Abstract
The use of animal models of human cytomegalovirus (HCMV) infection is critical to refine HCMV vaccine candidates. Previous reports have demonstrated that immunization of rhesus monkeys against rhesus cytomegalovirus (RhCMV) can reduce both local and systemic replication of RhCMV following experimental RhCMV challenge. These studies used prime/boost combinations of DNA expression plasmids alone or DNA priming and boosting with either inactivated virion particles or modified vaccinia virus Ankara (MVA) expressing the same antigens. Viral outcomes included reduced RhCMV replication at the site of subcutaneous inoculation and RhCMV viremia following intravenous inoculation. Since shedding of cytomegalovirus from mucosal surfaces is critical for horizontal transmission of the virus, DNA priming/MVA boosting was evaluated for the ability to reduce oral shedding of RhCMV following subcutaneous challenge. Of six rhesus monkeys vaccinated exclusively against RhCMV glycoprotein B (gB), phosphoprotein 65 (pp65), and immediate-early 1 (IE1), half showed viral loads in saliva that were lower than those of control monkeys by 1 to 3 orders of magnitude. Further, there was a strong association of memory pp65 T cell responses postchallenge in animals exhibiting the greatest reduction in oral shedding. These results highlight the fact that a DNA/MVA vaccination regimen can achieve a notable reduction in a critical parameter of viral replication postchallenge. The recently completed clinical trial of a gB subunit vaccine in which the rate of HCMV infection was reduced by 50% in the individuals receiving the vaccine is consistent with the results of this study suggesting that additional immunogens are likely essential for maximum protection in an outbred human population.
The nearly 40-year quest for a vaccine that confers protective efficacy against congenital infection with human cytomegalovirus (HCMV) remains unmet, although considerable progress has been made. Complexities in HCMV's natural history, incompletely defined correlates of immune protection, and financial and logistical factors in designing sufficiently powered clinical trials all contribute to the absence of a licensed HCMV vaccine(s). Animal model studies with rhesus monkey, mouse, and guinea pig systems have demonstrated that multiple vaccine strategies, including approaches based on those proposed for HCMV, are effective at limiting the extent of challenge virus replication. Immunization of HCMV-negative women with recombinant gB has been clinically evaluated (50, 51). A recently completed phase II trial assessed the efficacy of the vaccine to decrease cases of maternal HCMV infection (34). The endpoint of this study was the time to HCMV infection (50, 51), and the trial ended earlier than planned because vaccine efficacy exceeded goals. The results offer strong encouragement that vaccination regimes directed at prominent neutralizing epitopes can significantly decrease the rate of primary infection in HCMV-negative women. The impressive, but less than 100%, level of protection observed in this clinical study further indicates that augmentation of the gB-based vaccine is required to achieve universal protection. The specific attributes of a putative vaccine augmentation are speculative, but animal models can serve an essential role in identifying promising modalities that can be translated into clinical trials.
Debate about the possible constituents of an HCMV vaccine includes whether a vaccine should target only the prominent neutralizing antibody (NAb) target, gB, or whether both cellular and NAb viral immunogens should be part of a vaccine cocktail. The current study was undertaken to test whether there are significant differences between a vaccine directed against just rhesus CMV (RhCMV) gB, which is the predominant but not exclusive target of NAb in RhCMV-infected rhesus monkeys (49), and a vaccine directed against both gB and two RhCMV proteins bearing epitopes recognized in the context of cell-mediated immunity (CMI), pp65, and immediate-early 1 (IE1) (47). The rationale is based on the premise that, once RhCMV infection becomes systemic, immune responses beyond those that are solely humoral will be required for clearance before latency develops. To increase the biological relevance of our assessments of protective efficacy, an epithelial cell-tropic variant of RhCMV (UCD52) was used to as a challenge virus for the different treatment groups. Previous work has demonstrated that RhCMV UCD52 (i) is pathogenic in fetal rhesus monkeys following direct fetal inoculation (43), (ii) contains a full-length UL/b′ region (GenBank accession number GU552456), (iii) is epithelial cell tropic (Y. Yue and P. Barry, unpublished data), and (iv) is persistently shed in bodily fluids following subcutaneous inoculation (unpublished observations). The results of the current study demonstrate that while both gB- and gB-pp65-IE1-based vaccines significantly reduced RhCMV copy numbers in plasma following challenge, only the combined gB-pp65-IE1 vaccine profoundly limited oral shedding in 50% of the vaccinated monkeys. This study shows for the first time that vaccination can dramatically limit an important component of RhCMV's natural history, i.e., shedding of virus in the oral cavity. The absence of protection in 100% of the vaccinated animals highlights the finding that additional vaccine immunogens are likely required to achieve greater protective efficacy (11, 34, 37, 38). The results also emphasize the importance of the availability of a cohort of non-RhCMV-infected rhesus macaques to enable further studies with this nonhuman primate model of HCMV persistence and pathogenesis (7).
MATERIALS AND METHODS
Animals.
Healthy, genetically outbred rhesus macaques (Macaca mulatta) (n = 21; 13 females, 8 males) from the California National Primate Research Center, repeatedly confirmed by serology to be uninfected with RhCMV, were used for these studies. Their ages ranged from ∼2 years 11 months (n = 20) to 4 years 10 months (n = 1). All animals were housed together, away from any RhCMV-infected monkeys, in a single outdoor housing unit during immunization and after RhCMV challenge. The Institutional Animal Care and Use Committee of the University of California, Davis (UC Davis), which is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, approved all animal protocols in advance of any procedures.
Immunizations.
Animals were divided into four vaccine treatment groups as shown in Table 1: group 1 (n = 6), mock-immunized controls; group 2 (n = 6), animals immunized against RhCMV gB; group 3 (n = 6), animals immunized against RhCMV gB, pp65-2, and IE1; and group 4 (n = 3), animals immunized against RhCMV gB, pp65-2, and IE1 and boosted with formalin-inactivated RhCMV (FI-RhCMV) virions. Although group 4 was comprised of a smaller number of monkeys than the other groups (n = 3 versus n = 6), the goal was to provide a comparison of viral outcomes after challenge in this study with our previous study using RhCMV 68-1 as the challenge virus (2). The backbone pND expression vector and the expression plasmids for RhCMV pp65-2 (pND/65-2), the transmembrane deletion version of gB (pND/gBΔTM), and RhCMV IE1 (exons 2 to 4) have been described previously (2, 27, 46, 49). All expression plasmids were purified from bacteria using Endo-free plasmid extraction kits (Qiagen). DNA was resuspended in phosphate-buffered saline (PBS) buffer (Invitrogen) at a concentration of 1 mg/ml and stored at −20°C. FI-RhCMV was prepared as previously described (2). Briefly, 22 mg of pelleted RhCMV virions (∼6 × 109 PFU total) were formalin inactivated and resuspended at a concentration of 550 μg/ml. All DNA immunizations were performed at week 0 (Table 1) by a combination of intramuscular (i.m.) and intradermal (i.d.) injections as previously described (2, 27, 46). Modified vaccinia virus Ankara (MVA) vaccine vectors expressing RhCMV gB, pp65, and IE were constructed, purified, and expanded into amounts suitable for rhesus monkey studies as previously described (48). MVA booster immunizations (groups 1 to 3 [Table 1]) were performed at weeks 6 and 12 by i.m. injection of 5 × 108 PFU of each MVA construct, as previously described (48). Group 4 monkeys were boosted i.m. at weeks 6 and 12 with 100 μg of FI-RhCMV adjuvanted with Montanide ISA 720 (Seppic Inc., Fairfield, NJ), as previously described (2).
TABLE 1.
Immunization/challenge schedulea
| Group (no. of monkeys) | Immunization/challenge (dose) at: |
|||
|---|---|---|---|---|
| 0 wk | 6 wk | 12 wk | 20 wk | |
| I (6) | pND (450 μg i.m./150 μg i.d.) | MVA (5 × 108 PFU i.m.) | MVA (5 × 108 PFU i.m.) | RhCMV UCD52 (1 × 106 PFU s.c.) |
| II (6) | pND/gB (150 μg i.m./50 μg i.d.); pND (300 μg i.m./100 μg i.d.) | MVA/gB (5 × 108 PFU i.m.) | MVA/gB (5 × 108 PFU i.m.) | RhCMV UCD52 (1 × 106 PFU s.c.) |
| III (6) | pND/gB (150 μg i.m./50 μg i.d.); pND/pp65 (150 μg i.m./50 μg i.d.); pND/IE1 (150 μg i.m./50 μg i.d.) | MVA/gB-pp65 (5 × 108 PFU i.m.); MVA/IE1 (5 × 108 PFU i.m.) | MVA/gB-pp65 (5 × 108 PFU i.m.); MVA/IE1 (5 × 108 PFU i.m.) | RhCMV UCD52 (1 × 106 PFU s.c.) |
| IV (3) | pND/gB (150 μg i.m./50 μg i.d.); pND/pp65 (150 μg i.m./50 μg i.d.); pND/IE1 (150 μg i.m./50 μg i.d.) | FI-RhCMV (100 μg i.m. in Montanide ISA 720) | FI-RhCMV (100 μg i.m. in Montanide ISA 720) | RhCMV UCD52 (1 × 106 PFU s.c.) |
Abbreviations: pND, plasmid expression vector; i.m., intramuscular vaccination; i.d., intradermal vaccination; gB, glycoprotein B; pp65, phosphoprotein 65; IE1, immediate-early 1 (0 weeks); MVA, modified vaccinia virus Ankara; FI-RhCMV, formalin-inactivated RhCMV (6 and 12 weeks); RhCMV UCD52, RhCMV strain UCD52; s.c.: subcutaneous vaccination (20 weeks).
RhCMV challenge.
All of the monkeys were challenged by a subcutaneous route with 1 × 105 PFU of RhCMV variant UCD52, which is an epithelial cell-tropic variant of RhCMV that is persistently shed in the urine and saliva of inoculated monkeys at levels comparable to those observed in animals naturally exposed to wild-type RhCMV (Yue and Barry, unpublished observations, and see Fig. 2). RhCMV UCD52 contains a full-length UL/b′ region of the genome (GenBank accession number GU552456; referred to elsewhere as RhCMV 21252), including those genes encoding proteins involved in endothelial/epithelial cell tropism and neutrophil chemotaxis (26, 32). RhCMV UCD52 is pathogenic in inoculated rhesus fetuses (43). Sequence analysis of the gB gene amplified from RhCMV UCD52 showed that there was 88% amino acid identity (95% similarity) with the gB gene of the prototypical 68-1 variant of RhCMV (RhCMV 68-1) (GenBank accession number GU552457). Amino acid substitutions were confined mostly to the extracellular domain of the gB protein. Since the gB immunogen in the plasmid and MVA expression vectors was derived from that of RhCMV 68-1, inoculation of the vaccinated monkeys with RhCMV UCD52 represented a heterologous challenge with respect to the divergence of the gB protein. RhCMV UCD52 was delivered into four separated and shaved sites on the back, as previously described (2). Longitudinal blood draws and oral swabs were collected to monitor viral and immune parameters of RhCMV challenge, and a skin biopsy specimen of one of the inoculation sites was collected either 6 or 7 days postchallenge, as previously described (2).
DNA extraction and real-time PCR.
DNA was extracted from plasma and oral swab samples using the QIASymphony automated DNA processor (Qiagen) according to the manufacturer's instructions and published protocols (23). The final elution volume was 300 μl. Extracted DNA was stored at −80°C until real-time PCR analysis was performed. RhCMV DNA copies in plasma and oral swabs were detected by a previously described real-time PCR assay for RhCMV 68-1 (40).
Neutralization assays.
The neutralizing antibody (NAb) titer of monkey plasma (EDTA anticoagulant) was measured in the absence of complement using an engineered variant of RhCMV strain 68-1 that expressed the enhanced green fluorescent protein (EGFP) (10), as previously described (2). Briefly, 2.5 × 104 PFU of RhCMV/EGFP was incubated with serial half-log dilutions (1:31 to 1:31,000) of heat-inactivated (56°C, 30 min) plasma in a final volume of 500 μl (Dulbecco's modified Eagle's medium [DMEM]-10% fetal bovine serum). A pooled mixture of plasma from eight non-RhCMV-infected rhesus monkeys was included as a negative control. The virus and plasma mixture was incubated for 2 h (37°C) and then added in triplicate (100 μl/well) to monolayers of Telo-RF cells (9). The cells had been seeded the day before at a density of 2 × 104 cells/well in Optilux black-walled, clear-bottom, 96-well plates (BD Falcon). Three wells of cells were incubated in growth medium only. The virus-plasma mixture was removed after 4 h, the cells were washed twice with Hanks balanced salt solution (HBSS) (37°C), and 100 μl of complete medium was added to each well. The mean fluorescent intensity of each well was measured 48 h later on a SpectraMax M5 plate reader (Molecular Devices Corporation, Sunnyvale, CA) using the SoftMax Pro software (version 4.8). Nine individual readings were obtained per well (using a bottom read), and values were summed across the well. The excitation, emission, and cutoff wavelengths were 472, 507, and 495 nm, respectively. Data generated by the Softmax Pro software were imported into Excel, and the mean background fluorescence (measured from the wells treated with media only) was subtracted from the mean fluorescence for each sample analyzed (i.e., both immune and seronegative control samples). This generated the relative fluorescent unit (RFU) for each sample. The percent neutralization at each dilution of plasma was calculated as follows: [1 − (RFUimmune/RFUseroneg)] × 100, where RFUimmune is the RFU of subjects with immunity and RFUseroneg is the RFU of seronegative subjects. The 50% neutralization titer (NT50) was calculated by plotting the percent neutralization (y axis) versus the logarithm of the dilution (x axis). A trendline equation was established for the linear portion of the data, from which the NT50 was calculated (expressed as the reciprocal of the dilution).
Intracellular cytokine (ICC) staining.
Cryopreserved peripheral blood mononuclear cells (PBMC) were assayed for their ability to secrete gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), or interleukin 2 (IL-2) during in vitro restimulation with overlapping peptide pools (15-mers overlapping by 11 amino acids) representing the entire amino acid sequence of either pp65-2 or IE1 obtained by a previously published protocol (1, 21). Briefly, aliquots of 1 million PBMC were stimulated for 6 h with a peptide pool corresponding to either pp65 or IE1 at a final concentration of 1.0 μg/ml of each peptide or mock stimulated in the presence of antibodies to CD28 and CD49d (0.5 μg/ml, clones 9F10 and 28.2, respectively; BD Biosciences). Brefeldin A (BD Biosciences) at 10 μg/ml was added to the culture for the final 5 h of stimulation. After stimulation, the cells were surface stained with conjugated antibodies to CD3 and CD4 for 20 min at room temperature (CD3-PerCP-Cy5.5; CD4-fluorescein isothiocyanate [FITC]; BD Biosciences). Subsequently, the cells were fixed and permeabilized with fluorescence-activated cell sorting (FACS) permeabilizing solution (BD Biosciences). Permeabilized cells were then incubated with fluorochrome-conjugated antibodies to IFN-γ (IFN-γ-Alexa 700), TNF-α (TNF-α-phycoerythrin [PE]-Cy7) or IL-2 (IL-2-allophycocyanin [APC]), all from BD Biosciences, for 20 min at room temperature, washed, and fixed in 1% paraformaldehyde. A minimum of 300,000 events within a T lymphocyte gate were collected on a FACS Aria (BD Biosciences), and the data were analyzed with FlowJo software (TreeStar, Ashland, OR).
Data analysis.
Viral load copy numbers were summarized for each animal as the total area under the curve (AUC) for 21 weeks postchallenge. The AUC between two successive time points (T1 and T2, in weeks) was calculated as the area of the trapezoid formed by the viral loads (VL) at those two time points, according to the following formula: AUC (between T1 and T2) = 1/2(VLT1 + VLT2) × (T2 − T1). The sum of individual AUC measurements represented the total AUC for each animal. Logarithms of AUC values were used in analyses of variance to compare group means, and Levine's test was used to compare variances. Additional tests are quoted in context in Results.
RESULTS
Vaccine induction of RhCMV-specific immune responses.
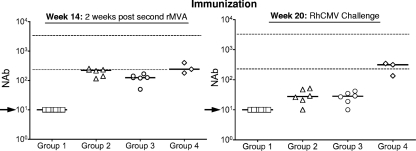
Consistently with our previous studies (2, 48), both the DNA prime/MVA boost and DNA prime/FI-RhCMV boost immunization strategies stimulated antibodies that neutralized RhCMV infection of fibroblasts. NAbs were not detected in any of the groups after the single DNA immunization and prior to the first MVA boost at week 6 but were detectable in the majority of immunized animals 2 weeks after the first booster immunization with either MVA or FI-RhCMV (data not shown). All of the vaccinated monkeys in groups 2, 3, and 4 had demonstrable NAb titers 2 weeks after the second booster immunization (week 14) (Fig. 1) that were either near or just below the lower end of the normative range of titers observed in naturally infected animals (indicated in Fig. 1). The NAb titers declined in all group 2 and 3 animals by the time of challenge at week 20, although five animals in each group had detectable titers (>10) at the time of challenge (week 20) (Fig. 1). In contrast, the NAb titers in group 4 monkeys remained relatively constant during this 6-week interval between the second FI-RhCMV boost and RhCMV challenge. All but one of the animals in group 3 developed either CD4 and/or CD8 T cell responses to either pp65 or IE1, although the responses were mostly transient during the immunization schedule (data not shown).
FIG. 1.
Fifty percent neutralizing antibody (NAb) titers for the four vaccine treatment groups at 14 weeks (2 weeks after the second booster MVA immunization) and 20 weeks (the time of RhCMV challenge). The median NAb titer for each group is represented by the solid line. The upper and lower dashed lines represent the normative range of NAb titers in long-term RhCMV-infected macaques (231 to 3,348; n = 24; mean = 973; median = 833 [unpublished data]). The arrows indicate the limit of detection (NAb < 10).
Detection of challenge virus in plasma.
To determine whether the immunization treatments altered the course of viral infection, the animals in each treatment group were challenged at week 20 with 1 × 105 PFU of RhCMV variant UCD52 (RhCMV UCD52) delivered by a subcutaneous route of inoculation. Oral swabs and blood samples were collected weekly from the challenged animals and processed for DNA to quantify RhCMV genome copy numbers. Following RhCMV UCD52 challenge, all of the monkeys in each treatment group remained cohoused with the monkeys from the other treatment groups. Naïve monkeys inoculated subcutaneously with RhCMV UCD52 routinely exhibit persistent, high-titer excretion of virus in saliva and urine (our unpublished observations). Consequently, all animals in this study were exposed to RhCMV both by subcutaneous challenge and repeated exposure to horizontally transmitted RhCMV from the other cohoused, virus-excreting animals. Horizontal transmission is the normal route of spread of RhCMV within large groups of monkeys (6), and this design recapitulates a key aspect of repeated exposure to horizontally transmitted virus in HCMV vaccine trials.
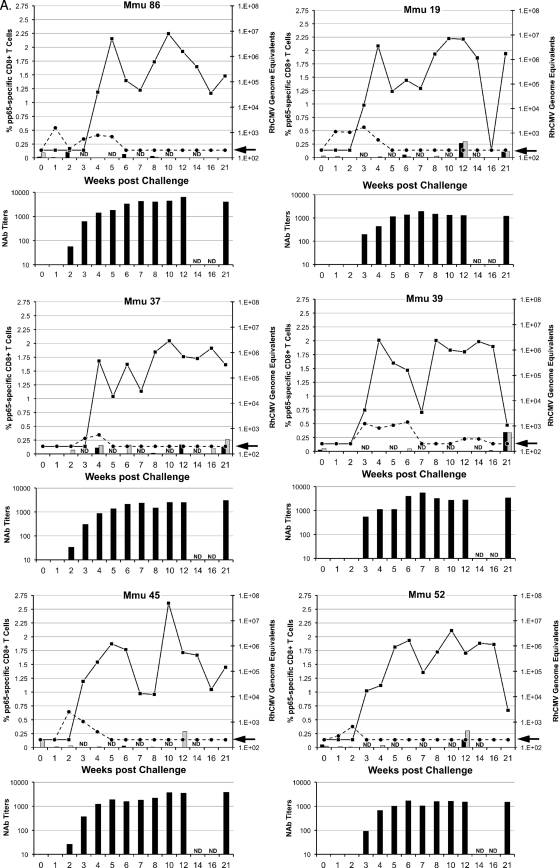
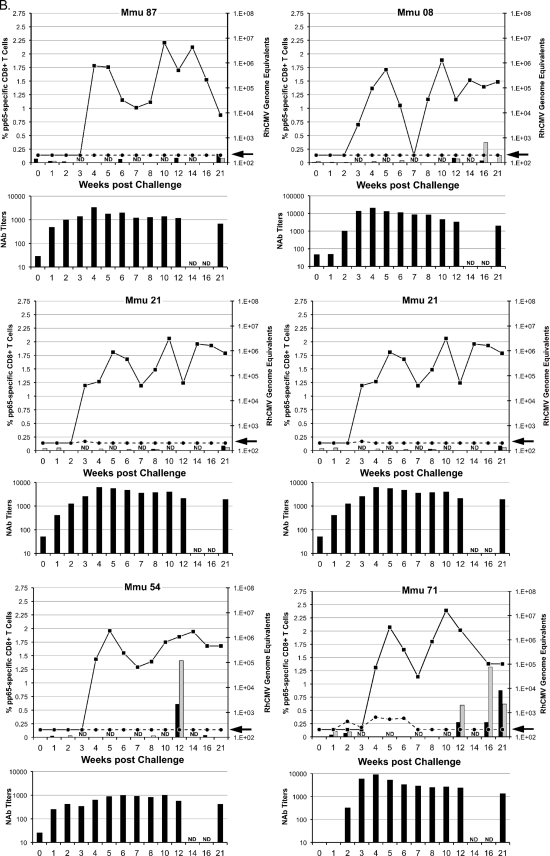
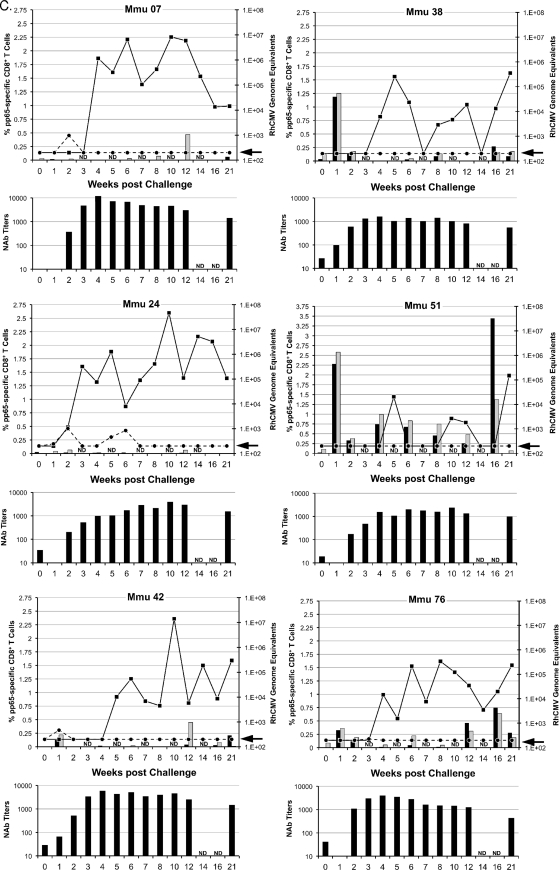
Two parameters of challenge were analyzed to determine whether the vaccine treatments in groups 2 to 4 significantly altered viral replication and dissemination from that in the unimmunized controls (group 1): detection of viral DNA in plasma and in oral (saliva) swabs (Fig. 2). RhCMV DNA was detected in plasma at multiple times for each of the six group 1 control monkeys (Fig. 2A) (limit of detection, 200 RhCMV genomes/ml of plasma). In contrast, the presence of NAb at the time of challenge in groups 2 to 4 greatly restricted the presence of detectable RhCMV DNA in plasma (Fig. 2B to D). Only one monkey in each of the three vaccine groups was positive more than once for RhCMV DNA in plasma, and almost half of all vaccine animals (7 of 15) had no detectable RhCMV DNA (compare Fig. 2A with 2B to D; Table 2). The presence of gB-directed NAb (groups 2 and 3) at the time of challenge correlated with both an absence and a reduced frequency (≤1 time) of detectable RhCMV DNA in plasma (P = 0.013 and 0.003, respectively; Fisher's exact test) (Table 2). While the presence of NAb at the time of challenge for group 4 was not associated with a complete absence of RhCMV in plasma, the detection of RhCMV was less frequent than in controls (P = 0.026, one-sided rank sum test). Clearance of RhCMV from plasma coincided with the development of de novo NAb titers in group 1 monkeys (Fig. 2A) and with a rebound of NAb titers in monkeys in groups 2 to 4 with detectable RhCMV DNA in their plasma (Fig. 2B to D).
FIG. 2.
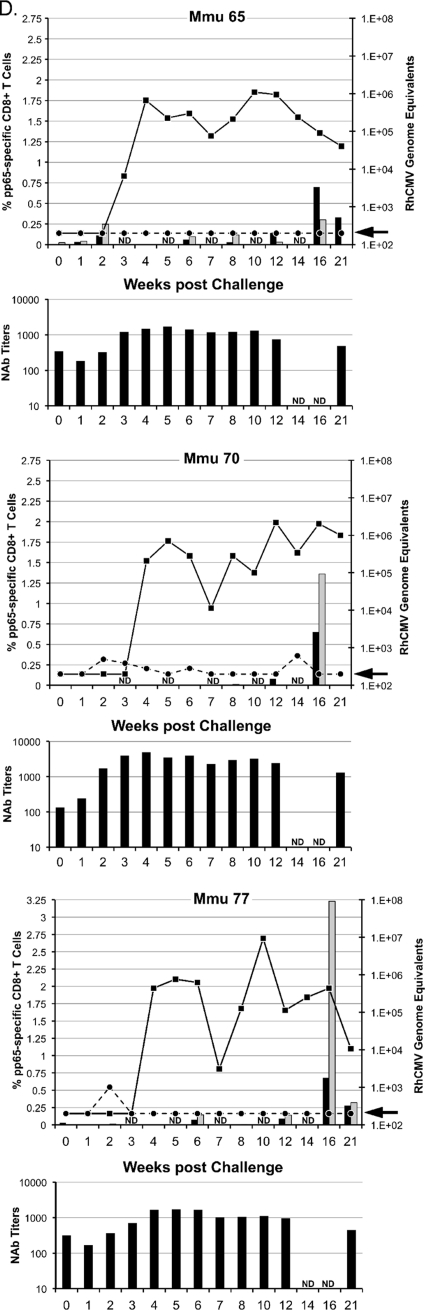
Viral and immune parameters in group 1 (A), 2 (B), 3 (C), and 4 (D) monkeys postchallenge. The upper graph for each animal in a group presents the RhCMV genome copy numbers per ml of plasma (dashed line) and saliva (solid line) postchallenge; the units for the RhCMV genome copy numbers are presented on the right y axis, relative to the time postchallenge at time zero (in weeks [x axis]). The arrow on each right y axis represents the minimum level of detection of RhCMV genomes/ml (200 copies). The percentages of pp65-specific CD8+ T cells (left y axis) expressing IFN-γ (black columns) and TNF-α (gray columns) were quantified by ICC at the time of challenge through 21 weeks after RhCMV challenge (x axis). ICC was not done (ND) at weeks 3, 5, 7, 10, and 14. The scale for the left y axis for Mmu 51 (group 3) (C) and Mmu 77 (group 4) (D) is different from that for all other animals. The lower graph for each animal represents the NAb titers (y axis) after RhCMV challenge (x axis). Titers are expressed as the reciprocal dilution achieving 50% neutralization of infection in fibroblasts (see Materials and Methods for details). NAb titers were not determined (ND) at weeks 14 and 16. The minimum level of NAb detection was 10. The three animals in group 3 (C) with poor control of RhCMV shedding in saliva are presented on the left half of panel C. The three animals of group 3 with more effective control of RhCMV shedding are presented on the right half of panel C (see the text for details).
TABLE 2.
Summary of the frequency of RhCMV DNA detected in plasma in relation to the presence or absence of NAb at the time of RhCMV challenge at week 20
| Group | No. of animals with indicated NAb result at wk 20 |
No. of animals with RhCMV DNA in plasmac |
|||
|---|---|---|---|---|---|
| Yesa | Nob | 0 times | 1 time | >1 time | |
| 1 (Control) | 0 | ||||
| 6 | 6 | ||||
| 2 (gB) | 5 | 4 | 1 | ||
| 1 | 1 | ||||
| 3 (gB-pp65-IE1) | 5 | 2 | 2 | 1 | |
| 1 | 1 | ||||
| 4 (FI-RhCMV) | 3 | 1 | 1 | 1 | |
| 0 | |||||
Number of animals in each group with detectable NAb (≥31) at week 20, the time of RhCMV challenge.
Number of animals in each group lacking detectable NAb (≤31) at week 20.
Number of animals in each group either with (yes) or without (no) NAb for which RhCMV DNA was detected in plasma 0 times, 1 time, or >1 time over the course of 21 weeks of observation after RhCMV challenge. The limitation of detection was 200 RhCMV genome copy numbers/ml.
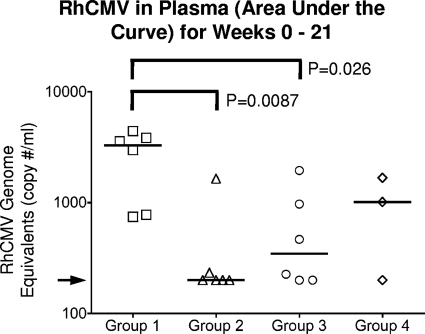
To further assess the importance of vaccination on the magnitude of challenge virus replication, the cumulative infectious burden of RhCMV in plasma was calculated over the entire period of observation as an area under the curve (AUC) (see Materials and Methods) (Fig. 3). The geometric mean plasma AUC values in the vaccinated groups were 13% (group 2), 21% (group 3), and 32% (group 4) of those of controls (data not shown). The longitudinal plasma burden (AUC) varied significantly across the four groups (P = 0.03, Kruskal-Wallis test, 3 degrees of freedom). Both groups 2 and 3 had significant reductions in plasma viral loads over the 21-week observation compared to group 1 control monkeys (P = 0.0087 and 0.026, respectively, for geometric mean AUCs; analysis of variance [ANOVA] using a logarithmic scale). The geometric mean AUC for RhCMV for the three animals of group 4 was lower than that of group 1 but less accurately determined (P = 0.095).
FIG. 3.
Area under the curve (AUC) of RhCMV in plasma over the course of 21 weeks of postchallenge observation. The median AUC for each group is represented by the solid line. The arrow indicates the level of detection for RhCMV DNA (200 RhCMV genomes/ml of saliva).
Detection of RhCMV UCD52 in saliva.
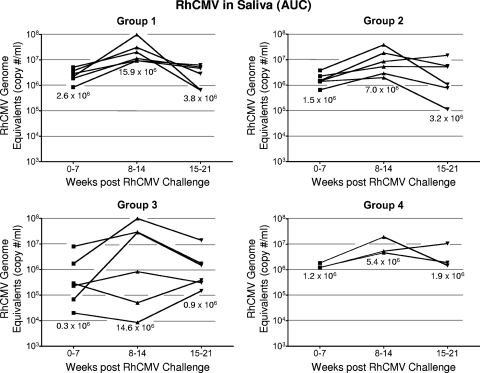
Oral shedding of RhCMV was detected in all control and vaccinated monkeys 3 to 5 weeks after subcutaneous inoculation of challenge virus (Fig. 2). Whereas the presence of NAb at the time of challenge was associated with a reduction in the frequency and magnitude of RhCMV in plasma, there were no similar protective effects related to either the magnitude or the pattern of RhCMV DNA in saliva. Once the group 1 control monkeys became positive for RhCMV DNA in saliva, they remained persistently positive through 21 weeks of observation, except at a single time point for one animal, 16 weeks postinoculation (Fig. 2A). Titers ranged from 103 to ∼5 × 107 RhCMV genomes/ml of saliva. The range of shedding for group 1, calculated as the AUC, remained within ∼1 log for each time interval (0 to 7, 8 to 14, and 15 to 21 weeks), with the peak of shedding occurring 8 to 14 weeks after inoculation (Fig. 4). Animals in groups 2 and 4 (gB and FI-RhCMV immunized, respectively) exhibited shedding profiles similar to those of controls in both the kinetics and magnitude of RhCMV in saliva, independently of their NAb responses (Fig. 2 and 4). While the median AUC for each time interval was lower for both group 2 and group 4 animals than for group 1 animals (geometric mean AUCs = 47 and 51% of the control AUC, respectively) (Fig. 4), the differences were not statistically significant (P = 0.4 and 0.55, respectively, for geometric mean AUCs). The geometric mean for RhCMV in saliva in group 3 was 19% of that for controls, but this large difference had only weak statistical significance (P = 0.084) due to the large variation in group 3 (Fig. 4). When AUCs were calculated on a logarithmic scale, there was less variance, and group 3 had significantly lower average oral shedding (P < 0.02, two-sided t test).
FIG. 4.
Longitudinal AUCs of RhCMV in saliva for weeks 0 to 7, 8 to 14, and 15 to 21. The median AUC of RhCMV for each group during each interval of time is indicated. The genome copies are expressed as copy number/ml on the indicated left axis.
Shedding in group 3 monkeys was noted for a distinction between animals in their magnitudes of shedding (Fig. 2C and 4). Three animals shed RhCMV in saliva at cumulative levels that were almost indistinguishable from levels in the control monkeys at all three time intervals, although one of these three monkeys had an AUC for the 0-to-7-week interval that was 1.6 logs lower than the median shedding AUC for group 1. In contrast, RhCMV shedding in the other three group 3 animals consistently remained 1 to 3.1 logs lower than the median AUC for group 1. This was particularly evident from weeks 8 to 14, when individual AUC values were 1.3 to 3.3 logs lower than the median AUC for group 1 during the same period of observation. Two of three animals had undetectable RhCMV DNA in their saliva for at least two sampling times (Fig. 2C). Although the reduction in the geometric mean AUC for group 3 was not statistically different from that for the group 1 controls during any time interval, the prominent reduction in shedding for three animals was especially notable compared to the shedding of all of the animals in the three other groups. The total infectious burden for all monkeys spanned 2.8 logs over the course of 21 weeks, and the AUCs for these three animals in group 3 were 1 to 1.9 logs lower than that for the animal in group 1 with the lowest total AUC for group 1 animals (Table 3). In addition, their shedding titers were the lowest of any of the animals in any of the four treatment groups. The variation in log-scale oral AUCs among the animals in group 3 was larger than the variation within group 1, generating a detectable inhomogeneity of variance (P = 0.00043, Levene's test for homogeneity of variance). This result was consistent with the interpretation that the gB-pp65-IE1 immunization regimen elicited a statistically significant reduction in oral shedding in a subset of group 3 animals. This measurable level of protection was independent of NAb titers, since there was no apparent association between viral loads in saliva and the magnitude of the NAb titers (Fig. 2C).
TABLE 3.
Numbers of animals in each group within specified intervals of total infectious burdens
| Group (challenge) | No. of animals with the following total infectious burden (AUC) of RhCMV DNA in saliva (RhCMV genome copy no. [106]/ml)a: |
||||
|---|---|---|---|---|---|
| <2 × 106 | 2 × 106-10 × 106 | >10 × 106-20 × 106 | >20 × 106-100 × 106 | >100 × 106 | |
| 1 (control) | 3 (15-19.6) | 2 (26.1-37.9) | 1 (101.6) | ||
| 2 (gB) | 2 (3.5-4.4) | 1 (13) | 3 (24.9-43.5) | ||
| 3 (gB-pp65-IE1) | 3 (0.2-1.4) | 2 (29.9-43.5) | 1 (115) | ||
| 4 (IE1/FI-RhCMV) | 1 (7.8) | 1 (17.2) | 1 (22.8) | ||
Ranges of AUCs in numbers of copies of RhCMV/ml. Data represent oral shedding over the 21 weeks of observation following RhCMV challenge.
Anti-RhCMV CMI responses after RhCMV challenge.
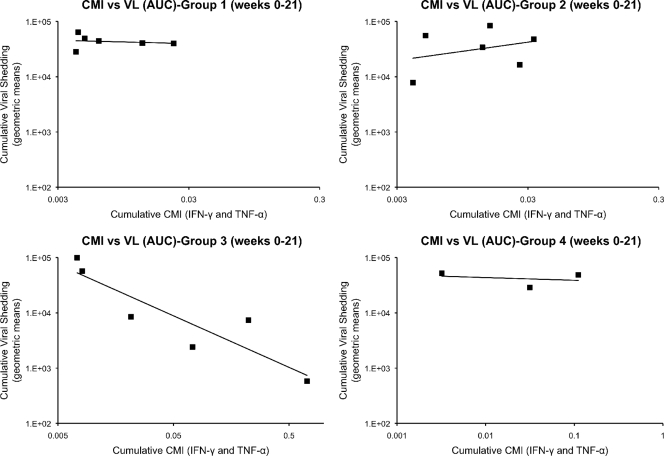
Longitudinal peripheral blood samples were obtained from all vaccinated and control monkeys over 21 weeks of postchallenge observation. PBMC were obtained and analyzed by ICC using overlapping peptide libraries for RhCMV pp65 and IE1. Only pp65-specific CD8+ T cells expressing either IFN-γ or TNF-α showed an association with control of RhCMV shedding in a subset of group 3 monkeys (Fig. 2C). No association of viral load with any measured T cell cytokine response was found for any monkey in any of the other 3 treatment groups (Fig. 2A, B, and D). In particular, the frequency of pp65-specific CD8+ T cells in group 3 monkeys was inversely related to the cumulative level of RhCMV in saliva (AUC). The three monkeys in group 3 whose pp65-specific T cell responses were <0.25% at every time point during the first 10 weeks of challenge (M. mulatta 07 [Mmu 07], 24, and 42) were characterized by the highest cumulative RhCMV burdens in saliva (Fig. 2 and 4). In contrast, all three monkeys in group 3 with prominent pp65-specific responses (>0.25%) 1 week after RhCMV challenge (Mmu 38, 51, and 76) were noted for shedding levels below those of the other monkeys in group 3 and the control monkeys (Fig. 2C). There was no relationship between the magnitude of oral shedding and IE1-specific immune responses (data not shown). The potential importance of the magnitude of the cell-mediated immunity (CMI) to pp65 and the control of oral shedding was highlighted by plotting the geometric means of the cumulative AUC of RhCMV in saliva over 21 weeks of observation against the geometric means of the cumulative CMI responses (IFN-γ and TNF-α) to pp65 (Fig. 5). Only in group 3 was there a statistically significant inverse association of CMI and oral shedding (P = 0.015, 1 df, F = 16.9). Similar observations were made by plotting cumulative AUCs against either just IFN-γ or TNF-α responses to pp65 (data not shown). Together, these results support the conclusion that vaccine-induced CMI to RhCMV (as measured against pp65 in this case), and not NAb, was an immune correlate of protection against oral shedding of RhCMV.
FIG. 5.
RhCMV viral loads (VL) versus cell-mediated immunity (CMI) responses to RhCMV pp65. The geometric means of the RhCMV viral loads in saliva for weeks 0 to 21 after RhCMV challenge and the cellular immune responses to pp65 (both IFN-γ and TNF-α) were plotted for each animal group. The geometric means (mean of logs, transformed back to the original scale) were calculated after a limit of detection was added to establish a value for zeroes. The limit of detection for RhCMV DNA in saliva was 200 and one-half of the smallest nonzero value for IFN-γ and TNF-α (0.0001 and 0.0005, respectively). A statistically significant association of CMI and oral shedding was observed only in group 3 (P = 0.015, 1 degree of freedom, F = 16.9).
DISCUSSION
This study considerably expands our previous RhCMV immunization/challenge studies in rhesus monkeys by evaluating whether immunization against prominent RhCMV antigens can significantly reduce two important parameters of challenge virus replication, viremia and oral shedding (2, 46, 48). This study is the first use of, as a challenge virus, an RhCMV variant (UCD52) that is more related to RhCMV natural strains in terms of both genetic coding content (32), particularly within UL/b′, and the capacity for high-level, persistent shedding in bodily fluids, such as saliva (5, 23). Coupled with the genetic drift within the gB coding region between the vaccine antigen and the challenge virus, this study design more closely represents the obstacles facing HCMV vaccine trials than those of previous animal studies. Generation of vaccine-induced protective immunity against HCMV will require protection against repeated exposure to antigenically diverse HCMV variants and, in the case of preventing vertical transmission, strict limitation of the dissemination of the challenge virus from the site of infection to distal sites, including the maternal-fetal interface.
Vaccination of rhesus monkeys with either gB alone (group 2) or gB-pp65-IE1 (group 3) significantly reduced RhCMV DNA in plasma, compared to levels in the unvaccinated group (group 1). However, only group 3 exhibited any quantifiable reduction in oral shedding of RhCMV postchallenge in 50% of the vaccinees. These results suggest that different vaccine endpoints are dependent on the composition of the vaccine. Our data are consistent with the interpretation that the generation of gB-specific immune responses alone is sufficient to significantly minimize the presence of RhCMV in plasma. It is worth noting that there was an apparent disparity between NAb titers and plasma viral loads. Group 4 animals, which were immunized with both DNA and FI-RhCMV, generally had higher NAb titers than the other two vaccinated groups at the time of challenge yet exhibited slightly higher plasma viral loads than either group 2 or group 3. The plasma AUC for group 4 was not significantly different from the AUC of either group 2 or 3, and the intergroup differences for groups 2 to 4 are probably a reflection of the smaller size of group 4 (3 animals versus 6 in each of groups 2 and 3). Whereas NAbs are critical for controlling plasma viral load, restriction of oral shedding requires additional antigens besides gB. Our data suggest that pp65-specific cellular immune responses contribute to protection. In support of that contention, we found that only vaccinees in group 3 that exhibited a pp65-specific memory T cell response at 1 week postchallenge and were still responsive during the following 15-week period showed better control of RhCMV DNA copy number in oral swabs postchallenge. Only animals in which there was evidence for control of shedding showed significant early T cell responses both to TNF-α and to IFN-γ. In groups 1, 2, and 4, minimal early T cell responses to either cytokine were noted. In contrast, all animal groups had evidence of late T cell responses (week 12 and later) to both TNF-α and IFN-γ. A likely explanation is the generation of an adaptive immune response to the challenge virus, since enough time had transpired since the time of challenge to expect de novo immunity to evolve. Of note, we never detected levels of IL-2 in either CD4+ or CD8+ T cells in any animal at any time point during the monitoring of cellular immunity either before or after challenge. While we can only speculate about the reasons for a lack of stimulation of IL-2, others have found that, in MVA-vaccinated humans, IFN-γ, TNF-α, and MIP-1β (not analyzed in our study) responses are the dominant cytokine responses in antigen-specific CD8 T cells, far exceeding the frequency of expression of IL-2 (35; data not shown).
The reasons for incomplete control of oral shedding in all animals in group 3 are unknown. The absence of reduced oral shedding in 50% of the group 3 monkeys suggests that a more robust pp65-specific immune response must be stimulated in some vaccinees to attain protective function. A similar conclusion can be drawn from the recently completed clinical trial in which vaccination with HCMV gB conferred a significant level (50%) of protective efficacy (34). The breeding cohort of rhesus monkeys at the CNPRC is comprised of outbred animals with defined parentage. All of the animals in this study were borne by different dams, and all but two (in different groups) were sired by different males. Consequently, the outbred monkeys have diverse haplotypes, which is analogous to the diversity in patients enrolled in the gB clinical trial (34). Since most vaccines have an efficacy distribution that has been thought to be related to haplotype (30, 31), then in an analogous manner it is possible that the failure of some animals to control viral load at the same level as others is related to differences in haplotype. This is especially relevant if there are missing antigens that might provide broader immunity and that cover haplotypes missed by the components of the vaccine. In fact, there may be redundancy in protection mechanisms, and once the fibroblast and endocytic complexes are accounted for in the vaccine, a higher frequency of responders may be enabled. Previous studies have demonstrated that gB immunization stimulates viral NAb titers that exceed those found in HCMV-infected individuals (14, 33). Nonetheless, the elevation of high-level gB NAb titers was transitory, suggesting that other factors are needed to maintain durable immunity (14, 33). Accordingly, HCMV antigens in addition to gB should be included in the vaccine to achieve a protective efficacy that approaches 100%.
The specificity of the additional antigens to augment the HCMV gB vaccine remains to be determined empirically, but HCMV pp65 is an obvious candidate, based on solid-organ and hematopoietic stem cell transplant (HCT) studies and our current study. HCMV pp65 T cells have been associated with control of viremia in HCT recipients and contribute a large proportion of the cellular immune response in many healthy adults and children (12, 18, 42). A prior role for cellular immunity to pp65 involved in the control of viral shedding had not been identified until the results of this study. We provided evidence of the necessity of systemic pp65-specific CD8+ T cell responses 1 week after RhCMV challenge for local control of RhCMV replication following the subcutaneous route of challenge used in this study. The presence of pp65-specific CD8+ T cell responses 1 week after RhCMV challenge was not clearly associated with local control of RhCMV replication following the subcutaneous route of challenge used for this study. Cells with nuclear and cytoplasmic inclusions, pathognomonic for RhCMV infection, were detected at the same time point in biopsy specimens of the inoculation site from all animals (data not shown). Other antigens that merit consideration for an HCMV vaccine include viral proteins mediating cell tropism to nonfibroblast cells. The UL128, 130, and 131a proteins, which together form a complex (UL128 complex) with glycoprotein H (gH) and gL, are essential for epithelial/endothelial cell tropism via an endocytic pathway (11, 16, 17, 28, 36-38, 45). Critically from a vaccine perspective, sera from HCMV-seropositive individuals can neutralize the infectivity of epithelial and endothelial cells infected with clinical isolates of HCMV which express the UL128 complex (11). In contrast, sera from gB-vaccinated individuals or those immunized with the Towne vaccine, which does not express the full complex, inefficiently block the infectivity in these two cell types.
A broad cell tropism for HCMV infection appears to be essential for the ability of HCMV to successfully complete its replicative cycle, particularly in endothelial and epithelial cells, by enabling efficient host-to-host spread. Multiple studies have documented the fact that virus can be excreted in saliva and urine long after resolution of primary infection and in breast milk during successive pregnancies and lactations (3, 8, 13, 15, 20, 22, 24, 29, 39, 41). An individual who sheds virus could expose pregnant women without preconceptional immunity to HCMV who are most susceptible to the consequences of horizontal transmission. Since the frequency of congenital infection in the United States is ∼0.7% (25), the overwhelming factor for the high rate of HCMV seroprevalence is the result of horizontal transfer of virus in bodily fluids from an infected to an uninfected individual. The probability of primary infection, and by extension the probability of vertical infection, is a direct function of the frequency of close personal contact (i.e., mucosal exposure to infectious bodily fluids) between an infected individual and another person. By extension, vaccines that reduce the viral load that would otherwise be shed in mucosal fluids of infected individuals would correspondingly reduce the risk of viral exposure to their close contacts (19).
Based on this scenario, the shedding of RhCMV in saliva was evaluated as a parameter of challenge virus dissemination, since it is likely that spread of the virus from the site of subcutaneous inoculation to sites of oral shedding involves multiple rounds of replication in multiple cell types. Our approach greatly extends the scope of our previous assessments of vaccine-induced protective efficacy by evaluating protection under conditions that reflect those in humans (2, 46, 48). This includes the use of a challenge virus that (i) recapitulates the pattern of long-term shedding in bodily fluids observed with RhCMV endemic in breeding colonies of rhesus monkeys (4, 5, 44) and (ii) expresses antigens divergent from those used for vaccination. Further, this study is the first to measure the potential for vaccination to interrupt the critical step of shedding in RhCMV's natural history. The prominent reductions in oral shedding exhibited by the three animals following RhCMV UCD52 challenge occurred in the face of repeated exposure from horizontally transmitted virus from the 18 cohoused animals that did not control shedding. Direct inoculation of vaccinated animals likely presents an extrarigorous test of vaccine-induced protective efficacy above that presented by natural exposure to virus across a mucosal surface. Future studies in which vaccinated animals are exposed to challenge virus by cohousing them with RhCMV-secreting animals can be designed to provide a model where the protective efficacy of the vaccine is tested in the face of natural titers of virus, natural routes of exposure, and natural frequencies of exposure. These will represent in a nonhuman primate model the challenges faced in optimizing HCMV vaccine modalities in clinical trials by recapitulating the frequencies of exposure and titers of horizontally transmitted, antigenically diverse variants of RhCMV. The results also highlight the critical importance of sustainable populations of RhCMV-free rhesus monkeys to enable expansion of this model (7).
Acknowledgments
This work was supported by funding from the National Institutes of Health (grant AI063356) and the California National Primate Research Center (grant RR000169) to P.A.B. and D.J.D., by grants CA030206 and CA77544 to D.J.D., and by the Margaret M. Deterding Infectious Disease Research Support Fund to P.A.B. The City of Hope Comprehensive Cancer Center is partially supported by grant CA033572.
We are grateful to Linda Wyatt and Bernard Moss (Laboratory of Viral Diseases, National Institute of Allergy and Infectious Diseases) for the gifts of plasmid and viral vectors and instructions on their use.
Footnotes
Published ahead of print on 29 December 2010.
REFERENCES
- 1.Abel, K., et al. 2006. Rapid virus dissemination in infant macaques after oral simian immunodeficiency virus exposure in the presence of local innate immune responses. J. Virol. 80:6357-6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abel, K., et al. 2008. A heterologous DNA prime/protein boost immunization strategy for rhesus cytomegalovirus. Vaccine 26:6013-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora, N., Z. Novak, K. B. Fowler, S. B. Boppana, and S. A. Ross. 2010. Cytomegalovirus viruria and DNAemia in healthy seropositive women. J. Infect. Dis. 202:1800-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asher, D. M., J. C. J. Gibbs, D. J. Lang, D. C. Gadjusek, and R. M. Chanock. 1974. Persistent shedding of cytomegalovirus in the urine of healthy rhesus monkeys. Proc. Soc Exp. Biol. Med. 145:794-801. [DOI] [PubMed] [Google Scholar]
- 5.Asher, D. M., C. J. Gibbs, Jr., and D. J. Lang. 1969. Rhesus monkey cytomegaloviruses: persistent asymptomatic viruses. Bacteriol. Proc. 69:191. [Google Scholar]
- 6.Barry, P. A., and W.-L. W. Chang. 2007. Primate Betaherpesviruses, p. 1051-1075. In A. Arvin, G. Campadielli, P. Moore, E. Mocarski, B. Roizman, R. Whitley, and K. Yamanishi (ed.), Human herpesviruses: biology, therapy and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed]
- 7.Barry, P. A., and L. Strelow. 2008. Development of breeding populations of rhesus macaques that are specific pathogen free for rhesus cytomegalovirus. Comp. Med. 58:43-46. [PMC free article] [PubMed] [Google Scholar]
- 8.Britt, W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr. Top. Microbiol. Immunol. 325:417-470. [DOI] [PubMed] [Google Scholar]
- 9.Chang, W. L., V. Kirchoff, G. S. Pari, and P. A. Barry. 2002. Replication of rhesus cytomegalovirus in life-expanded rhesus fibroblasts expressing human telomerase. J. Virol. Methods 104:135-146. [DOI] [PubMed] [Google Scholar]
- 10.Chang, W. L., A. F. Tarantal, S. S. Zhou, A. D. Borowsky, and P. A. Barry. 2002. A recombinant rhesus cytomegalovirus expressing enhanced green fluorescent protein retains the wild-type phenotype and pathogenicity in fetal macaques. J. Virol. 76:9493-9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui, X., B. P. Meza, S. P. Adler, and M. A. McVoy. 2008. Cytomegalovirus vaccines fail to induce epithelial entry neutralizing antibodies comparable to natural infection. Vaccine 26:5760-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cwynarski, K., et al. 2001. Direct visualization of cytomegalovirus-specific T-cell reconstitution after allogeneic stem cell transplantation. Blood 97:1232-1240. [DOI] [PubMed] [Google Scholar]
- 13.Dworsky, M., M. Yow, S. Stagno, R. F. Pass, and C. Alford. 1983. Cytomegalovirus infection of breast milk and transmission in infancy. Pediatrics 72:295-299. [PubMed] [Google Scholar]
- 14.Frey, S. E., et al. 1999. Effects of antigen dose and immunization regimens on antibody responses to a cytomegalovirus glycoprotein B subunit vaccine. J. Infect. Dis. 180:1700-1703. [DOI] [PubMed] [Google Scholar]
- 15.Gautheret-Dejean, A., et al. 1997. Detection of human Betaherpesvirinae in saliva and urine from immunocompromised and immunocompetent subjects. J. Clin. Microbiol. 35:1600-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerna, G., et al. 2005. Dendritic-cell infection by human cytomegalovirus is restricted to strains carrying functional UL131-128 genes and mediates efficient viral antigen presentation to CD8+ T cells. J. Gen. Virol. 86:275-284. [DOI] [PubMed] [Google Scholar]
- 17.Gerna, G., et al. 2008. Human cytomegalovirus serum neutralizing antibodies block virus infection of endothelial/epithelial cells, but not fibroblasts, early during primary infection. J. Gen. Virol. 89:853-865. [DOI] [PubMed] [Google Scholar]
- 18.Gibson, L., et al. 2004. Human cytomegalovirus proteins pp65 and immediate early protein 1 are common targets for CD8+ T cell responses in children with congenital or postnatal human cytomegalovirus infection. J. Immunol. 172:2256-2264. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths, P. D. 2002. Strategies to prevent CMV infection in the neonate. Semin. Neonatol. 7:293-299. [DOI] [PubMed] [Google Scholar]
- 20.Hamprecht, K., et al. 1998. Detection of cytomegaloviral DNA in human milk cells and cell free milk whey by nested PCR. Virol. Methods 70:167-176. [DOI] [PubMed] [Google Scholar]
- 21.Hartigan-O'Connor, D. J., K. Abel, and J. M. McCune. 2007. Suppression of SIV-specific CD4+ T cells by infant but not adult macaque regulatory T cells: implications for SIV disease progression. J. Exp. Med. 204:2679-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howard, M. R., et al. 1997. Detection of human herpesvirus 8 DNA in semen from HIV-infected individuals but not healthy semen donors. AIDS 11:F15-F19. [DOI] [PubMed] [Google Scholar]
- 23.Huff, J. L., R. Eberle, J. Capitanio, S.-S. Zhou, and P. A. Barry. 2003. Differential detection of B virus and rhesus cytomegalovirus in rhesus macaques. J. Gen. Virol. 84:83-92. [DOI] [PubMed] [Google Scholar]
- 24.Kashiwagi, Y., S. Nemoto, Hisashi, Kawashima, K. Takekuma, T. Matsuno, A. Hoshika, and J. Nozaki-Renard. 2001. Cytomegalovirus DNA among children attending two day-care centers in Tokyo. Pediatr. Int. 43:493-495. [DOI] [PubMed] [Google Scholar]
- 25.Kenneson, A., and M. J. Cannon. 2007. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev. Med. Virol. 17:253-276. [DOI] [PubMed] [Google Scholar]
- 26.Lilja, A. E., and T. Shenk. 2008. Efficient replication of rhesus cytomegalovirus variants in multiple rhesus and human cell types. Proc. Natl. Acad. Sci. U. S. A. 105:19950-19955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loomis-Huff, J. E., R. Eberle, K. M. Lockridge, G. Rhodes, and P. A. Barry. 2001. Immunogenicity of a DNA vaccine against herpes B virus in mice and rhesus macaques. Vaccine 19:4865-4873. [DOI] [PubMed] [Google Scholar]
- 28.Macagno, A., et al. 2010. Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128-131A complex. J. Virol. 84:1005-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mansat, A., et al. 1997. Cytomegalovirus detection in cryopreserved semen samples collected for therapeutic donor insemination. Hum. Reprod. 12:1663-1666. [DOI] [PubMed] [Google Scholar]
- 30.Ovsyannikova, I. G., et al. 2007. Relationship between HLA polymorphisms and gamma interferon and interleukin-10 cytokine production in healthy individuals after rubella vaccination. Clin. Vaccine Immunol. 14:115-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ovsyannikova, I. G., V. S. Pankratz, R. A. Vierkant, R. M. Jacobson, and G. A. Poland. 2006. Human leukocyte antigen haplotypes in the genetic control of immune response to measles-mumps-rubella vaccine. J. Infect. Dis. 193:655-663. [DOI] [PubMed] [Google Scholar]
- 32.Oxford, K. L., et al. 2008. Protein coding content of the U(L)b′ region of wild-type rhesus cytomegalovirus. Virology 373:181-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pass, R. F., et al. 1999. A subunit cytomegalovirus vaccine based on recombinant envelope glycoprotein B and a new adjuvant. J. Infect. Dis. 180:970-975. [DOI] [PubMed] [Google Scholar]
- 34.Pass, R. F., et al. 2009. Vaccine prevention of maternal cytomegalovirus infection. N. Engl. J. Med. 360:1191-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Precopio, M. L., et al. 2007. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J. Exp. Med. 204:1405-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryckman, B. J., M. C. Chase, and D. C. Johnson. 2010. Human cytomegalovirus TR strain glycoprotein O acts as a chaperone promoting gH/gL incorporation into virions but is not present in virions. J. Virol. 84:2597-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryckman, B. J., M. A. Jarvis, D. D. Drummond, J. A. Nelson, and D. C. Johnson. 2006. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J. Virol. 80:710-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryckman, B. J., et al. 2008. Characterization of the human cytomegalovirus gH/gL/UL128-131 complex that mediates entry into epithelial and endothelial cells. J. Virol. 82:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schleiss, M. R. 2006. Role of breast milk in acquisition of cytomegalovirus infection: recent advances. Curr. Opin. Pediatr. 18:48-52. [DOI] [PubMed] [Google Scholar]
- 40.Sequar, G., et al. 2002. Experimental coinfection of rhesus macaques with rhesus cytomegalovirus and simian immunodeficiency virus: pathogenesis. J. Virol. 76:7661-7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stagno, S., et al. 1975. Cervical cytomegalovirus excretion in pregnant and nonpregnant women: suppression in early gestation. J. Infect. Dis. 131:522-527. [DOI] [PubMed] [Google Scholar]
- 42.Sylwester, A. W., et al. 2005. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 202:673-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tarantal, A. F., et al. 1998. Neuropathogenesis induced by rhesus cytomegalovirus in fetal rhesus monkeys (Macaca mulatta). J. Infect. Dis. 177:446-450. [DOI] [PubMed] [Google Scholar]
- 44.Vogel, P., B. J. Weigler, H. Kerr, A. Hendrickx, and P. A. Barry. 1994. Seroepidemiologic studies of cytomegalovirus infection in a breeding population of rhesus macaques. Lab. Anim. Sci. 44:25-30. [PubMed] [Google Scholar]
- 45.Wille, P. T., A. J. Knoche, J. A. Nelson, M. A. Jarvis, and D. C. Johnson. 2010. A human cytomegalovirus gO-null mutant fails to incorporate gH/gL into the virion envelope and is unable to enter fibroblasts and epithelial and endothelial cells. J. Virol. 84:2585-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yue, Y., et al. 2007. Immunogenicity and protective efficacy of DNA vaccines expressing rhesus cytomegalovirus glycoprotein B, phosphoprotein 65-2, and viral interleukin-10 in rhesus macaques. J. Virol. 81:1095-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yue, Y., A. Kaur, S. S. Zhou, and P. A. Barry. 2006. Characterization and immunological analysis of the rhesus cytomegalovirus homologue (Rh112) of the human cytomegalovirus UL83 lower matrix phosphoprotein (pp65). J. Gen. Virol. 87:777-787. [DOI] [PubMed] [Google Scholar]
- 48.Yue, Y., et al. 2008. Evaluation of recombinant modified vaccinia Ankara virus-based rhesus cytomegalovirus vaccines in rhesus macaques. Med. Microbiol. Immunol. 197:117-123. [DOI] [PubMed] [Google Scholar]
- 49.Yue, Y., S. S. Zhou, and P. A. Barry. 2003. Antibody responses to rhesus cytomegalovirus glycoprotein B in naturally infected rhesus macaques. J. Gen. Virol. 84:3371-3379. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, C., H. Buchanan, W. Andrews, A. Evans, and R. F. Pass. 2006. Detection of cytomegalovirus infection during a vaccine clinical trial in healthy young women: seroconversion and viral shedding. J. Clin. Virol. 35:338-342. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, C., and R. F. Pass. 2004. Detection of cytomegalovirus infection during clinical trials of glycoprotein B vaccine. Vaccine 23:507-510. [DOI] [PubMed] [Google Scholar]