classes of multiply spliced viral RNA transcripts, including those which code for Nef (38). Our analysis found that there were no significant differences between any of the viruses (P = 0.43) (Fig. 1c). Thus, the levels of multiply spliced transcripts produced in the absence of integration are generally equivalent to those produced following integration.
Western blot analysis next confirmed that the introduction of stop codons into the first and third codons of nef was sufficient to knock out Nef expression. For both the D116N mutated integrase virus and the wt integrase virus, in the presence of inhibitory concentrations of raltegravir, Nef expression was readily detectable despite an absence of measurable integration as detected by Alu-HIV qPCR (Fig. 1d).
HLA-ABC is downregulated by Nef in the absence of integration.
We next sought to determine whether Nef expressed in the absence of integration was sufficient to reduce overall cell surface MHC-I levels. Rev-CEM cells were again infected with VSV-G pseudotyped virus, bearing either wt integrase or D116N mutated integrase, either with or without nef. For some infections with wt integrase, 1 μM raltegravir was added.
Rev-CEM cells have previously been transduced with the vector pNL-GFP-RRE-SA; this construct contains HIV-1 LTRs, a Rev response element (RRE), and a GFP gene, in conjunction with various splice donor and acceptor sites (55). In the presence of Tat, transcription from the LTR is transactivated, but in the absence of Rev, the GFP coding sequence is spliced out of the transcript. However, the GFP coding sequence is retained in the presence of both Tat and Rev, and GFP is expressed in the cell. In our experiments, infected Rev-CEM cells were detectable as GFP positive by flow cytometry, even when integration was not detectable by qPCR. This was because there was sufficient translation of Tat and Rev to induce GFP expression. The GFP-positive infected cells were stained with an antibody capable of detecting the HLA-A, HLA-B, and HLA-C allotypes (i.e., HLA-ABC).
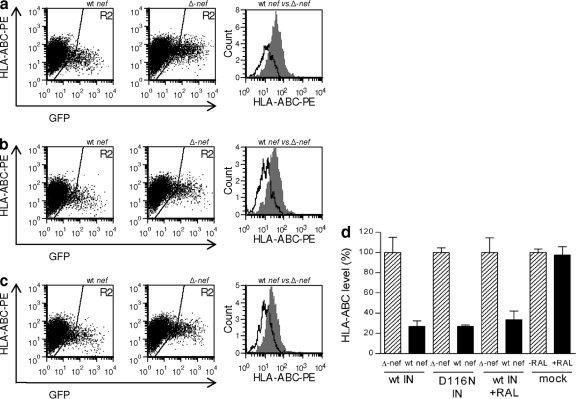
For wt integrating virus, Nef mediated downregulation of HLA-ABC to 27% of the levels seen with Δnef virus (P < 0.0001) (Fig. 2 a and d). When integration was blocked either through the integrase D116N mutation or through the use of raltegravir, the extents of Nef-mediated downregulation were similar, i.e., to 27% and 34%, respectively (P < 0.0001 and P = 0.0002) (Fig. 2b, c, and d). An analysis of the viruses bearing wild-type nef genes found no significant differences between them (P = 0.16); hence, the ability of Nef to downregulate HLA-ABC was not mediated by the capacity of the virus to integrate (Fig. 2d).
FIG. 2.
Nef-mediated downregulation of HLA-ABC by nonintegrating virus. Analysis of infected GFP-positive cells (gate R2) for cell surface HLA-ABC expression. Representative results for wt-integrase-bearing virus (a). Virus with wt nef downregulates HLA-ABC to a greater degree than Δnef virus. The results are overlaid in the histogram; wt virus is displayed in white and Δnef virus in shaded gray. A similar pattern of results is seen for virus bearing the integrase-deficient D116N mutation (b) or with virus bearing wt integrase but in the presence of inhibitory concentrations of raltegravir (c). The bar chart shows combined results of 3 to 5 independent experiments with duplicate infections with wt or defective D116N integrase (IN) with raltegravir (RAL) (d). Geometric means of fluorescence for each receptor are expressed relative to the level after infection by the Δnef virus (100%). Error bars indicate standard deviations (SD).
We were also able to demonstrate that raltegravir itself did not affect expression of the epitopes recognized by the HLA-ABC antibody, as both untreated and treated cells had similar HLA-ABC expression levels (P > 0.5) (Fig. 2d).
HLA-A is downregulated by Nef in the absence of integration.
In productive HIV-1 infections, Nef selectively downregulates the HLA class I allotypes recognized by cytotoxic T cells (HLA-A and HLA-B), while leaving those recognized by NK cells (HLA-C and HLA-E) unperturbed on the cell surface (10). Thus, our data for HLA-ABC were a compound result of Nef-mediated reductions of cell surface HLA-A and HLA-B, but with unaffected levels of HLA-C.
To more specifically measure Nef-mediated downregulation of HLA allotypes recognized by CTLs, we assayed HLA-A31 cell surface expression levels in Rev-CEM cells following infection with integrating and nonintegrating pseudovirus.
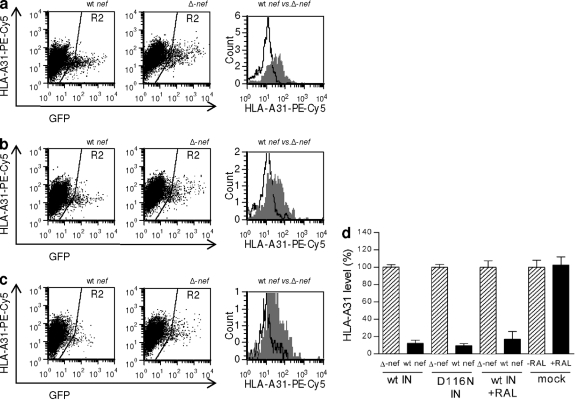
With integrating infections, we observed that Nef mediated a reduction in levels of HLA-A31 to about 12% of the usual cell surface expression levels (P < 0.0001), relative to those seen with Δnef virus (Fig. 3 a and d). When integration was prevented, either through the integrase D116N mutation or through the use of raltegravir, the same degree of downregulation was observed, i.e., cell surface expression of HLA-A31 was reduced to 9% (P < 0.0001) or 17% (P < 0.0001), respectively (Fig. 3b, c, and d). We then compared levels of HLA-A31 following infection by all wt viruses and were unable to show any significant differences among them (P > 0.5). Thus, the capacity to reduce cell surface HLA-A31 via Nef is not linked to the ability of the infecting virus to integrate.
FIG. 3.
Nef-mediated downregulation of HLA-A31 by nonintegrated virus. Analysis of infected GFP-positive cells (gate R2) for cell surface HLA-A31 expression. Representative results for wt-integrase-bearing virus (a). Virus with wild-type nef downregulates HLA-A31 to a greater degree than Δnef virus. The results are overlaid in the histogram; wt virus is displayed in white and Δnef virus in shaded gray. A similar pattern of results is seen for virus bearing the integrase-deficient D116N mutation (b) or with virus bearing wt integrase but in the presence of inhibitory concentrations of raltegravir (c). The bar chart shows combined results of 3 independent experiments with duplicate infections with wt or defective D116N integrase (IN) with raltegravir (RAL) (d). Geometric means of fluorescence for each receptor are expressed relative to the level after infection by the Δnef virus or that for a no-drug control (100%). Error bars indicate standard deviations (SD).
As with HLA-ABC, we saw no evidence of altered HLA-A31 expression levels due to the influence of raltegravir (P > 0.5) (Fig. 3d).
HLA-E is unaffected by Nef irrespective of viral integration.
We next examined the specificity of our findings by assaying cell surface HLA-E levels, which can affect NK cell recognition of target cells (10, 46). HLA-E is ubiquitously expressed on B cells and T cells, as well as placental cells and trophoblasts, at low levels (48), and we were also able to identify it at low levels in the Rev-CEM cell line.
We found that neither integrating infections with wt virus nor nonintegrating infections that had been blocked by defective integrase or raltegravir resulted in any measurable Nef-mediated downregulation of cell surface HLA-E levels in Rev-CEM cells (P > 0.5) (Fig. 4). Following infection by all viruses studied, HLA-E levels were commonly around 2.25-fold higher than in uninfected controls, irrespective of the status of nef or integrase or the use of raltegravir (Fig. 4).
FIG. 4.
Lack of Nef-mediated downregulation of HLA-E by both integrating and nonintegrating virus. Analysis of infected GFP-positive cells (gate R2) for cell surface HLA-E expression. Representative results for wt-integrase-bearing virus (a). Neither wt virus nor Δnef virus downregulates HLA-E. The results are overlaid in the histogram; wt virus is displayed in white and Δnef virus in shaded gray. The same pattern of results is seen for virus bearing the integrase-deficient D116N mutation (b) or with virus bearing wt integrase but in the presence of inhibitory concentrations of raltegravir (c). The bar chart shows combined results of 3 independent experiments with duplicate infections with wt or defective D116N integrase (IN) with raltegravir (RAL) (d). Geometric means of fluorescence for each receptor are expressed relative to the level after infection by the Δnef virus (100%). Error bars indicate standard deviations (SD).
HLA-ABC is downregulated by Nef in primary CD4+ T cells in the absence of integration.
Given our findings with the Rev-CEM GFP cell line, we next wished to examine the effect of Nef on total MHC-I (HLA-ABC) cell surface expression in the absence of integration in a natural infection model using primary CD4+ T cells infected with replication-competent NL4-3 virus.
Our initial attempts to use a pNL4-3-derived GFP reporter virus to specifically measure HLA-ABC expression on infected cells were unsuccessful (data not shown). Infections with pNL4-3-ΔE-eGFP-derived virus (61), which encodes enhanced GFP (eGFP) in place of Env, and infections using virus derived from the pBRNL4-3-nef-IRES-eGFP construct (43), which encodes eGFP under an internal ribosome entry site (IRES) downstream from nef, both resulted in cells infected with nonintegrating virus being nondetectable by eGFP, despite the fact that integrating virus commonly leads to detection levels of about 12% in our system.
We therefore performed our analysis using nonfluorescent wt NL4-3 virus and analyzed HLA-ABC expression on the total cell population rather than only on infected cells. We controlled the capacity of the virus to integrate through the use of 2 μM raltegravir, a concentration that we showed can block integration to below levels of detection by Alu-LTR qPCR. Thus, we were able to compare the capacities of Nef to mediate downregulation of cell surface HLA-ABC both in the presence and in the absence of integration.
We found that the nef-bearing wt virus could mediate an 18% reduction in HLA-ABC expression relative to the level for the Δnef virus (P = 0.0086) (Fig. 5). In parallel infections in which integration was blocked with raltegravir, we again found evidence of cell surface HLA-ABC downregulation; a reduction in expression of 26% for wt virus relative to that for Δnef virus (P = 0.0113) was observed (Fig. 5).
FIG. 5.

Nef-mediated downregulation of HLA-ABC in activated primary CD4+ T cells by nonintegrated virus. Analysis of infected live cells for cell surface HLA-ABC expression. Representative histogram plot for wt-integrase-bearing virus (white) versus Δnef virus (shaded gray) (a). Virus with wt nef downregulates HLA-ABC to a greater extent than Δnef virus. Representative histogram plot for wt-integrase-bearing virus (white) versus Δnef virus (shaded gray) in the presence of 2 μM raltegravir (b). Virus with wild-type nef downregulates HLA-ABC to a greater extent than Δnef virus. Combined results of two independent experiments of duplicate infections with wt integrase virus in the absence of drug (wt IN) or in the presence of 2 μM raltegravir (wt IN + RAL) (c). Geometric means of fluorescence are expressed relative to the level after infection by the Δnef virus (100%). Error bars indicate standard deviations (SD). Values for wt and Δnef viruses were tested for statistical significance using an unpaired two-tailed t test (*; P < 0.05; **, P < 0.01).
As these findings are based on HLA-ABC analysis of the bulk population, they cannot be directly compared to our findings in the Rev-CEM cell line. Additionally, infection with Δnef virus typically results in infection levels that are reduced compared to those for wt virus (19). This was also observed when we infected cells with either wt or Δnef pBRNL4-3-nef-IRES-eGFP-derived virus, as we detected infection levels of 12% and 5%, respectively, when assaying for eGFP expression. Given these expected differences in infection rates, the 18% and 26% downregulation levels that we observed for integrating and nonintegrating virus, based on a comparison of wt nef to Δnef virus, are likely an underestimate. Our results also demonstrate that Nef-mediated HLA-ABC downregulation by nonintegrating virus in primary CD4+ T cells is readily detectable and occurs to a degree similar to that for integrated virus.
DISCUSSION
It is evident that transcription and translation of HIV-1 genes prior to integration are common features of viral replication (8, 57). Studies of transcription prior to integration in resting T cells demonstrated an advantage to the virus, as nef and tat translation increased the activation state of the infected cell, which was thereby more likely to yield a productive infection (58). Equally, translation of nef prior to integration is particularly apparent in macrophages (25). Studies of transcription from preintegrated cDNA can be modeled by blocking integration, either through the use of defective integrase or by integrase strand transfer inhibitors, as the patterns of transcription and translation observed in these contexts are similar to those seen in studies of transcription prior to integration (21, 56, 58). Such studies demonstrated that preintegration translation of nef variously downregulates CD4, CXCR4, and CCR5 (18, 45), which may serve to limit signal transduction through these receptors (59) and possibly limit viral superinfection (31, 45, 49, 50). Similar studies have also demonstrated that preintegration translation of rev might limit superinfection of cells at the level of integration (29, 30). We therefore suspected that transcription of preintegrated cDNA could also result in modulation of cell surface MHC-I expression by Nef.
The experiments presented herein confirm our hypothesis and demonstrate that transcription from unintegrated HIV-1 can selectively downregulate HLA-ABC and HLA-A31 in the absence of any effect on HLA-E expression. In every instance, both for the Rev-CEM cell line and for primary CD4+ T cells, the extent of downregulation observed was independent of the capacity of the virus to integrate its genome into the host cell. Surprisingly, we found that in Rev-CEM cells the levels of multiply spliced mRNA transcripts were equivalent either with or without integration. Though the transcription levels described are supportive of our findings for MHC-I expression, we were surprised that our attempts to analyze cells expressing eGFP via unintegrated eGFP reporter virus BRNL4-3-nef-IRES-eGFP or NL4-3-ΔE-eGFP (both for primary cells and for cell lines) were unsuccessful. Although levels of viral multiply spliced RNA transcripts were equivalent for both integrated and nonintegrated virus, it is clear, given the paucity of eGFP expression from the reporter viruses, that there are fundamental differences in transcription from these reporter viruses in the presence and absence of integration.
Our data for HLA-A31 downregulation in Rev-CEM cells are consistent with levels of downregulation mediated by Nef for the HLA-A allotype in T cell lines, though more extensive downregulation is typically seen in primary CD4+ T cells (24). The results demonstrate that transcription from preintegrated virus both in T cell lines and in activated primary CD4+ T cells yields fully functional levels of Nef that are capable of mediating MHC-I modulation. Given that the pattern of MHC-I modulation seen with integrating virus is already known to protect from CTLs (11) and that the same pattern is produced with nonintegrating virus, we conjecture that cells bearing unintegrated cDNA should be equally protected from CTL responses. We propose that synthesis of Nef prior to integration leads to comprehensive and selective HLA-A and HLA-B downregulation and subsequent CD8+ CTL evasion, without affecting NK cell-mediated lysis, since levels of HLA-C and HLA-E cell surface expression are not affected. The fact that HLA-E levels were elevated after infection, independent of Nef, is consistent with observations that an epitope in p24 can lead to an upregulation of HLA-E cell surface levels (34).
During acute infection, the viral load usually diminishes, concomitant with the emergence of HIV-specific CTLs (6). Typically, CTL levels remain high throughout untreated HIV infection (1) and yet can decrease following administration of therapy that leads to a reduced viral load (35). Downmodulation of MHC-I is thought to be an important feature of CTL evasion by HIV, as individuals infected with Nef-defective HIV-1 show remarkably potent CTL responses (13). Equally, work with macaques infected by simian immunodeficiency virus SIVmac showed that introduction of a point mutation in Nef that disrupted MHC-I downmodulation quickly reverted within 4 weeks (32). These findings suggest that there is a strong selective pressure for HIV to initiate Nef-mediated immune evasion. We now show that this can occur even prior to attainment of complete HIV integration.
For activated CD4+ T cells, MHC-I downregulation as a result of preintegration transcription might occur several hours prior to integration, giving the virus added time to evade immune responsiveness. For infections of resting T cells, which have slower viral replication kinetics than activated T cells, there might be a delay of 2 to 3 days prior to integration during which preintegration transcription can occur, representing a further advantage for the virus (58). An even longer delay could apply to macrophages, e.g., up to 30 days (25). However, this depends on the same pattern of MHC-I modulation occurring in both resting T cells and macrophages, as we have shown here for activated primary CD4+ T cells.
Nef produced from preintegrated DNA seems to be fully functional in terms of MHC-I modulation, and it is likely that other aspects of Nef functions are also intact. Previous studies showed that preintegration synthesis of Nef can modulate CD4, CXCR4, and CCR5 expression as well as T-cell activation (8, 45, 58). In SIV, downregulation of the T-cell receptors (TCR) CD3 and CD28 is considered a critical function for most nef alleles (42); such downregulation allows SIV to interfere with the formation of the immunological synapses between infected and antigen-presenting cells, thereby limiting chronic immune activation (3, 27). Thus, transcription of nef prior to integration may also be beneficial for this aspect of immune control in SIV disease.
Given the clear benefit of avoiding CTL- and NK cell-mediated lysis, transcription of nef from preintegrated cDNA might be a key mechanism in immune evasion following HIV infection of a host cell.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) and Merck Canada, Inc. R.D.S. is the recipient of a postdoctoral fellowship jointly funded by the CIHR Canadian HIV Trials Network (CTN) and the Canadian Foundation for AIDS Research (CANFAR). D.A.D. is the recipient of a predoctoral fellowship from CIHR.
We thank Daria Hazuda of Merck, Inc., for helpful comments. We also thank Yuntao Wu and Jon Marsh for provision of the Rev-CEM cell line, Robert Siliciano for provision of the pNL4-3-ΔE-eGFP construct, and Jan Münch, Michael Schindler, and Frank Kirchhoff for provision of the pBRNL4-3-nef-IRES-eGFP construct, all via the NIH AIDS Research and Reference Reagent Program. We also thank Cesar Collazos, Susan Colby-Germinario, and Maureen Oliviera of the McGill AIDS Centre and Christian Young of the Lady Davis Institute flow cytometry core facilities for valuable technical assistance.
Footnotes
Published ahead of print on 5 January 2011.
REFERENCES
- 1.Altman, J., et al. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science 274:94-96. [PubMed] [Google Scholar]
- 2.Ansari-Lari, M., L. Donehower, and R. Gibbs. 1995. Analysis of human immunodeficiency virus type 1 integrase mutants. Virology 211:332-335. [DOI] [PubMed] [Google Scholar]
- 3.Arhel, N., et al. 2009. The inability to disrupt the immunological synapse between infected human T cells and APCs distinguishes HIV-1 from most other primate lentiviruses. J. Clin. Invest. 119:2965-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bar-Magen, T., et al. 2010. Identification of novel mutations responsible for resistance to MK-2048, a second-generation HIV-1 integrase inhibitor. J. Virol. 84:9210-9216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berke, G. 1995. The CTL's kiss of death. Cell 81:9-12. [DOI] [PubMed] [Google Scholar]
- 6.Borrow, P., H. Lewicki, B. Hahn, G. Shaw, and M. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Brussel, A., and P. Sonigo. 2004. Evidence for gene expression by unintegrated human immunodeficiency virus type 1 DNA species. J. Virol. 78:11263-11271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cara, A., and M. Klotman. 2006. Retroviral E-DNA: persistence and gene expression in nondividing immune cells. J. Leukoc. Biol. 80:1013-1017. [DOI] [PubMed] [Google Scholar]
- 9.Cocchi, F., et al. 1995. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811-1815. [DOI] [PubMed] [Google Scholar]
- 10.Cohen, G., et al. 1999. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10:661-671. [DOI] [PubMed] [Google Scholar]
- 11.Collins, K., B. Chen, S. Kalams, B. Walker, and D. Baltimore. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397-401. [DOI] [PubMed] [Google Scholar]
- 12.Donahue, D., et al. 2010. Stage-dependent inhibition of HIV-1 replication by antiretroviral drugs in cell culture. Antimicrob. Agents Chemother. 54:1047-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dyer, W., et al. 1999. Strong human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte activity in Sydney Blood Bank Cohort patients infected with nef-defective HIV type 1. J. Virol. 73:436-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman, A., G. Englund, J. Orenstein, M. Martin, and R. Craigie. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farnet, C., and W. Haseltine. 1991. Circularization of human immunodeficiency virus type 1 DNA in vitro. J. Virol. 65:6942-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foley, G., et al. 1965. Continuous culture of human lymphoblasts from peripheral blood of a child with acute leukemia. Cancer 18:522-529. [DOI] [PubMed] [Google Scholar]
- 17.Gelderblom, H., et al. 2008. Viral complementation allows HIV-1 replication without integration. Retrovirology 5:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillim-Ross, L., A. Cara, and M. Klotman. 2005. Nef expressed from human immunodeficiency virus type 1 extrachromosomal DNA downregulates CD4 on primary CD4+ T lymphocytes: implications for integrase inhibitors. J. Gen. Virol. 86:765-771. [DOI] [PubMed] [Google Scholar]
- 19.Goldsmith, M. A., M. T. Warmerdam, R. E. Atchison, M. D. Miller, and W. C. Greene. 1995. Dissociation of the CD4 downregulation and viral infectivity enhancement functions of human immunodeficiency virus type 1 Nef. J. Virol. 69:4112-4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen, T., and M. Bouvier. 2009. MHC class I antigen presentation: learning from viral evasion strategies. Nat. Rev. Immunol. 9:503-513. [DOI] [PubMed] [Google Scholar]
- 21.Hazuda, D., et al. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 22.Iyer, S., D. Yu, A. Biancotto, L. Margolis, and Y. Wu. 2009. Measurement of human immunodeficiency virus type 1 preintegration transcription by using Rev-dependent Rev-CEM cells reveals a sizable transcribing DNA population comparable to that from proviral templates. J. Virol. 83:8662-8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kantor, B., H. Ma, J. Webster-Cyriaque, P. Monahan, and T. Kafri. 2009. Epigenetic activation of unintegrated HIV-1 genomes by gut-associated short chain fatty acids and its implications for HIV infection. Proc. Natl. Acad. Sci. U. S. A. 106:18786-18791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasper, M., and K. Collins. 2003. Nef-mediated disruption of HLA-A2 transport to the cell surface in T cells. J. Virol. 77:3041-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly, J., et al. 2008. Human macrophages support persistent transcription from unintegrated HIV-1 DNA. Virology 372:300-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khiytani, D., and N. Dimmock. 2002. Characterization of a human immunodeficiency virus type 1 pre-integration complex in which the majority of the cDNA is resistant to DNase I digestion. J. Gen. Virol. 83:2523-2532. [DOI] [PubMed] [Google Scholar]
- 27.Kirchhoff, F. 2009. Is the high virulence of HIV-1 an unfortunate coincidence of primate lentiviral evolution? Nat. Rev. Microbiol. 7:467-476. [DOI] [PubMed] [Google Scholar]
- 28.Le Gall, S., et al. 2000. Distinct trafficking pathways mediate Nef-induced and clathrin-dependent major histocompatibility complex class I down-regulation. J. Virol. 74:9256-9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levin, A., et al. 2010. A novel role for the viral Rev protein in promoting resistance to superinfection by human immunodeficiency virus type 1. J. Gen. Virol. 91:1503-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levin, A., J. Rosenbluh, Z. Hayouka, A. Friedler, and A. Loyter. 2010. Integration of HIV-1 DNA is regulated by interplay between viral rev and cellular LEDGF/p75 proteins. Mol. Med. 16:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michel, N., I. Allespach, S. Venzke, O. Fackler, and O. Keppler. 2005. The Nef protein of human immunodeficiency virus establishes superinfection immunity by a dual strategy to downregulate cell-surface CCR5 and CD4. Curr. Biol. 15:714-723. [DOI] [PubMed] [Google Scholar]
- 32.Münch, J., N. Stolte, D. Fuchs, C. Stahl-Hennig, and F. Kirchhoff. 2001. Efficient class I major histocompatibility complex down-regulation by simian immunodeficiency virus Nef is associated with a strong selective advantage in infected rhesus macaques. J. Virol. 75:10532-10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nara, P., et al. 1987. Simple, rapid, quantitative, syncytium-forming microassay for the detection of human immunodeficiency virus neutralizing antibody. AIDS Res. Hum. Retroviruses 3:283-302. [DOI] [PubMed] [Google Scholar]
- 34.Nattermann, J., et al. 2005. HIV-1 infection leads to increased HLA-E expression resulting in impaired function of natural killer cells. Antivir. Ther. 10:95-107. [DOI] [PubMed] [Google Scholar]
- 35.Ogg, G., et al. 1999. Longitudinal phenotypic analysis of human immunodeficiency virus type 1-specific cytotoxic T lymphocytes: correlation with disease progression. J. Virol. 73:9153-9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poon, B., M. Chang, and I. Chen. 2007. Vpr is required for efficient Nef expression from unintegrated human immunodeficiency virus type 1 DNA. J. Virol. 81:10515-10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poon, B., and I. Chen. 2003. Human immunodeficiency virus type 1 (HIV-1) Vpr enhances expression from unintegrated HIV-1 DNA. J. Virol. 77:3962-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purcell, D., and M. Martin. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 67:6365-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quan, Y., C. Liang, B. Brenner, and M. Wainberg. 2009. Multidrug-resistant variants of HIV type 1 (HIV-1) can exist in cells as defective quasispecies and be rescued by superinfection with other defective HIV-1 variants. J. Infect. Dis. 200:1479-1483. [DOI] [PubMed] [Google Scholar]
- 40.Sastry, L., Y. Xu, R. Cooper, K. Pollok, and K. Cornetta. 2004. Evaluation of plasmid DNA removal from lentiviral vectors by benzonase treatment. Hum. Gene Ther. 15:221-226. [DOI] [PubMed] [Google Scholar]
- 41.Schaefer, M., E. Wonderlich, J. Roeth, J. Leonard, and K. Collins. 2008. HIV-1 Nef targets MHC-I and CD4 for degradation via a final common beta-COP-dependent pathway in T cells. PLoS Pathog. 4:e1000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schindler, M., et al. 2006. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 125:1055-1067. [DOI] [PubMed] [Google Scholar]
- 43.Schindler, M., et al. 2003. Down-modulation of mature major histocompatibility complex class II and up-regulation of invariant chain cell surface expression are well-conserved functions of human and simian immunodeficiency virus nef alleles. J. Virol. 77:10548-10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartz, O., V. Maréchal, S. Le Gall, F. Lemonnier, and J. Heard. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 2:338-342. [DOI] [PubMed] [Google Scholar]
- 45.Sloan, R., D. Donahue, B. Kuhl, T. Bar-Magen, and M. Wainberg. 2010. Expression of Nef from unintegrated HIV-1 DNA downregulates cell surface CXCR4 and CCR5 on T-lymphocytes. Retrovirology 7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevenson, M., et al. 1990. Integration is not necessary for expression of human immunodeficiency virus type 1 protein products. J. Virol. 64:2421-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swann, S., et al. 2001. HIV-1 Nef blocks transport of MHC class I molecules to the cell surface via a PI 3-kinase-dependent pathway. Virology 282:267-277. [DOI] [PubMed] [Google Scholar]
- 48.Tripathi, P., and S. Agrawal. 2007. The role of human leukocyte antigen E and G in HIV infection. AIDS 21:1395-1404. [DOI] [PubMed] [Google Scholar]
- 49.Venzke, S., N. Michel, I. Allespach, O. Fackler, and O. Keppler. 2006. Expression of Nef downregulates CXCR4, the major coreceptor of human immunodeficiency virus, from the surfaces of target cells and thereby enhances resistance to superinfection. J. Virol. 80:11141-11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wildum, S., M. Schindler, J. Münch, and F. Kirchhoff. 2006. Contribution of Vpu, Env, and Nef to CD4 down-modulation and resistance of human immunodeficiency virus type 1-infected T cells to superinfection. J. Virol. 80:8047-8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams, M., et al. 2002. Direct binding of human immunodeficiency virus type 1 Nef to the major histocompatibility complex class I (MHC-I) cytoplasmic tail disrupts MHC-I trafficking. J. Virol. 76:12173-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiskerchen, M., and M. Muesing. 1995. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 69:376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, Y. 2008. The second chance story of HIV-1 DNA: unintegrated? Not a problem! Retrovirology 5:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu, Y., M. Beddall, and J. Marsh. 2007. Rev-dependent indicator T cell line. Curr. HIV Res. 5:394-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, Y., M. Beddall, and J. Marsh. 2007. Rev-dependent lentiviral expression vector. Retrovirology 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, Y., and J. Marsh. 2003. Early transcription from nonintegrated DNA in human immunodeficiency virus infection. J. Virol. 77:10376-10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, Y., and J. Marsh. 2003. Gene transcription in HIV infection. Microbes Infect. 5:1023-1027. [DOI] [PubMed] [Google Scholar]
- 58.Wu, Y., and J. Marsh. 2001. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science 293:1503-1506. [DOI] [PubMed] [Google Scholar]
- 59.Wu, Y., and A. Yoder. 2009. Chemokine coreceptor signaling in HIV-1 infection and pathogenesis. PLoS Pathog. 5:e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu, J., et al. 2008. A more precise HIV integration assay designed to detect small differences finds lower levels of integrated DNA in HAART treated patients. Virology 379:78-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, H., et al. 2004. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J. Virol. 78:1718-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]