Abstract
For intracellular survival it is imperative that viruses have the capacity to manipulate various cellular responses, including metabolic and biosynthetic pathways. The unfolded protein response (UPR) is induced by various external and internal stimuli, including the accumulation of misfolded proteins in the endoplasmic reticulum (ER). Our previous studies have indicated that the replication and assembly of the flavivirus West Nile virus strain Kunjin virus (WNVKUN) is intimately associated with the ER. Thus, we sought to determine whether the UPR was induced during WNVKUN infection. WNVKUN induces UPR signaling during replication, which is coordinated with peak replication. Interestingly, signaling is biased toward the ATF6/IRE-1 arm of the response, with high levels of Xbp-1 activation but negligible eukaryotic translation initiation factor 2α phosphorylation and downstream transcription. We show that the PERK-mediated response may partially regulate replication, since external UPR stimulation had a limiting effect on early replication events and cells deficient for PERK demonstrated increased replication and virus release. Significantly, we show that the WNVKUN hydrophobic nonstructural proteins NS4A and NS4B are potent inducers of the UPR, which displayed a high correlation in inhibiting Jak-STAT signaling in response to alpha interferon (IFN-α). Sequential removal of the transmembrane domains of NS4A showed that reducing hydrophobicity decreased UPR signaling and restored IFN-α-mediated activation. Overall, these results suggest that WNVKUN can stimulate the UPR to facilitate replication and that the induction of a general ER stress response, regulated by hydrophobic WNVKUN proteins, can potentiate the inhibition of the antiviral signaling pathway.
The unfolded protein response (UPR) is a cellular stress response that is induced upon accumulation of misfolded proteins within the endoplasmic reticulum (ER). This can occur through treatment with glycosylation inhibitors (e.g., tunicamycin), changes in calcium homeostasis, nutrient depletion, overexpression of abnormal proteins, or virus infection (9). Virus infection is especially significant, since viral protein translation, modification, and sometimes virion assembly can all place significant stress on the organelle (15). The cell attempts to alleviate this stress by activating three signaling pathways which act to increase chaperone expression, protein degradation, and ER volume and decrease protein input by inhibiting translation (4). Unfolded proteins are recognized by the chaperone molecule immunoglobulin heavy-chain binding protein (BiP) (22), which dissociates from three transmembrane proteins: PKR-like ER kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme 1 (IRE-1). PERK and IRE-1 then are able to dimerize and undergo autophosphorylation and activation. PERK phosphorylates eukaryotic translation initiation factor 2α (eIF2α) on Ser51 (12, 28), leading to an inhibition of general translation and a paradoxical increase in activating transcription factor 4 (ATF4) (3), which upregulates expression of many redox and metabolic proteins to aid in ER stress recovery (14). It also induces expression of growth arrest and DNA damage molecule 34 (GADD34), which then forms a complex with protein phosphatase 1 (PP-1) to dephosphorylate eIF2α as a negative-feedback mechanism (6, 31) to resume protein translation. However, in times of extreme ER stress, ER-associated caspases such as C/EBP-homologous protein (CHOP) are also upregulated, leading to apoptosis (13, 30). This arm of the UPR is also a component of the integrated stress response, which responds to nutrient deficiency (1, 12), hypoxia (14, 24) and double-stranded RNA (dsRNA) (8), as well as ER stress.
In contrast, the ATF6 and IRE-1 pathways are specific to the UPR. Activated IRE-1 splices a 26-nucleotide (nt) region from X box binding protein 1 (Xbp-1) mRNA causing a frameshift which allows expression of the full-length transcription factor Xbp-1 (7). Xbp-1 then upregulates transcription of mRNAs encoding degradative factors (e.g., ER degradation enhancing α-mannosidase-like protein 1 [EDEM-1]) and some chaperones (26) involved in ER-associated degradation (ERAD), transporting misfolded proteins out of the ER for ubiquitination and proteasomal degradation (36, 42). Xbp-1 has also been shown to increase transcription of genes involved in lipid biosynthesis and thus increase the volume of the ER (45) to cope with ER stress. Upon dissociation of BiP from the luminal domain of ATF6, this transmembrane protein is incorporated into COPII vesicles and translocated to the Golgi body, where it undergoes proteolytic processing by Site-1 and Site-2 proteases (53). It then translocates to the nucleus and upregulates transcription of ER chaperone molecules such as BiP and calnexin (56), facilitating refolding of misfolded proteins (10). ATF6 expression has also been observed to upregulate transcription of Xbp-1 (55), indicating some cross talk between the two pathways.
Virus infection is a strong inducer of UPR signaling; however, some downstream effectors are not necessarily beneficial for viral replication, e.g., the induction of apoptosis or production of degradative proteins. As such, many viruses regulate the UPR to create an environment more favorable for replication. Studies with hepatitis C virus (HCV) have shown that both infection and expression of viral nonstructural (NS) proteins can stimulate ATF6 cleavage, chaperone upregulation, and protein translation (47), while suppressing the IRE-1/Xbp-1 arm of the UPR (46). Further work identified NS4B of HCV as a strong regulator of UPR signaling (59) which, interestingly, is the major membrane-inducing protein (11). Other members of the Flaviviridae family have also been shown to induce UPR components; Japanese encephalitis virus (JEV) and dengue virus (DENV) infection increase Xbp-1 signaling and induction of the downstream molecules EDEM-1, ERdj4, and p58(IPK) (58), which may be important in upregulating membrane biogenesis (45) for the formation of viral membrane structures. An increase in BiP (GRP78) expression was also observed in DENV-infected cells (50). In addition, studies with the New York 99 strain of West Nile virus (WNVNY99) have shown that all three pathways of the UPR were activated upon infection of neuronal cells; in particular, downstream apoptotic factors such as CHOP, GADD34, caspase-3, and PARP were upregulated and may play a role in limiting virus replication (37).
Recent studies have also demonstrated a link between UPR signaling and immune regulation, in particular the Jak-STAT cascade in response to type I interferon (IFN) stimulation. UPR induction by treatment with thapsigargin or other stress inducers was shown to cause serine phosphorylation of the type I IFN receptor (IFNAR), targeting it for ubiquitination and degradation by proteasomal components (28). The interactions between immune signaling and UPR activation have interesting ramifications for these pathways during virus infection, since they are both targets for viral manipulation during replication, and suggest a combined mechanism for viral regulation of cellular processes.
In the present study we show that the Kunjin strain of WNV (WNVKUN) induces a robust ER stress response that coincides with peak viral RNA and protein production. This response is particularly skewed toward ATF6 and IRE-1 activation, which results in high levels of Xbp-1 transcription and splicing. We also show that the hydrophobic, membrane-bound viral proteins NS4A and NS4B strongly induced Xbp-1 transcription and processing when individually expressed. We also demonstrate a correlation between UPR signaling and the inhibition of type I IFN signaling. Finally, we show that this cross talk between the UPR and IFN signaling pathways is regulated by the hydrophobicity of viral proteins, suggesting a general mechanism for the inhibition of immune signaling during WNV replication.
MATERIALS AND METHODS
Cells and virus stocks.
Vero C1008 cells were maintained in Dulbecco modified Eagle medium (DMEM; Gibco) supplemented with 5% fetal calf serum (FCS; Lonza), penicillin-streptomycin (100 IU/ml and 100 μg/ml, respectively; Gibco), and 200 μM Glutamax (Gibco). PERK knockout mouse embryonic fibroblasts (MEFs) were kindly provided by David Ron (New York University) and maintained in DMEM supplemented with penicillin-streptomycin and Glutamax as described above, plus 10% FCS, 1% nonessential amino acids (Gibco), and 1 μM 2-mercaptoethanol (Gibco). All cells were grown at 37°C in a 5% CO2 incubator. WNVKUN stocks were propagated from an existing MRM61C secondary stock in Vero C1008 cells at 37°C in DMEM supplemented with 0.2% (wt/vol) bovine serum albumin (BSA) for 32 h. After infection, the virus-containing supernatant was collected and centrifuged at 4,800 rpm for 5 min to remove cell debris, and then aliquots were stored at −80°C. The virus titer was determined by plaque assay as described below.
Antibodies and reagents.
Thapsigargin and tunicamycin (containing homologues A, B, C, and D; Sigma) were dissolved in dimethyl sulfoxide (DMSO; Sigma) at concentrations of 1 and 5 mM, respectively. Interferon-2α (IFN-2α) was sourced from Roche Pharmaceuticals and diluted in 0.1% BSA and phosphate-buffered saline (PBS). Mouse anti-NS1 and NS5 monoclonal antibodies were kindly provided by Roy Hall (University of Queensland). WNVKUN-specific rabbit anti-NS3 polyclonal antisera has been described previously (52). Mouse anti-dsRNA (clone J2) antibodies were purchased from English & Scientific Consulting Bt. (Hungary). Rabbit anti-eIF2a and rabbit anti-p-eIF2a [Ser51] polyclonal antibodies (Invitrogen) were kindly provided by Bryan Williams (Monash Institute of Medical Research). Rabbit anti-STAT1 polyclonal antibody was sourced from Santa Cruz, and mouse anti-p-STAT1[Tyr701] monoclonal antibody was obtained from BD Transduction Laboratories. The cell markers rabbit anti-BiP polyclonal antibody and rabbit anti-green fluorescent protein (anti-GFP) polyclonal antibody were obtained from Sigma and Invitrogen, respectively.
Plaque assay.
Vero C1008 cells were seeded in DMEM complete medium in six-well plates, followed by incubation at 37°C overnight. The virus stock was diluted 10-fold in 0.2% BSA-DMEM, and cells were infected with 300 μl of stock dilutions (in duplicate), followed by incubation at 37°C for 60 min. Then, 2 ml of a semisolid overlay containing 0.3% (wt/vol) low-melting-point agarose, 2.5% (wt/vol) FCS, penicillin-streptomycin, Glutamax, HEPES, and NaCO3 was added to the cells, and the mixture was solidified at 4°C for 30 min. The cells were incubated at 37°C for 3 days, fixed in 4% (vol/vol) formaldehyde (in PBS) for 1 h, and stained in 0.4% crystal violet (with 20% [vol/vol] methanol and PBS) at room temperature for 1 h. Plaques were manually counted, and the PFU per ml were determined.
GFP-NS fusion plasmid cloning.
Amplicons containing GFP fused to nonstructural (NS) proteins NS3, NS4A, NS4B, and NS5 were amplified from previously constructed plasmids (21) by using sequence-specific primers (see Table S1 in the supplemental material) and cloned into pcDNA3.1+ (Invitrogen) using the restriction enzymes EcoRV and XhoI (Promega). Clones were isolated from transformed Escherichia coli JM109 by using a Qiagen Hi-Speed Midiprep kit as indicated by the manufacturer. Prior to transfection, plasmid preparations were further purified by using phenol-chloroform extraction and re-eluted in 0.1× Tris-EDTA (10 mM Tris, 1 mM EDTA).
Infection and time-course experiments.
Vero cells were infected with WNVKUN at the indicated multiplicity of infection (MOI) in 300 μl of 0.2% BSA-DMEM for 60 min, after which the infection medium was further supplemented with 0.2% BSA-DMEM to a final volume of 1.5 ml. The cells were incubated at 37°C for the indicated time and then collected for further processing. For ER stress treatments, thapsigargin, tunicamycin, or the drug vehicle (DMSO) was added directly to cell media to a final concentration of 300 nM and incubated at 37°C until 24 h postinfection (hpi).
Transfection and cell sorting.
Vero cells were seeded into 60-mm dishes, transfected with plasmid DNA previously incubated with Lipofectamine 2000 or Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instructions, and incubated at 37°C for a further 18 h. Cells were collected using 0.25% trypsin-EDTA (Gibco), pelleted at 2,000 rpm, and washed in PBS twice. After the washes, the cells were resuspended in MACS buffer (PBS, 0.5% FCS, 2 mM EDTA) and sorted on a MoFlo fluorescence-activated cell sorting (FACS) according to GFP expression. Approximately 50,000 GFP-positive cells per sample were pooled, pelleted, and lysed in TRIzol reagent.
Western blotting.
Transfected or WNVKUN-infected cells were aspirated in PBS and then lysed in sodium dodecyl sulfate (SDS) lysis buffer (0.5% SDS, 1 mM EDTA, 50 mM Tris-HCl) or coimmunoprecipitation (COP) buffer (10 mM Tris, 150 mM NaCl, 5 mM EDTA, 1% NP-40) containing protease inhibitors leupeptin (1 μg/ml), phenylmethylsulfonyl fluoride (0.5 mM), and the phosphatase inhibitors sodium orthovanadate (25 mM), sodium fluoride (25 mM), and β-glycerophosphate (25 mM) (Sigma). Lysates were diluted in LDS loading buffer (Invitrogen), heated at 70°C for 5 min, and separated on a 4 to 12% Bis-Tris polyacrylamide gel (Invitrogen). Proteins were transferred to Hi-Bond ECL nitrocellulose membrane (Amersham Biosciences), and the membrane was blocked with 5% (wt/vol) skim milk (Diploma) in Tris-buffered saline with 0.05% Tween (PBS-T) or 5% (wt/vol) BSA (Sigma) in TBS-Tween (TBS-T). Primary antibodies were incubated at 4°C with membrane overnight in blocking solution as described above. After primary incubation, the membrane was washed in TBS-T and then incubated with secondary antibodies conjugated to either Cy5 (Amersham Biosciences) or Alexa Fluor 647 or Alexa Fluor 488 (Molecular Probes) in TBS-T at room temperature for 2 h. The membrane was washed twice in TBS-T and then in TBS, and the proteins were visualized on the Storm Fluorescent scanner (Amersham Biosciences) on either a 635-nm or a 430-nm emission channel.
Immunofluorescence.
The cells were fixed and permeabilized on coverslips with acetone-methanol (1:1) at −20°C for 5 min. The cells were washed twice in PBS and then incubated in primary antibodies diluted in 1% BSA-PBS at room temperature for 60 min. Cells were then washed twice in 0.1% BSA-PBS for 5 min, followed by incubation with secondary antibodies conjugated to either Alexa Fluor 488 or Alexa Fluor 594 (Molecular Probes) in 1% BSA-DMEM at room temperature for 45 min. Cells were washed in PBS then mounted on coverslides using Ultramount mounting media (Fronine). Immunofluorescent staining was visualized on a Leica confocal microscope and pictures assembled by using Adobe Photoshop.
RNA extraction, qPCR, and Xbp-1 splicing assays.
RNA was extracted from plasmid-transfected or WNVKUN-infected cells with TRIzol reagent (Invitrogen) as indicated by the manufacturer. Total RNA was then treated with RQ1 DNase (Promega) at 37°C for 30 min to remove any contaminating DNA. cDNA was synthesized with Superscript III reverse transcriptase (Invitrogen) with gene-specific reverse primers (see Table S1 in the supplemental material) at 50°C for 50 min. After heat inactivation at 65°C, reverse transcription (RT) reactions were diluted to 10-fold in diethyl pyrocarbonate-treated deionized water. cDNA levels were quantified by using quantitative PCR (qPCR) with Sybr GreenER (Invitrogen) on an ICycler PCR cycling machine (Bio-Rad). Primers were designed to internal control ribosomal protein 13A (RPL13A), UPR genes Xbp-1, EDEM-1, ATF4, and GADD34, as well as the WNVKUN genome (see Table S2 in the supplemental material). Fold induction of each gene was calculated by comparing threshold cycle values (CT) to the internal control RPL13A. Splicing of the Xbp-1 mRNA was studied by using RT-PCR across the splice site, followed by PstI (Promega) digestion. Briefly, RNA was treated with DNase as indicated above and then amplified with Xbp-1 primers using a Superscript III Platinum Taq One-Step PCR kit (Invitrogen) under the following conditions: 1 cycle of 50°C for 50 min and 94°C for 2 min; followed by 40 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min; followed by a final extension of 72°C for 10 min. Amplicons were directly digested with PstI at 37°C overnight, separated on a 2% agarose gel containing 50 ng of ethidium bromide/ml, and visualized by UV light.
RESULTS
WNVKUN replication induces UPR signaling.
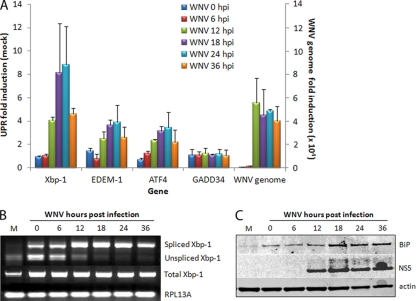
Many viruses have been observed to upregulate UPR signaling during replication and in particular manipulate downstream signaling to benefit virus replication. To investigate the extent of UPR signaling during WNVKUN replication, RNA and protein samples of infected cells were collected at different time points postinfection and analyzed for transcriptional and translation upregulation of UPR components by qPCR or Western blotting. As can be observed in Fig. 1A, the mRNA levels of Xbp-1, EDEM-1, and ATF4 were elevated in the WNVKUN-infected cells. The observed increase in UPR gene mRNA appeared to correlate with the increase in WNVKUN genomic RNA, observed at the end of the latent period (ca. 12 to 15 hpi). Accordingly, an increase in BiP protein levels was also observed (Fig. 1C). Xbp-1 mRNA splicing is used to assess activation of the IRE-1-mediated UPR response (58), and we observed an increase in Xbp-1 splicing from 12 hpi (Fig. 1B) that agreed with the increase in EDEM-1 mRNA expression observed in Fig. 1A. Interestingly, Xbp-1 mRNA expression was also significantly upregulated, up to 10-fold during replication, while both EDEM-1 and ATF4 transcription only increased 3-fold. This indicated a partial activation of all three UPR sensors, with particular emphasis on ATF6 activation, as shown by both BiP and Xbp-1 upregulation. Given the high levels of spliced (activated) Xbp-1 transcript, it would be expected that downstream transcriptional targets would be upregulated accordingly. However, EDEM-1 transcription was only modestly upregulated, suggesting that downstream UPR signaling mediated via IRE-1 activation is possibly manipulated by WNVKUN replication. A similar level of manipulation was also observed via PERK-mediated activation, whereupon although ATF4 transcription was modestly upregulated, downstream transcription of GADD34 was not significantly induced (Fig. 1A).
FIG. 1.
WNVKUN induces UPR signaling. Vero C1008 cells were infected with WNVKUN at an MOI of 3, and samples collected at 0, 6, 12, 18, 24, and 36 hpi. (A) RNA extracted from infected cells was quantified for upregulation of UPR genes Xbp-1, EDEM-1, ATF4, and GADD34 by using qPCR, and the fold induction was calculated compared to mock cells at the same time point. Error bars indicate +1 standard deviations from replicate assays of two independent experiments. (B) RNA samples from above were also analyzed for spliced Xbp-1 mRNA by using RT-PCR and endonuclease digestion as previously described (58). (C) Protein samples from the time course were analyzed by Western blotting for the synthesis of BiP compared to the infection control NS5 and the internal control actin.
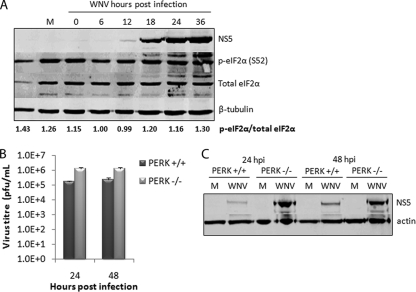
To investigate PERK-mediated signaling during WNVKUN replication further, we also analyzed the activation of eIF2α, which is phosphorylated by PERK on Ser51 (p-eIF2α) following BiP dissociation. Consistent with the qPCR data, p-eIF2α did not increase significantly throughout infection, as is shown by the quantitative ratio of p-eIF2α to total eIF2α (Fig. 2A). A slight increase in p-eIF2α was observed late in replication (36 hpi), although this could be attributed to higher levels of total protein.
FIG. 2.
WNVKUN does not induce or require PERK-mediated signaling for replication. (A) Vero C1008 cells were infected with WNVKUN at an MOI of 3, and protein samples collected at 0, 6, 12, 18, 24, and 36 hpi. Cell lysates were analyzed by Western blotting for p-eIF2α (Ser 51), total eIF2α, NS5, and the internal control tubulin. The ratio of p-eIF2α to eIF2α was quantified by using Bio-Rad Quantity One software. (B) PERK−/− and PERK+/+ MEFs were infected with WNVKUN at an MOI of 3 for 24 and 48 h, and supernatant and protein samples were collected. (C) Plaque assays were used to analyze the release of infectious virus, and Western blotting for NS5 was used to detect viral protein synthesis. Error bars indicate +1 standard deviations for replicate assays of two independent experiments.
Overall, downstream PERK signaling was not strongly induced throughout WNVKUN replication, and is suggested to be dispensable for efficient replication. To further assess this hypothesis, PERK−/− MEFs were infected with WNVKUN and analyzed for protein and virion production. Interestingly, replication was slightly enhanced in PERK−/− cells with up to 10-fold more infectious particles produced (Fig. 2B) and significantly higher viral protein levels (Fig. 2C) at 24 and 48 hpi. This suggests that overall UPR signaling is biased toward Xbp-1/ATF6 activation, with only minimal PERK signaling, during WNVKUN replication.
Overstimulation of the UPR limits virus replication.
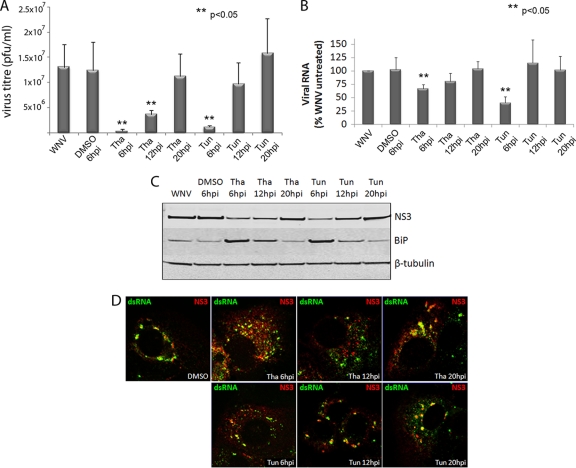
To further elucidate the effect of UPR signaling on WNVKUN replication, we induced the activation of this pathway using drugs that rapidly activate UPR signaling. Thapsigargin is a SERCA-pump inhibitor that causes the efflux of calcium from the ER, thus inhibiting calcium-dependent chaperones and increasing protein misfolding. Tunicamycin inhibits N-linked glycosylation within the ER, also inducing the UPR by increasing the accumulation of misfolded proteins in the lumen. Previous studies with human cytomegalovirus (HCMV) have shown that thapsigargin and tunicamycin treatment during replication severely retarded viral gene expression and infectious virion release (17). Given that UPR signaling was strongly induced during WNVKUN replication, infected cells were treated with thapsigargin or tunicamycin at various time points, and replication levels were assessed for viral RNA, protein, and infectious virions at 24 hpi. Treatment of uninfected cells with the drugs alone induced strong UPR signaling by 6 h, with transcriptional upregulation of Xbp-1, EDEM-1, and ATF4; Xbp-1 mRNA splicing; and increased translation of BiP by 12 h of treatment (see Fig. S1A to C in the supplemental material). Interestingly, chemical induction of the UPR after the latent period (i.e., 20 hpi) did not significantly alter virus replication, other than a slight upregulation of RNA production. However, UPR induction early during the infection cycle (i.e., 6 hpi) had a strong limiting effect on WNVKUN RNA, protein, and virus production (Fig. 3A to C), as well as disrupting colocalization of viral proteins with dsRNA (Fig. 3D). Interestingly, at 12 hpi chemical induction of the UPR had an intermediate effect on replication, moderately affecting RNA and protein levels. These observations suggest that induction of the UPR can regulate WNVKUN replication, if activated before the latent period when downstream effectors are able to limit virus replication. It is also interesting that thapsigargin in general had a stronger effect on virus replication than tunicamycin, perhaps also suggesting a role for ER calcium stores in virus replication.
FIG. 3.
Synthetic UPR activation during WNVKUN infection inhibits replication. Vero C1008 cells were infected with WNVKUN at an MOI of 3 and then treated with 300 nM tunicamycin, thapsigargin, or the drug vehicle (DMSO) at 6, 12, or 20 hpi. At 24 hpi, viral supernatants, protein, and RNA samples were collected, as well as fixed for immunofluorescence, and analyzed for viral replication. (A and B) Virus titers were determined by plaque assays (A), and RNA samples were analyzed for viral RNA expression by qPCR (compared to internal control RPL13A) (B). (C) Cell protein lysates were analyzed for viral protein expression and BiP upregulation by Western blotting, compared to the internal control actin. (D) Cells were fixed using acetone-methanol and labeled for NS3 (in red) and dsRNA (in green) to study formation of replication complexes after treatment. Error bars indicate +1 standard deviations of two independent experiments. Statistical analysis was performed by using the Student t test.
WNVKUN NS proteins differentially regulate UPR signaling.
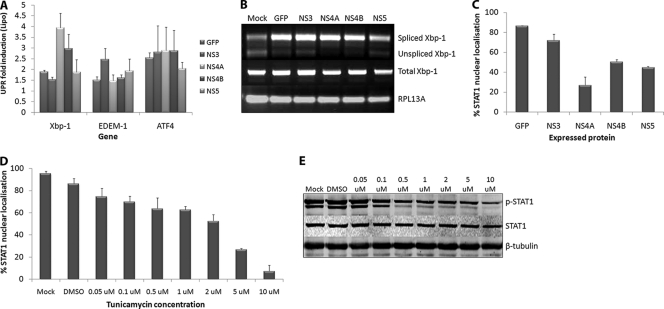
The manipulation of UPR signaling by virus replication has also been observed with the expression of individual viral proteins. NS4B of HCV was shown to exert similar effects on the UPR as in during infection with the HCV replicon (59), as did US11 of HCMV (48). We were interested in investigating whether any of the WNVKUN NS proteins was able to induce UPR signaling comparable to that observed during infection. NS3, NS4A, NS4B, and NS5 were fused to GFP and expressed in Vero cells (see Fig. S2 in the supplemental material) and then sorted according to GFP fluorescence to create a pool of transiently expressing cells. qPCR analyses of RNA collected from expressing cells showed that GFP alone did not significantly induce UPR transcriptional activation (Fig. 4A). However, expression of GFP-NS4A or NS4B-GFP significantly upregulated Xbp-1 transcription, whereas NS3-GFP expression induced EDEM-1 transcription. ATF4 transcription was modestly upregulated in all samples, perhaps suggesting a nonspecific response to the transfection procedure. RT-PCR studies also showed that all of the GFP fusion proteins induced some degree of Xbp-1 splicing, including the GFP control itself; however, only cells expressing GFP-NS4A or NS4B-GFP showed a complete absence of unspliced Xbp-1 (Fig. 4B). This is significant since several studies have shown that unspliced Xbp-1 is a potent inhibitor of UPR signaling and may act as negative feedback following IRE-1 activation (57). Interestingly, however, the production of spliced Xbp-1 in these cells did not result in EDEM-1 transcription as would be expected. We observed that EDEM-1 mRNA levels remained relatively low in all samples, suggesting that the modest EDEM-1 upregulation during WNVKUN replication may be due to the combined effects of all viral proteins, whereas the Xbp-1 activation could be attributed to the expression of GFP-NS4A alone.
FIG. 4.
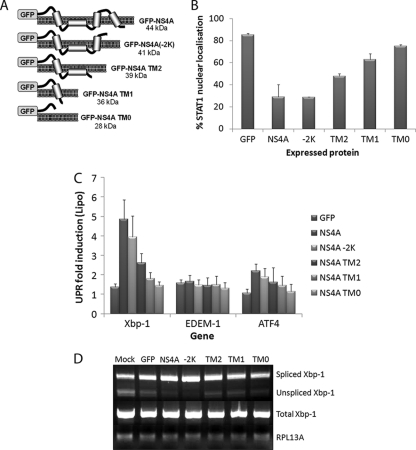
WNV NS proteins differentially regulate UPR and IFN signaling. Vero C1008 cells were transfected with GFP fusion constructs of NS3, NS4A, NS4B, and NS5 and expressed for 18 h. The cells were then sorted according to GFP expression, and a pool of GFP-positive cells was collected. A total of 5 × 104 cells were lysed, and extracted RNA was analyzed for the upregulation of UPR genes Xbp-1, EDEM-1, and ATF4 (A) and the presence of spliced Xbp-1 mRNA (B). Error bars indicate +1 standard deviations of replicate assays of two independent experiments. (C) Cells were also transfected with the above constructs, stimulated with 1,000 U of IFN-α (Roche)/ml at 24 h posttransfection (hpt) for 30 min, and then fixed and labeled with STAT1 and DAPI (4′,6′-diamidino-2-phenylindole) to detect nuclear trafficking. Approximately 100 expressing cells per sample were scored on nuclear or cytoplasmic localization. Error bars indicate +1 standard deviations from two independent experiments. Vero C1008 cells were treated with different concentrations of tunicamycin for 12 h at 37°C and then stimulated with 1,000 U of IFN-α (Roferon; Roche)/ml for 30 min at the specified tunicamycin concentration. The cells were then fixed for immunofluorescence or lysed for Western blot analysis. (D) Fixed cells were labeled with a STAT1 antibody, and the nuclear stain DAPI to detect nuclear localization. Approximately 100 cells per concentration were quantified for nuclear localization. Error bars indicate +1 standard deviations from two independent experiments. (E) Cell lysates were probed for phospho-STAT1 and STAT1 to detect STAT1 phosphorylation and compared to the internal control actin.
Expression of viral proteins has also been shown to regulate the type I IFN signaling and Jak-STAT pathways (16, 27, 29). In particular, the small hydrophobic proteins of WNV and DENV have been demonstrated to prevent STAT protein phosphorylation and nuclear trafficking (29, 40, 41). Since NS4A and NS4B also upregulated Xbp-1 signaling (Fig. 4A and B) and UPR signaling has been demonstrated to inhibit IFN responses (28), we hypothesized that these proteins elicit their effect on IFN signaling via UPR activation. The expression of GFP-NS4A resulted in a dramatic decrease in the percentage of cells that showed STAT1 nuclear trafficking (Fig. 4C) compared to GFP and NS3-GFP expression, which correlated with their ability to activate UPR signaling. Interestingly, NS4B-GFP and NS5-GFP also showed some inhibitory effects on STAT1 activation. The ability of NS4B and NS5 to prevent IFN signaling been previously reported (2, 25, 29, 40), although the mechanisms of action appear to vary with the different proteins and viruses. We also confirmed that UPR activation could inhibit IFN signaling in Vero cells (which are deficient in IFN-β and thus only respond to exogenously supplied IFN) by treating them with increasing amounts of tunicamycin and assessing STAT1 nuclear trafficking (Fig. 4D) and phosphorylation (Fig. 4E). This showed a dose-dependent effect of tunicamycin on IFN signaling, with concentrations above 1 μM showing strong inhibition of STAT1 phosphorylation and nuclear trafficking. Interestingly, STAT1 expression was unaffected, suggesting an alteration of phosphorylation, and not general translational inhibition is inhibiting downstream signaling.
Hydrophobic domains of viral proteins mediate UPR activation and IFN inhibition.
We have shown that the hydrophobic NS proteins (NS4A and NS4B) of WNVKUN are important in inducing and manipulating the UPR and in the inhibition of IFN signaling (29, 40). Given that we and others have shown that UPR activation can inhibit IFN signaling (28, 39), we suggest that the hydrophobic nature of these proteins induce the UPR and thus inhibit IFN signaling during replication. To investigate this further, we created mutants of GFP-NS4A (a highly hydrophobic NS protein with four transmembrane [TM] domains) (38), sequentially removing each from the C terminus of NS4A (Fig. 5A). These constructs were based on similar truncations used to determine the correct membrane topology of the DENV NS4A protein (38). These were expressed in Vero C1008 cells (see Fig. S3 in the supplemental material) and analyzed for UPR activation and IFN inhibition. Figure 5B shows that a progressive reduction in the hydrophobicity of NS4A actually restores IFN signaling (as measured by STAT1 nuclear localization), almost to the background effect of GFP expression alone. Consistent with our hypothesis, the sequential TM deletions of GFP-NS4A also resulted in reduced UPR signaling. Expression of GFP-NS4A and GFP-NS4A(−2K) strongly induced Xbp-1 splicing (Fig. 5D) as well as Xbp-1 transcription (Fig. 5C). Sequential removal of the hydrophobic regions of NS4A then showed a stepwise decrease in Xbp-1 mRNA levels and splicing. Accordingly, cells expressing the cytoplasmic N terminus of NS4A (NS4A TM0) exhibited similar levels of Xbp-1 activation as GFP expression alone. Transcript levels of EDEM-1 remained low in all samples, while ATF4 transcription was only slightly upregulated, similar to NS4A/NS4B expression. Overall, expression of GFP-NS4A(−2K) resulted in similar levels of Xbp-1 activation and IFN inhibition as full-length GFP-NS4A. This correlates with earlier studies which suggest that NS4A(−2K) is the major species of NS4A present during infection (due to a cleavage site upstream of the C-terminal hydrophobic domain) and thus suggests a particular role for this protein during replication, in addition to its role in remodeling cellular membranes (38, 43). The remaining mutants (TM2, TM1, and TM0) show a progressive restoration or activation of IFN and UPR signaling, respectively, showing that the decrease in hydrophobicity, not a specific domain of NS4A, is responsible for the manipulation of these two pathways.
FIG. 5.
Deletion of TM regions from NS4A results in decreased regulation of UPR and IFN signaling. (A) TM deletion mutants of NS4A were created (based upon TM predictions by Miller et al. [38]) with GFP at the N terminus. Vero C1008 cells were transfected with full-length NS4A and the TM mutants (as well as a GFP control). (B) At 24 hpt, the cells were fixed and labeled with STAT1 and the nuclear stain DAPI to detect nuclear localization in expressing cells. Approximately 100 cells for each fusion protein were scored for STAT localization as either complete nuclear, partial nuclear, or cytoplasmic localization, and percentage nuclear trafficking calculated. Error bars indicate +1 standard deviations from duplicate experiments. (C) At 18 hpt, cells were collected in MACS buffer and sorted by using FACS to create a pool of expressing cells, and then 5 × 104 cells were lysed for RNA extraction and qPCR analyses. RNA was analyzed for the upregulation of Xbp-1, EDEM-1, and ATF4 compared to the internal control RPL13A. Error bars indicate +1 standard deviations from duplicate experiments. (D) RNA samples were also analyzed for splicing of the Xbp-1 mRNA.
DISCUSSION
Viral manipulation of UPR signaling has been well documented in the literature. Nonbeneficial downstream effects, such as inhibition of translation initiation, apoptosis induction, and ER-associated degradation are proposed to be regulated to allow increased replication during HCMV and HCV infection (18, 46, 47). We show here that WNVKUN mediates a similar effect, skewing the UPR toward Xbp-1 transcription and mRNA splicing and chaperone production while only modestly inducing ATF4 and EDEM-1 transcription and PERK-mediated eIF2α phosphorylation. We also observed an increase in WNVKUN replication in PERK−/− MEFs, suggesting that while signaling through PERK is controlled during replication, it may also have a limiting effect early in replication. Studies with vesicular stomatitis virus have also shown increased replication in PERK−/− MEFs (3), which was attributed to the prevention of translation inhibition and induction of apoptosis. Our results conflict with earlier UPR studies with the pathogenic WNVNY99 strain, which showed that all three arms, in particular PERK and downstream apoptotic mediators, were activated during replication (37). We believe these differences may be due to different strains utilized (i.e., pathogenic [NY99] versus attenuated [KUNV]), cell types (neuronal versus epithelial), and time points used in the respective studies.
In the present study we observed that UPR manipulation was most prominent at 18 to 24 hpi, when viral protein and RNA synthesis is greatest. This is a paradoxical observation; the increasing levels of viral proteins are activating the UPR as the viral polyprotein is translated and processed in the ER membrane; however, the same proteins are in turn modulating downstream signaling to regulate PERK but activate Xbp-1-associated signaling. This correlates with data from Yoshida et al. (54), who showed that the ATF6 (and hence Xbp-1 and BiP expression) arm is upregulated earlier in the UPR, to attempt to fold misfolded proteins before committing to ER-associated degradation (via IRE-1 signaling) (54). This shift between ATF6 and IRE-1 activation is also observed in WNVKUN-infected cells with Xbp-1 transcription significantly upregulated before EDEM-1 or GADD34. However, later in infection, the virus is then able to exert its own effect on the pathway and regulate any further signaling through to ER-associated degradation.
The strong upregulation of Xbp-1 transcription and splicing in WNVKUN-infected cells suggests a particular role for Xbp-1 in replication. Recent independent studies by Sriburi et al. (45) and Shaffer et al. (44) have demonstrated that the active transcription factor Xbp-1 can induce both phospholipid biosynthesis enzymes and proliferation of cellular membranes, respectively (44, 45). This would be highly beneficial for WNVKUN replication since the ER is a source of membranes for formation of viral membrane structures such as convoluted membranes and vesicle packets (32, 35). The spliced form of Xbp-1 is also able to upregulate both chaperone molecules and enzymes involved in ER-associated degradation. However, the degradative effectors downstream of Xbp-1 were not significantly upregulated in infected cells, suggesting that WNVKUN replication manipulates Xbp-1 to upregulate membrane proliferation but prevent ER-associated degradation signaling. Interestingly, Yu et al. (58) showed that reducing Xbp-1 expression in JEV and DENV-infected cells did not greatly affect virus release but did make cells more susceptible to virus-induced cytopathic effects (58). It could be implied that Xbp-1-associated signaling may protect cells from ER stress-induced apoptosis and thus is upregulated in WNVKUN-infected cells to allow further replication.
Although we demonstrated that the differential regulation of the UPR by WNVKUN may benefit replication, the exact mechanism(s) remained unknown. To further study this, the UPR was overstimulated by ER stress drugs thapsigargin and tunicamycin during infection, reasoning that the overall effect on replication may be determined. Interestingly, general UPR signaling elicited by these drugs was only inhibitory at early stages of replication (6 to 12 hpi). This was interesting for a number of reasons. (1). The detrimental aspects described earlier (translational inhibition and ERAD) had a stronger inhibitory effect than the positive contributions of the more beneficial components (chaperone production and membrane biosynthesis) of UPR signaling. This could not be directly contributed to the inhibition of translation because RNA replication and association of replicative components were also affected. In fact, colocalization of replication components (presumably with virus-induced membranes) was completely ablated in cells treated before induction of peak replication. (2). The levels of protein and RNA synthesis were examined to determine whether synthetic UPR induction was able to affect virus replication. Therefore, at later stages of replication (12 to 24 hpi) when protein and RNA levels peaked, the addition of thapsigargin or tunicamycin to infected cells had little or no effect. Indeed, treatment at 20 hpi resulted in marginally higher genomic RNA and virion production. One explanation for this is that by 18 hpi, viral proteins (such as NS4A) are expressed at high enough levels to modulate UPR signaling as described earlier. Also, by this time viral membrane structures have formed and most likely protect viral replication components from external interference. Overall, these experiments suggest that UPR signaling can have a limiting effect on WNV replication, but only in the early stages of replication when the level of viral components is not enough to exert significant effects on downstream effectors.
Many viral proteins are able to individually regulate UPR signaling in the absence of virus replication. A good example of this is NS4B of HCV; expression of this hydrophobic protein causes splicing of Xbp-1 but not downstream transcription of EDEM-1 (59), similar to the modulation observed in HCV- and WNVKUN-infected cells. Expression of GFP-fusion WNV proteins in cells also initiated differential effects on UPR signaling. Unexpectedly, GFP alone elicited a modest response, suggesting that transfection and ectopic expression of a foreign protein can induce some cellular stress responses. However, expression of GFP-NS4A and to a lesser extent NS4B-GFP significantly upregulated Xbp-1 transcription and splicing to a greater extent than the GFP control. Similar to UPR signaling observed during infection, neither EDEM-1 nor ATF4 were strongly upregulated, suggesting that the hydrophobic proteins of WNVKUN are responsible for the manipulation of UPR signaling observed during infection. The parallels observed between HCV and WNVKUN are remarkable, for not only are NS4B and NS4A (of HCV and WNVKUN, respectively) mainly responsible for UPR signaling in infected cells, but they can also induce membrane proliferation and recruitment for virus replication (11, 43). This supports the theory that viral control of the UPR can aid in the production of membrane-associated replication complexes and that these membranes can in part be induced by Xbp-1 signaling, which has links to membrane proliferation. However, we cannot discount that a portion of the expressed proteins may be misfolded and promote induction of the UPR, although we have observed a selective activation of the ER sensors rather than a global induction of the UPR with the individual proteins, suggesting that each is affecting the system differently rather a general cellular response.
There are many reports of cross talk between UPR signaling and other stress pathways, in particular with the downstream effectors of PERK, such as amino acid depletion (via GCN2), dsRNA detection (via PKR), and oxidative stress pathways. Interestingly, studies have also shown an association of the antiviral response (via type I IFN) with UPR signaling, where induction of the UPR can lead to hyperphosphorylation and ubiquitin-mediated degradation of the IFNARs (28). This prompted the investigation of this interaction during virus replication, as we and others have shown that similar WNVKUN NS proteins are able to regulate both the IFN and UPR signaling pathways. We showed that the highly hydrophobic NS4A and NS4B can manipulate the UPR, skewing the response toward Xbp-1 signaling. Correspondingly, these proteins, together with NS2B (also hydrophobic), are also strong inhibitors of the late IFN signaling pathway (29). We hypothesized that the hydrophobic domains of these proteins are able to manipulate the UPR, which in turn then inhibits IFN signaling, as modeled in Fig. 6. Expression of NS4A mutants with sequential hydrophobic deletions confirmed this hypothesis, since we showed that decreasing the hydrophobicity of viral proteins caused a concomitant decrease in UPR (Xbp-1) signaling and restoration of Jak-STAT signaling. The exact mechanisms via which these hydrophobic regions exert their effects are not yet understood. Given that these proteins are also associated with ER membranes, a direct interaction between viral hydrophobic proteins and the UPR sensors (also membrane bound) could be proposed, allowing direct control over UPR signaling and perhaps downstream targets.
FIG. 6.
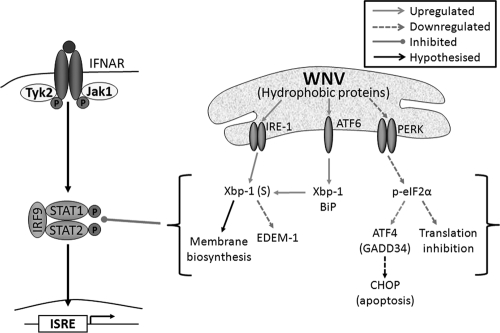
WNV replication and/or hydrophobic NS protein expression regulates the UPR in order to inhibit Jak-STAT signaling. WNV replication and/or expression of hydrophobic proteins (NS2B, NS4A, and NS4B) regulates UPR signaling to benefit replication. The ATF6/IRE-1 pathways are strongly activated, resulting in Xbp-1 transcription and splicing which may aid in the proliferation of ER membranes for the replication complex. In contrast, the PERK arm of the UPR is not strongly induced, potentially avoiding inhibitory downstream effects, such as translation inhibition and apoptosis, which may limit replication. Downstream UPR signaling during WNV replication can also inhibit STAT1 phosphorylation and nuclear trafficking; however, this is dependent on expression of hydrophobic proteins, since the removal of hydrophobic domains resulted in decreased Xbp-1 activation and the restoration of STAT1 signaling.
We also showed a progressive restoration of IFN signaling and decrease in Xbp-1 signaling, rather than a sudden change in signaling, as each transmembrane domain was removed. This negates the possibility that a single domain in NS4A (or other viral hydrophobic proteins) is responsible for the manipulation of these two signaling pathways, suggesting instead that the general hydrophobic nature of viral proteins is sufficient. Since membrane proteins also have a higher tendency to aggregate and/or misfold in the ER, a broad mechanism of manipulation could be proposed (as depicted in Fig. 6), via which overexpression of any hydrophobic protein may induce a partial UPR, thus allowing manipulation of other pathways by a myriad of viral proteins. This hypothesis has not yet been explored in current literature; however, there are examples of hydrophobic proteins inhibiting STAT phosphorylation and nuclear translocation. The herpes simplex virus type 1 immediate-early protein ICP27 has been shown to inhibit STAT phosphorylation both during infection and when expressed individually (19). This protein also has a hydrophobic C terminus (20) which may be inducing UPR signaling. In addition, Marburg virus VP40 is a hydrophobic, peripheral membrane protein (23) that has also been demonstrated to inhibit STAT1 and Jak1 phosphorylation (49). Interestingly, our data also showed that NS5 (a cytoplasmic protein) is also able to modestly inhibit IFN signaling but does not elicit significant UPR signaling, suggesting an alternative mechanism by which the antiviral response may be regulated (5). It would be beneficial to virus replication to have numerous regulatory mechanisms of overlapping pathways, thus ensuring control over host cell components during replication.
The exact mechanisms by which WNVKUN can manipulate different aspects of UPR are still to be determined. Given the differential effects of the individual nonstructural proteins on separate pathways, a direct interaction of these proteins (in particular NS4A) with downstream components could be hypothesized. In general, however, it can be shown that WNVKUN and possibly other viruses not only induce UPR signaling as a consequence of viral protein expression and processing but also utilize this stress response as a means of controlling a myriad of other cellular pathways, such as membrane biosynthesis, apoptosis signaling, and host immune responses.
Supplementary Material
Acknowledgments
We thank David Ron (New York University) for kindly providing PERK−/− and PERK+/+ MEFs and Roy Hall for his generous contribution of antibodies. We also thank Roleen Lata (Ludwig Centre for Cancer Research) and Geza Paukovics (Macfarlane Burnett Institute for Public Health) for their assistance with the FACS analysis.
This study was supported by NHMRC project grant 511123 awarded to J.M.M.
Footnotes
Published ahead of print on 29 December 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Anthony, T. G., B. C. McGrath, D. R. Cavener, and R. C. Wek. 2003. The eIF2a kinase GCN2 is necessary for adaptation to dietary essential amino acid (EAA) deprivation in growing mice. FASEB J. 17:A811-A811. [Google Scholar]
- 2.Ashour, J., M. Laurent-Rolle, P. Y. Shi, and A. Garcia-Sastre. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 83:5408-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baltzis, D., et al. 2004. Resistance to vesicular stomatitis virus infection requires a functional cross talk between the eukaryotic translation initiation factor 2 alpha kinases PERK and PKR. J. Virol. 78:12747-12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernales, S., F. R. Papa, and P. Walter. 2006. Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 22:487-508. [DOI] [PubMed] [Google Scholar]
- 5.Best, S. M., et al. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 79:12828-12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brush, M. H., D. C. Weiser, and S. Shenolikar. 2003. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1α to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol. Cell. Biol. 23:1292-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calfon, M., et al. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92-96. [DOI] [PubMed] [Google Scholar]
- 8.Dar, A. C., T. E. Dever, and F. Sicheri. 2005. Higher-order substrate recognition of elF2α by the RNA-dependent protein kinase PKR. Cell 122:887-900. [DOI] [PubMed] [Google Scholar]
- 9.Dorner, A. J., L. C. Wasley, and R. J. Kaufman. 1989. Increased synthesis of secreted proteins induces expression of glucose-regulated proteins in butyrate-treated Chinese hamster ovary cells. J. Biol. Chem. 264:20602-20607. [PubMed] [Google Scholar]
- 10.Du, Z., et al. 2005. Inhibition of IFN-α signaling by a PKC- and protein tyrosine phosphatase SHP-2-dependent pathway. Proc. Natl. Acad. Sci. U. S. A. 102:10267-10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egger, D., et al. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez, J., I. Yaman, P. Sarnow, M. D. Snider, and M. Hatzoglou. 2002. Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2α. J. Biol. Chem. 277:19198-19205. [DOI] [PubMed] [Google Scholar]
- 13.Groenendyk, J., and M. Michalak. 2005. Endoplasmic reticulum quality control and apoptosis. Acta Biochim. Polonica 52:381-395. [PubMed] [Google Scholar]
- 14.Harding, H. P., et al. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11:619-633. [DOI] [PubMed] [Google Scholar]
- 15.He, B. 2006. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 13:393-403. [DOI] [PubMed] [Google Scholar]
- 16.Heim, M. H., D. Moradpour, and H. E. Blum. 1999. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J. Virol. 73:8469-8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isler, J. A., T. G. Maguire, and J. C. Alwine. 2005. Production of infectious human cytomegalovirus virions is inhibited by drugs that disrupt calcium homeostasis in the endoplasmic reticulum. J. Virol. 79:15388-15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isler, J. A., A. H. Skalet, and J. C. Alwine. 2005. Human cytomegalovirus infection activates and regulates the unfolded protein response. J. Virol. 79:6890-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, K. E., B. Song, and D. M. Knipe. 2008. Role for herpes simplex virus 1 ICP27 in the inhibition of type I interferon signaling. Virology 374:487-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, M. A., C. T. Prideaux, K. Kongsuwan, S. G. Tyack, and M. Sheppard. 1995. Icp27 immediate-early gene, glycoprotein-K (Gk) and DNA helicase homologs of infectious laryngotracheitis virus (Gallid herpesvirus-1) Sa-2 strain. Arch. Virol. 140:623-634. [DOI] [PubMed] [Google Scholar]
- 21.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimata, Y., D. Oikawa, Y. Shimizu, Y. Ishiwata-Kimata, and K. Kohno. 2004. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire 1. J. Cell Biol. 167:445-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolesnikova, L., H. Bugany, H. D. Klenk, and S. Becker. 2002. VP40, the matrix protein of Marburg virus, is associated with membranes of the late endosomal compartment. J. Virol. 76:1825-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koumenis, C., et al. 2002. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2α. Mol. Cell. Biol. 22:7405-7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laurent-Rolle, M., et al. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J. Virol. 84:3503-3515. [DOI] [PMC free article] [PubMed]
- 26.Lee, A. H., N. N. Iwakoshi, and L. H. Glimcher. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23:7448-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, R. J., B. L. Chang, H. P. Yu, C. L. Liao, and Y. L. Lin. 2006. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 80:5908-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, J., et al. 2009. Virus-induced unfolded protein response attenuates antiviral defenses via phosphorylation-dependent degradation of the type I interferon receptor. Cell Host Microbe 5:72-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, W. J., et al. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 79:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma, Y. J., J. W. Brewer, J. A. Diehl, and L. M. Hendershot. 2002. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J. Mol. Biol. 318:1351-1365. [DOI] [PubMed] [Google Scholar]
- 31.Ma, Y. J., and L. M. Hendershot. 2003. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J. Biol. Chem. 278:34864-34873. [DOI] [PubMed] [Google Scholar]
- 32.Mackenzie, J. M., M. K. Jones, and E. G. Westaway. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 73:9555-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackenzie, J. M., M. T. Kenney, and E. G. Westaway. 2007. West Nile virus strain Kunjin NS5 polymerase is a phosphoprotein localized at the cytoplasmic site of viral RNA synthesis. J. Gen. Virol. 88:1163-1168. [DOI] [PubMed] [Google Scholar]
- 34.Mackenzie, J. M., A. A. Khromykh, M. K. Jones, and E. G. Westaway. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245:203-215. [DOI] [PubMed] [Google Scholar]
- 35.Mackenzie, J. M., A. A. Khromykh, and R. G. Parton. 2007. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2:229-239. [DOI] [PubMed] [Google Scholar]
- 36.McCracken, A. A., and J. L. Brodsky. 2005. Recognition and delivery of ERAD substrates to the proteasome and alternative paths for cell survival, p. 17-40. Curr. Top. Microbiol. Immunol. 300:17-40. [DOI] [PubMed] [Google Scholar]
- 37.Medigeshi, G. R., et al. 2007. West Nile virus infection activates the unfolded protein response, leading to CHOP induction and apoptosis. J. Virol. 81:10849-10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, S., S. Kastner, J. Krijnse-Locker, S. Buhler, and R. Bartenschlager. 2007. Non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 282:8873-8882. [DOI] [PubMed] [Google Scholar]
- 39.Minakshi, R., et al. 2009. The SARS coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One 4:e8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munoz-Jordan, J. L., et al. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. U. S. A. 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romisch, K. 2005. Endoplasmic reticulum-associated degradation. Annu. Rev. Cell Dev. Biol. 21:435-456. [DOI] [PubMed] [Google Scholar]
- 43.Roosendaal, J., E. G. Westaway, A. Khromykh, and J. M. Mackenzie. 2006. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J. Virol. 80:4623-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaffer, A. L., et al. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21:81-93. [DOI] [PubMed] [Google Scholar]
- 45.Sriburi, R., S. Jackowski, K. Mori, and J. W. Brewer. 2004. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J. Cell Biol. 167:35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tardif, K. D., K. Mori, R. J. Kaufman, and A. Siddiqui. 2004. Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. J. Biol. Chem. 279:17158-17164. [DOI] [PubMed] [Google Scholar]
- 47.Tardif, K. D., K. Mori, and A. Siddiqui. 2002. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J. Virol. 76:7453-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tirosh, B., et al. 2005. Human cytomegalovirus protein US11 provokes an unfolded protein response that may facilitate the degradation of class I major histocompatibility complex products. J. Virol. 79:2768-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valmas, C., et al. 2010. Marburg virus evades interferon responses by a mechanism distinct from Ebola virus. PLoS Pathog. 6:e1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wati, S., et al. 2009. Dengue virus infection induces upregulation of GRP78, which acts to chaperone viral antigen production. J. Virol. 83:12871-12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westaway, E. G., A. A. Khromykh, M. T. Kenney, J. M. Mackenzie, and M. K. Jones. 1997. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology 234:31-41. [DOI] [PubMed] [Google Scholar]
- 52.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye, J., et al. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6:1355-1364. [DOI] [PubMed] [Google Scholar]
- 54.Yoshida, H., et al. 2003. A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell 4:265-271. [DOI] [PubMed] [Google Scholar]
- 55.Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881-891. [DOI] [PubMed] [Google Scholar]
- 56.Yoshida, H., et al. 2000. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20:6755-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshida, H., A. Uemura, and K. Mori. 2009. pXBP1(U), a negative regulator of the unfolded protein response activator pXBP1(S), targets ATF6 but not ATF4 in proteasome-mediated degradation. Cell Struct. Funct. 34:1-10. [DOI] [PubMed] [Google Scholar]
- 58.Yu, C. Y., Y. W. Hsu, C. L. Liao, and Y. L. Lin. 2006. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J. Virol. 80:11868-11880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng, Y., et al. 2005. Hepatitis C virus nonstructural protein NS4B can modulate an unfolded protein response. J. Microbiol. 43:529-536. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.