Abstract
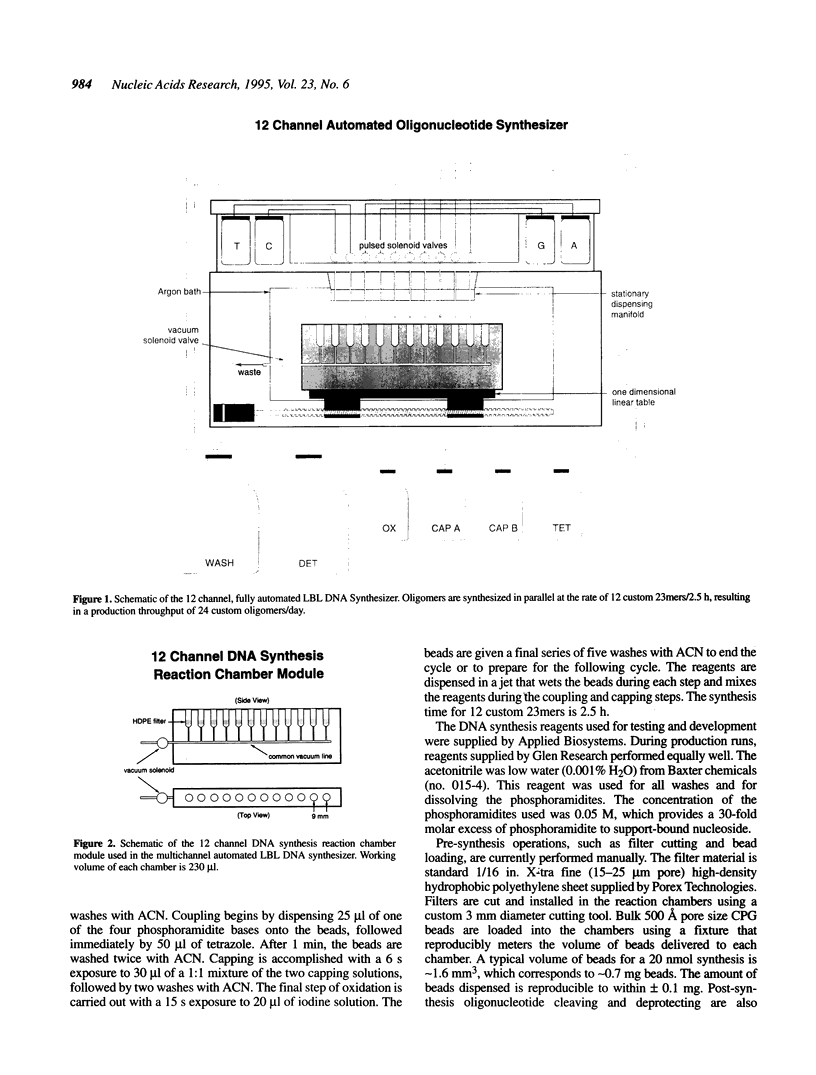
We describe an approach to high-throughput parallel DNA synthesis in which a multiwell format is used. The reactions are carried out in open wells using an argon ambient atmosphere to prevent reagent contamination. The controlled-pore glass beads which form the substrate for synthesis are held in individual wells with high-density polyethylene filter bottoms through which reagents are drawn into a vacuum manifold. The synthesis is carried out using direct reagent dispensing into the individual reaction wells. A computer controls the sequence in which reagents are dispensed and the timing of the periodic vacuum pulses required to synthesize the desired sequence. Experiments to date have demonstrated the viability of the approach for a variety of test sequences. Results obtained with HPLC analysis demonstrate coupling efficiencies as high as 99.5% under optimized conditions. Use of the oligomers for DNA sequencing templates and as PCR primers has been demonstrated in production applications. The current instrument design consists of a series of discrete reaction chambers in a 12 channel module which can be multiplexed in a 12 x n format where n can be 1-8, i.e. 96 wells. A projected time interval for 12 parallel syntheses is 2.5 h, with 96 syntheses in 3.5 h. Because of the reduced volume of reagents required in the open well format, significant cost savings are projected.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Itakura K., Rossi J. J., Wallace R. B. Synthesis and use of synthetic oligonucleotides. Annu Rev Biochem. 1984;53:323–356. doi: 10.1146/annurev.bi.53.070184.001543. [DOI] [PubMed] [Google Scholar]
- Olson M., Hood L., Cantor C., Botstein D. A common language for physical mapping of the human genome. Science. 1989 Sep 29;245(4925):1434–1435. doi: 10.1126/science.2781285. [DOI] [PubMed] [Google Scholar]
- Richard C. W., 3rd, Cox D. R., Kapp L., Murnane J., Cornelis F., Julier C., Lathrop G. M., James M. R. A radiation hybrid map of human chromosome 11q22-q23 containing the ataxia-telangiectasia disease locus. Genomics. 1993 Jul;17(1):1–5. doi: 10.1006/geno.1993.1275. [DOI] [PubMed] [Google Scholar]
- Shortle D., DiMaio D., Nathans D. Directed mutagenesis. Annu Rev Genet. 1981;15:265–294. doi: 10.1146/annurev.ge.15.120181.001405. [DOI] [PubMed] [Google Scholar]
- Weber J. L. Informativeness of human (dC-dA)n.(dG-dT)n polymorphisms. Genomics. 1990 Aug;7(4):524–530. doi: 10.1016/0888-7543(90)90195-z. [DOI] [PubMed] [Google Scholar]
- Weber J. L., May P. E. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet. 1989 Mar;44(3):388–396. [PMC free article] [PubMed] [Google Scholar]