Abstract
Gammaherpesviruses encode numerous immunomodulatory molecules that contribute to their ability to evade the host immune response and establish persistent, lifelong infections. As the human gammaherpesviruses are strictly species specific, small animal models of gammaherpesvirus infection, such as murine gammaherpesvirus 68 (γHV68) infection, are important for studying the roles of gammaherpesvirus immune evasion genes in in vivo infection and pathogenesis. We report here the genome sequence and characterization of a novel rodent gammaherpesvirus, designated rodent herpesvirus Peru (RHVP), that shares conserved genes and genome organization with γHV68 and the primate gammaherpesviruses but is phylogenetically distinct from γHV68. RHVP establishes acute and latent infection in laboratory mice. Additionally, RHVP contains multiple open reading frames (ORFs) not present in γHV68 that have sequence similarity to primate gammaherpesvirus immunomodulatory genes or cellular genes. These include ORFs with similarity to major histocompatibility complex class I (MHC-I), C-type lectins, and the mouse mammary tumor virus and herpesvirus saimiri superantigens. As these ORFs may function as immunomodulatory or virulence factors, RHVP presents new opportunities for the study of mechanisms of immune evasion by gammaherpesviruses.
A hallmark of gammaherpesvirus biology is the ability of the virus to evade the host immune response and establish persistent, lifelong infections. To accomplish this, gammaherpesviruses encode numerous molecules that enable them to escape, inhibit, or subvert multiple facets of the immune system. These immune evasion strategies include impairment of immune cell function, downregulation of antigen presentation, interference with cytokine and chemokine responses and signaling, inhibition of the complement pathway, hijacking of the DNA damage response pathway, and blockade of apoptosis and autophagy (76; reviewed in references 44 and 87). The human gammaherpesviruses, Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV), are associated with several malignancies, including Burkitt's lymphoma, Hodgkin's disease, and nasopharyngeal carcinoma for EBV (37, 62) and Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma for KSHV (52). Because these viruses are strictly species specific, however, it has been helpful to evaluate the contributions of virus-encoded immunomodulatory molecules to gammaherpesvirus pathogenesis using rodent models. Since the full spectrum of immune evasion strategies cannot be expected to be utilized by any single virus, it is useful to identify viruses that are experimentally manipulable, including via analysis of infection in laboratory mice, and that contain potentially novel immunomodulatory genes.
Studies of in vivo gammaherpesvirus infection and pathogenesis have relied on small animal model systems, particularly the mouse pathogen murine gammaherpesvirus 68 (γHV68; also known as MHV-68 or MuHV-4). γHV68 carries several immune evasion genes that are conserved in primate gammaherpesviruses, including an E3 ubiquitin ligase called K3, a complement regulatory protein, the open reading frame 36 (ORF 36) protein kinase, and homologs of cyclin D, Bcl-2, and a G protein-coupled receptor (84). The study of these genes during in vivo γHV68 infection has identified important roles for them in the establishment and maintenance of latency, persistent replication, reactivation, virulence, and induction of lymphoproliferative disease (21, 28, 34, 42, 53, 73, 75, 81, 82). In addition, γHV68 carries multiple unique ORFs, designated M ORFs, which are not shared with other gammaherpesviruses. Although many of the M ORFs do not have significant sequence similarity to other known viral or cellular genes, several have been found to have interesting immunomodulatory functions. M2 drives B cell proliferation and differentiation and has been shown to interfere with cellular pathways in several ways, including activating Vav1 and Vav2, downregulating STAT1 and STAT2, and inhibiting induction of apoptosis through the DNA damage pathway (45, 46, 63, 66). M1 stimulates and drives expansion of Vβ4+ CD8 T cells (17), and M3 functions as a chemokine decoy receptor that binds to a broad spectrum of chemokines and disrupts chemokine signaling (57, 80). Thus, the study of γHV68 as a mouse model for gammaherpesvirus infection has yielded insight into the roles of homologs of primate gammaherpesvirus immune evasion genes in in vivo infection and pathogenesis and has resulted in the discovery of novel immunomodulatory molecules and/or mechanisms employed by gammaherpesviruses.
We report here the characterization and genome sequence of a new rodent gammaherpesvirus, designated rodent herpesvirus Peru (RHVP), that was isolated from a lung homogenate of a pygmy rice rat (Oligoryzomys microtis) trapped in July 1996 in Peru. RHVP is related to previously identified rhadinoviruses but is distinct from γHV68. Like γHV68, RHVP carries numerous ORFs that are conserved across gammaherpesviruses. However, RHVP additionally carries several unique ORFs that have sequence similarity to primate gammaherpesvirus genes or to other viral or cellular genes but are not present in γHV68. As RHVP establishes both acute and latent infections in laboratory mice, the availability of this virus for study presents a new opportunity to examine the function of gammaherpesvirus genes during in vivo infection and potentially to identify novel mechanisms of immune evasion by gammaherpesviruses.
MATERIALS AND METHODS
Virus isolation and growth.
Vero cells and mouse embryonic fibroblasts (MEFs) were cultured in Dulbecco's modified Eagle medium (DMEM) containing 5% or 10% fetal calf serum (FCS), respectively (D5 or D10). RHVP was grown in Vero cells, and virus stocks were generated from infected-cell lysates subjected to two rounds of centrifugation to remove cell debris. Titers of the virus stocks were determined by a standard plaque assay using an adaptation of a previously described method (89), as follows. Plaque assays were done using Vero cells, and infected cells were overlaid with 0.5% noble agar in minimal essential medium (MEM) containing 10% FCS. At 7 days postinfection (dpi), cell monolayers were fixed with 2% paraformaldehyde, the noble agar was removed, and the monolayers were stained with 0.1% crystal violet to visualize viral plaques. For the generation of RHVP-L and RHVP-E isolates by limiting dilution cloning, serial 10-fold dilutions of RHVP were plated onto Vero cells in 96-well plates and allowed to incubate until cytopathic effect (CPE) developed. Infected-cell samples were collected from wells plated with a dilution of virus at which less than 40% of the total wells developed the CPE. These samples were subjected to a second round of limiting dilution cloning, and a single isolate of each was expanded in Vero cells to generate virus stocks, as described above.
Sequencing and assembly of the viral genome.
Viral genomic DNA was purified for sequencing as previously described (84), with the following modifications. Vero cells were infected with RHVP, and cell lysates were harvested when extensive CPE was observed, approximately 3 to 4 dpi. Following precipitation and resuspension of purified DNA, a sample was run on a low-melting-point agarose gel, and a large, distinct band corresponding to viral genomic DNA was gel purified by phenol extraction followed by ethanol precipitation. The resulting viral genomic DNA sample was sheared, size selected and adaptor ligated, and sequenced at the Washington University Genome Sequencing Center on the 454 GS-FLX platform (454 Life Sciences) according to the manufacturer's instructions.
We obtained 123,297 unique reads with an average read length of 399 bp. Because the coverage was too high for efficient assembly, we randomly selected sets containing different numbers of sequence reads for assembly with Newbler. A set of 19,000 randomly selected sequence reads generated the largest contig of 124,160 bp, which is in the expected range for a gammaherpesvirus genome. This contig incorporated 10,810 of the selected reads, giving an expected coverage of approximately 35-fold. The fraction of selected reads that was incorporated into the RHVP contig is consistent with BLAST analysis data indicating that 44% of unique reads from our data set are cellular (data not shown). To determine if there were additional reads corresponding to the ends of the viral genome that had not assembled into the contig, we searched for reads matching the left-most and right-most 50 bp of the contig's sequence. This identified another 121 bp and 54 bp of sequence corresponding to the left and right ends of the genome, respectively, for a total genome size of 124,335 bp.
The genome of the RHVP-L isolate was similarly assembled, using a randomly selected set of 12,000 sequence reads, into a contig of 123,913 bp with approximately 38-fold coverage. The RHVP and RHVP-L genomes were aligned using Geneious (version 4.7; A. J. Drummond, et al. [http://www.geneious.com/]), and nucleotide differences between the two, except for those within repeat regions, were resolved by PCR amplification and resequencing of the relevant genomic regions of both viruses. For resolution of nucleotide differences within repeat regions, all sequence reads corresponding to the relevant regions were extracted from the RHVP and RHVP-L datasets and reassembled to determine the consensus sequence.
Viral genome and phylogenetic analyses.
Predicted ORFs were identified using Geneious (see above), fgenesV (SoftBerry, Mount Kisco, NY) (13, 69), and GeneMark (6) and were analyzed using BLAST algorithms (3). ORFs that were predicted by both fgenesV and GeneMark and that contained canonical start and stop codons were annotated and queried against the GenBank nonredundant protein database, using BLASTP. Those that did not have any matches with E values of less than e−3 were further queried against the nucleotide database, using TBLASTX. Additionally, the RHVP genome was translated in all six reading frames, using “translate” from Sean Eddy's SQUID library, version 1.51 (ftp://selab.janelia.org/pub/software/squid/). ORFs that were at least 20 amino acids in length, with no initiator methionine required, were compared to the Pfam 24.0 database using the HMMER 3.0 program HMMscan (http://hmmer.org/) with the Pfam model trusted cutoff parameter settings. Matches to a Pfam model with E values of less than e−3 were deemed significant. Translated sequences of predicted ORFs were further examined for other features, including signal peptides, conserved protein domains, and structural similarity to other known proteins, through SignalP analysis (16), comparisons to the Pfam database (18), using HHpred (68), and protein structure modeling, using Phyre (36). Amino acid sequence alignments were performed using Geneious (see above) and ClustalW (77). Analysis of the genome for potential tRNA genes and repeat regions was performed using tRNAscan-SE 1.21 (49) and Tandem Repeats Finder (5), respectively, using the default parameters.
For phylogenetic analysis, sequences from gammaherpesvirus genomic segments encoding glycoprotein B and DNA polymerase were translated and concatenated. These sequences were aligned and phylogenetic analysis was performed with ClustalX, version 2.0 (41), using the neighbor-joining method with 1,000 bootstrap replicates. Phylogenetic trees were visualized using TreeView (56). The names and GenBank accession numbers of the viruses analyzed are murine cytomegalovirus (MCMV), NC_004065; murine gammaherpesvirus 68, U975530; Epstein-Barr virus, NC_007605; Kaposi's sarcoma-associated herpesvirus, U75698; herpesvirus saimiri (HVS), NC_001350; herpesvirus ateles (HVA), AF083424; alcelaphine herpesvirus 1 (AlHV-1), NC_002531; porcine lymphotropic herpesvirus 1 (PLHV-1), AF478169; equine herpesvirus 2 (EHV-2), NC_001650; bovine herpesvirus 4 (BoHV-4), NC_002665; callitrichine herpesvirus 3 (CalHV-3), NC_004367; and rhesus monkey rhadinovirus (RRV), NC_003401.
Mice and acute in vivo infection.
All mice were housed and bred in a specific-pathogen-free environment at Washington University School of Medicine in accordance with federal and university policies. Mice were intraperitoneally injected with various PFUs of virus in 0.3 ml of D5 medium. All studies were performed using age- and sex-matched mice between 8 and 12 weeks of age. To determine acute viral titers, mice were euthanized on various days postinfection and half of the spleen, half a lobe of liver, and one lobe of lung were harvested from each mouse. Tissues were mechanically disrupted using 1-mm silica beads, and viral titers in the tissues were determined by plaque assays on Vero cells as described above. Statistical significance was determined using the Mann-Whitney test.
Assessment of viral ex vivo reactivation from latency.
Mice were infected with 106 PFU of virus, and at 28 dpi, splenocytes and peritoneal cells were collected and assessed for ex vivo reactivation, using an adaptation of a previously described method (89). Briefly, splenocytes or peritoneal cells from 2 to 4 mice per experimental group were pooled, plated in serial 2-fold dilutions onto MEF monolayers, and incubated for 28 days to allow for viral reactivation and replication. The supernatant from each well was then transferred to Vero cell monolayers, which were assessed for CPE after 7 to 10 days. To determine whether preformed infectious virus was present in the cell samples, splenocytes and peritoneal cells that had been mechanically disrupted by bead beating were analyzed in parallel with nondisrupted cells.
Detection of viral genome in tissues from latently infected mice.
Splenocytes and peritoneal cells collected from uninfected mice and infected mice at 28 dpi as described above were stored at −80°C in D10 medium containing 10% dimethyl sulfoxide (DMSO). Total nucleic acid was prepared from thawed samples using a DNeasy blood and tissue kit (Qiagen), and PCR was performed on each sample using primers to ORF 50 to detect the viral genome and primers to the host alpha/beta interferon (IFN-αβ) receptor (IFNαβR) gene as a control. PCR products were visualized on a 1% agarose gel by ethidium bromide staining. Primer sequences used are as follows: ORF 50 primer 1, 5′-CCAGCAAAGTCTGGAGAGGG-3′; ORF 50 primer 2, 5′-GTCACGTGGGATAACATATGGGAG-3′; IFNαβR primer 1, 5′-TGCTTTGAGGAGCGTCTGGA-3′; and IFNαβR primer 2, 5′-CATGCACTACCACACCAGGCTTC-3′. Expected product sizes for the ORF 50 and IFNαβR primers are 409 bp and 250 bp, respectively.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the RHVP and RHVP-L genome sequences are HQ221963 and HQ698924, respectively.
RESULTS
Identification and sequencing of a novel rodent gammaherpesvirus.
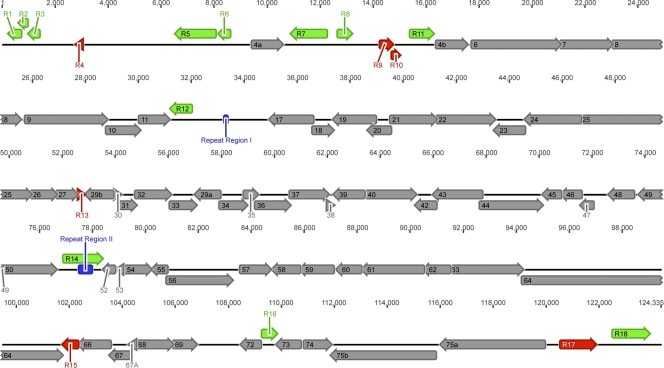
We initially isolated an unknown virus, designated HTN-0005, from a lung homogenate of a pygmy rice rat (Oligoryzomys microtis) trapped in July 1996 in a grassy field in Iquitos, Peru, during a search for new hantaviruses (61). Preliminary characterization indicated that the virus killed suckling mice on the fifth day after intracranial inoculation. Partial sequencing of total nucleic acid from lysates of infected Vero cells revealed significant amino acid sequence similarity, but not identity, to γHV68 (data not shown). We performed 454 mass sequencing on purified viral genomic DNA and assembled a single contig of approximately 124 kb with ∼35-fold coverage. We present here the annotated sequence of a novel rodent gammaherpesvirus (Fig. 1), designated rodent herpesvirus Peru (RHVP).
FIG. 1.
Schematic of the RHVP genome. The RHVP genome is represented by a black line, and genome coordinates are indicated above. Predicted ORFs are represented by block arrows. ORFs with similarity to conserved gammaherpesvirus genes are shown in gray and are labeled with the corresponding ORF numbers. All other ORFs are designated “R” ORFs and are numbered 1 through 18 from left to right of the genome. Those with similarity to other viral or cellular genes are shown in green, and those with no significant similarity to known genes are shown in red. Repeat regions are represented by blue bars.
A total of 82 ORFs were identified in the RHVP genome, using the following criteria: (i) sequence similarity to known viral or cellular genes, (ii) prediction by two different gene prediction algorithms, fgenesV (SoftBerry, Mount Kisco, NY) (13, 69) and GeneMark (6), and (iii) the presence of an initiating methionine and a stop codon. Translated sequences of these ORFs were analyzed by BLASTP, and those with significant similarity to conserved gammaherpesvirus ORFs were given ORF numbers according to the previously established convention for rhadinoviruses (2, 65, 84) and are shown as gray arrows in Fig. 1. ORFs with sequence similarity to other viral or cellular genes are shown as green arrows, and those with no significant similarity to any known gene are shown as red arrows. A description of all ORFs is given in Table 1. Overall, the RHVP genome is very similar to those of other gammaherpesviruses, and the arrangement of the conserved gammaherpesvirus ORFs in the RHVP genome is colinear with that in the γHV68, KSHV, and HVS genomes (2, 65, 84). Amino acid sequence identity between RHVP ORFs and the corresponding ones in other gammaherpesviruses ranges from less than 20% to over 60% (Table 1), which is consistent with RHVP being a novel member of the gammaherpesvirus subfamily.
TABLE 1.
Predicted open reading frames of RHVPa
| Open reading frame | Genome location (bp) | Coding strand | Size (aa) | % amino acid identity with indicated virus |
Possible function | ||
|---|---|---|---|---|---|---|---|
| γHV68 | KSHV | HVS | |||||
| R1 | 236-697 | L | 153 | Similar to that of MHC class I | |||
| R2 | 639-932 | L | 97 | Similar to that of MHC class I | |||
| R3 | 1000-1383 | L | 127 | Similar to that of MHC class I | |||
| R4 | 2769-3014 | L | 81 | ||||
| R5 | 6552-8027 | L | 491 | Ig domain-containing protein | |||
| R6 | 8175-8582 | L | 135 | Similar to that of IL-4/IL-13 | |||
| ORF 4a | 9438-10589 | R | 383 | 27 | 15 | 26 | Complement regulatory protein |
| R7 | 10913-12229 | L | 438 | Ig domain-containing protein | |||
| R8 | 12677-13183 | R | 168 | C-type lectin-like protein | |||
| R9 | 14271-14738 | R | 155 | ||||
| R10 | 14723-15001 | R | 92 | ||||
| R11 | 15403-16266 | R | 287 | 18 | Similar to that of HVS ORF 14 and MMTV superantigen | ||
| ORF 4b | 16391-17554 | R | 387 | 23 | 16 | 25 | Complement regulatory protein |
| ORF 6 | 17724-21074 | R | 1,116 | 47 | 42 | 43 | Single-stranded DNA binding protein |
| ORF 7 | 21077-23041 | R | 654 | 40 | 33 | 32 | Transport protein |
| ORF 8 | 23050-25569 | R | 839 | 63 | 51 | 46 | Glycoprotein B |
| ORF 9 | 25753-28812 | R | 1,019 | 61 | 54 | 55 | DNA polymerase |
| ORF 10 | 28805-30064 | R | 419 | 20 | 19 | 18 | |
| ORF 11 | 30036-31175 | R | 379 | 28 | 22 | 20 | |
| R12 | 31220-31996 | L | 258 | 15 | 10 | Similar to that of K3 (E3 ubiquitin ligase) | |
| ORF 17 | 34959-36587 | L | 542 | 36 | 32 | 36 | Capsid protein |
| ORF 18 | 36586-37359 | R | 257 | 38 | 40 | 40 | |
| ORF 19 | 37356-38948 | L | 530 | 42 | 36 | 36 | Tegument protein |
| ORF 20 | 38671-39519 | L | 282 | 29 | 22 | 27 | |
| ORF 21 | 39518-41251 | R | 577 | 25 | 21 | 20 | Thymidine kinase |
| ORF 22 | 41272-43452 | R | 726 | 40 | 29 | 28 | Glycoprotein H |
| ORF 23 | 43441-44583 | L | 380 | 36 | 28 | 16 | |
| ORF 24 | 44602-46695 | L | 697 | 49 | 40 | 41 | |
| ORF 25 | 46706-50830 | R | 1,374 | 59 | 54 | 55 | Major capsid protein |
| ORF 26 | 50834-51784 | R | 316 | 49 | 44 | 45 | Capsid protein |
| ORF 27 | 51787-52575 | R | 262 | 26 | 0 | 7 | |
| R13 | 52618-52854 | R | 78 | ||||
| ORF 29b | 52866-53915 | L | 349 | 58 | 53 | 27 | Cleavage and/or packaging protein |
| ORF 30 | 53984-54223 | R | 79 | 37 | 32 | 29 | |
| ORF 31 | 54184-54786 | R | 200 | 44 | 32 | 32 | |
| ORF 32 | 54768-56087 | R | 439 | 25 | 23 | 22 | Viral DNA cleavage or packaging protein |
| ORF 33 | 56071-57054 | R | 327 | 45 | 29 | 33 | |
| ORF 29a | 57008-57952 | L | 314 | 42 | 36 | 17 | Cleavage and/or packaging protein |
| ORF 34 | 57951-58958 | R | 335 | 38 | 28 | 33 | |
| ORF 35 | 58873-59370 | R | 165 | 29 | 21 | 24 | |
| ORF 36 | 59297-60607 | R | 436 | 27 | 22 | 24 | Kinase |
| ORF 37 | 60604-62055 | R | 483 | 54 | 40 | 44 | Alkaline exonuclease |
| ORF 38 | 62010-62243 | R | 77 | 36 | 23 | 18 | |
| ORF 39 | 62249-63400 | L | 383 | 60 | 42 | 45 | Glycoprotein M |
| ORF 40 | 63478-65340 | R | 620 | 28 | 18 | 18 | Helicase-primase |
| ORF 42 | 65332-66105 | L | 257 | 43 | 29 | 35 | |
| ORF 43 | 65990-67840 | L | 616 | 52 | 42 | 45 | Capsid protein |
| ORF 44 | 67776-70115 | R | 779 | 57 | 52 | 54 | Helicase |
| ORF 45 | 70134-70811 | L | 225 | 37 | 11 | 20 | |
| ORF 46 | 70828-71577 | L | 249 | 59 | 51 | 53 | Uracil DNA glycosidase |
| ORF 47 | 71562-72026 | L | 154 | 23 | 22 | 23 | Glycoprotein L |
| ORF 48 | 72630-73556 | L | 308 | 19 | 13 | 8 | |
| ORF 49 | 73761-74603 | L | 280 | 25 | 17 | 16 | |
| ORF 50 | 74655-76640 | R | 661 | 19 | 17 | 14 | Transcriptional activator; Rta homolog |
| R14 | 76918-78372 | R | 484 | 24 | Similar to that of γHV68 M7 (glycoprotein 150) | ||
| ORF 52 | 78415-78831 | L | 138 | 42 | 30 | 23 | |
| ORF 53 | 78903-79154 | L | 83 | 53 | 27 | 33 | |
| ORF 54 | 79256-80218 | R | 320 | 35 | 26 | 27 | dUTPase |
| ORF 55 | 80240-80812 | L | 190 | 48 | 32 | 40 | |
| ORF 56 | 80811-83288 | R | 825 | 43 | 36 | 37 | DNA replication protein |
| ORF 57 | 83584-84735 | R | 383 | 16 | 15 | 24 | |
| ORF 58 | 84794-85831 | L | 345 | 29 | 21 | 22 | |
| ORF 59 | 85837-87102 | L | 421 | 29 | 23 | 21 | DNA replication protein |
| ORF 60 | 87222-88145 | L | 307 | 62 | 57 | 57 | Ribonucleotide reductase, small subunit |
| ORF 61 | 88180-90498 | L | 772 | 49 | 44 | 46 | Ribonucleotide reductase, large subunit |
| ORF 62 | 90507-91496 | L | 329 | 34 | 32 | 26 | Assembly or DNA maturation protein |
| ORF 63 | 91507-94245 | R | 912 | 29 | 20 | 23 | Tegument protein |
| ORF 64 | 94223-101740 | R | 2,505 | 31 | 24 | 26 | Tegument protein |
| R15 | 101750-102325 | L | 191 | ||||
| ORF 66 | 102334-103554 | L | 406 | 41 | 28 | 32 | |
| ORF 67 | 103520-104260 | L | 246 | 48 | 35 | 44 | Tegument protein |
| ORF 67a | 104247-104498 | L | 83 | ||||
| ORF 68 | 104549-105919 | R | 456 | 38 | 27 | 33 | |
| ORF 69 | 105924-106814 | R | 296 | 53 | 35 | 36 | |
| ORF 72 | 108475-109221 | L | 248 | 21 | 23 | 20 | Cyclin D homolog |
| R16 | 109298-109816 | R | 172 | 21 | vBcl-2 homolog; inhibitor of apoptosis | ||
| ORF 73 | 109832-110716 | L | 294 | 20 | 5 | 13 | LANA homolog |
| ORF 74 | 110864-111886 | R | 340 | 27 | 22 | 18 | G protein-coupled receptor |
| ORF 75b | 111883-115824 | L | 1,313 | 35 | 26 | 26 | Tegument protein/FGARAT |
| ORF 75a | 116019-119936 | L | 1,305 | 29 | 25 | 25 | Tegument protein/FGARAT |
| R17 | 120546-121865 | R | 439 | ||||
| R18 | 122524-123879 | R | 451 | Ig domain-containing protein | |||
Abbreviations: L, left; R, right; FGARAT, N-formylglycinamide ribotide amidotransferase.
As γHV68 encodes several tRNA-like molecules containing 3′ stem-loops from which mature viral microRNAs (miRNAs) are generated (8, 58, 84), we searched the RHVP genome for the presence of tRNAs, using tRNAscan-SE 1.21 (49). No tRNAs were found in the RHVP genome, although the 8 γHV68 tRNAs were readily detected in the γHV68 genome using this method (data not shown). Thus, the expression of tRNA-miRNA structures by γHV68 does not appear to be conserved in RHVP. The RHVP genome was also analyzed using Tandem Repeats Finder (5) to identify repeat regions. We found two regions containing repeat sequences at coordinates 33272 to 33356 (repeat region I) and 77519 to 77960 (repeat region II), which are represented by blue bars in Fig. 1. Repeat region I contains a 42-bp sequence that is repeated twice. The precise repeat unit for repeat region II could not be determined because the region contains three overlapping sequences of 110 bp, 111 bp, and 222 bp, each of which is repeated 2 to 3 times. Potential repeat units for each region that were identified by this analysis are described in Table 2. This analysis did not, however, identify potential terminal-repeat sequences at either end of the genome, although it is possible that this was due to the incorporation of only one copy of the terminal-repeat unit into the assembled genome.
TABLE 2.
Repeat regions of RHVP
| Repeat region | Possible repeat units | Genome location (bp) | Length (nt) | No. of copies |
|---|---|---|---|---|
| I | CCATTGTGTGAGGGTCTTGGTCCCATTTTGAGGAGGGGTCAC | 33272-33313 | 42 | 2 |
| II | CCCCACTGAGGCTTCCCCCAAAGAAGGTTCTTCCACAGAAGCTCAACCAGAGCAGACCCTCGGCCCCACAGAAGTTACCCCCACAAATGCTAAACCAGAAGAGGTTCCTG | 77553-77662 | 110 | 3 |
| TTCTTCCACAGAAGCTCAACCAGAGCAGACCCTCGGCCCCACAGAAGTTACCCCCACAAATGCTAAACCAGAAGAGGTTCCTGCGCCCACAGATGCCCCCTCTAAAGAAAA | 77580-77690 | 111 | 3.5 | |
| CCCCCGCCCAAGACAAGTCAGCCGAGGGCTCTGTCCCCACTGAGGCTTCCCCCAAAGAAGGTTCTTCCACAGAAGCTCAACCAGAGCAGACCCTCGGCCCCACAGAAGTTACCCCCACAAATGCTAAACCAGAAGAGGTTCCTGCGCCCACAGATGCCCCCTCTAAAGAAAATTCTAATACAGAAGCTCAACCAGAACAGACCTCCATCCCCACAGAACTTG | 77519-77740 | 222 | 2 |
Interestingly, RHVP encodes two copies of the ORF 4 complement regulatory protein (designated ORFs 4a and 4b), whereas γHV68, KSHV, and HVS each encodes only one copy (2, 65, 84). RHVP ORFs 4a and 4b both have sequence similarity to ORF 4 from the other gammaherpesviruses (Table 1) as well as to cellular complement regulatory proteins, including membrane cofactor protein (CD46), decay accelerating factor (CD55), and complement component 4 binding protein (C4BP). A multiple-sequence alignment of RHVP ORFs 4a and 4b with mouse complement regulatory proteins as well as ORF 4 homologs from other gammaherpesviruses (Fig. 2) shows conservation of key residues—Cys2, Pro5, Tyr/Phe29, Cys31, Gly34, Cys45, Trp51, Ala/Pro56, and Cys58—that define the short consensus repeat (SCR) modules present in complement regulatory proteins (47). RHVP also has two copies of ORF 75, which encodes an N-formylglycinamide ribotide amidotransferase enzyme (84). Multiple copies of ORF 75 are also present in γHV68, which has 3 copies (84), and in HVS, which has a second copy called ORF 3 (2). The significance to the biology of gammaherpesviruses of having multiple copies of ORF 75 is unknown.
FIG. 2.
RHVP encodes two copies of the ORF 4 complement regulatory protein. The amino acid sequences of RHVP ORFs 4a and 4b were aligned with those of mouse membrane cofactor protein (CD46), decay accelerating factor (CD55), and complement component 4 binding protein (C4BP) and ORF 4 homologs from KSHV, HVS, and γHV68. Residues are shaded as follows: black, 100% similar; dark gray, 80 to 100% similar; light gray, 60 to 80% similar; and unshaded, <60% similar. Key residues that define the short consensus repeats (SCRs)—Cys2, Pro5, Tyr/Phe29, Cys31, Gly34, Cys45, Trp51, Ala/Pro56, and Cys58—are indicated by asterisks, and the four SCRs are indicated by black bars. The amino acids included in this alignment are CD46, 37 to 293; CD55, 86 to 295; C4BP, 50 to 284; KSHV ORF 4, 17 to 277; HVS ORF 4, 15 to 273; γHV68 ORF 4, 15 to 274; RHVP ORF 4a, 16 to 279; and RHVP ORF 4b, 25 to 289.
RHVP contains novel ORFs that may encode immune evasion molecules.
In addition to the 64 conserved gammaherpesvirus ORFs, the RHVP genome contains 18 ORFs that do not appear to be broadly conserved among gammaherpesviruses. Although a few of these ORFs have some sequence similarity to ORFs in other gammaherpesviruses, the sequence identities between them are low and the ORFs are generally not present across multiple members of the gammaherpesvirus family. Taking this and the naming of ORFs in previously sequenced gammaherpesviruses into consideration, we designated these ORFs “R” ORFs and numbered them from 1 through 18 from the left to the right of the genome (Fig. 1). Twelve R ORFs, shown as green arrows, have significant amino acid sequence similarity to known viral or cellular proteins.
The R1, R2, and R3 ORFs.
R1, R2, and R3 are all similar to major histocompatibility complex (MHC) class I molecules (E values of 2e−26, 8e−30, and 3e−20, respectively). A multiple-sequence alignment of these 3 ORFs with the mouse class I molecule, H-2Kb, and the human HLA-A2 molecule (Fig. 3 A) shows significant amino acid identity between each ORF and mouse and human class I molecules. In particular, many of the residues that contribute to peptide binding in the A and F pockets (indicated by “A” and “F” on the figure) (24) are conserved in the RHVP class I-like ORFs. The substitutions of serine and leucine in RHVP R2 at the Thr143 and Trp147 positions, which contribute to F pocket peptide binding, are also seen in HLA-E and in the nonclassical class I molecules H2-M3, CD1d, and MICA, respectively (24). Also conserved is the N-linked glycosylation site seen in all MHC class 1 α1 domains (Fig. 3A, asterisk) (7).
FIG. 3.
Novel RHVP ORFs with potential immunomodulatory function. RHVP-translated ORFs were aligned with other viral or cellular proteins, and residues were shaded as described in the legend to Fig. 2. (A) Alignment of RHVP R1, R2, and R3 with H-2Kb and HLA-A2. The predicted signal peptides, α1/α2 and α3 domains, and the region corresponding to the transmembrane (TM) domain in H-2Kb are indicated by black bars. The conserved MHC class I residues that contribute to peptide binding in the A or F pockets are indicated by “A” or “F,“ respectively, and the invariant glycosylation site is marked with an asterisk. Full-length sequences for each protein were used in this alignment. (B) Alignment of RHVP R6 with IL-13 from human, mouse, rat, and cotton rat (hIL-13, mIL-13, rIL-13, and crIL-13, respectively). The predicted signal peptides and regions corresponding to helices A to D in human IL-13 are indicated by black bars. Contact residues from the site I interface between IL-13 and IL-4Rα are indicated by “1,” and those from the site II and site III interfaces between IL-13 and IL-13Rα1 are indicated by “2” and “3,” respectively. Full-length sequences were used in this alignment for each protein, with the exception of hIL-13, for which amino acids 15 to 146 were used. (C) Alignment of RHVP R8 with mouse Clr-g (mClr-g) and the rat cytomegalovirus C-type lectin-like protein (RCTL). The 6 cysteine residues that define the consensus lectin-like domain are indicated by asterisks, and an arginine residue that is uniquely conserved among CLEC2 family members is indicated by a diamond. Amino acid residues included in this alignment are as follows: mClr-g, 78 to 217; RCTL, 43 to 180; and R8, 34 to 169. (D) Alignment of RHVP R11 with mouse mammary tumor virus superantigen (MMTV sAg) and ORF 14 from HVS and HVA. The C-terminal portion of MMTV sAg that is important for interactions with I-A molecules is indicated by a black line. Amino acid residues included in this alignment are as follows: MMTV sAg, 144 to 305; HVS ORF 14, 91 to 249; HVA ORF 14, 112 to 273; and RHVP R11, 111 to 287. (E) Alignment of RHVP R12 with KSHV K3 and K5, γHV68 K3, and HVS ORF 12. The RING-CH domain is indicated by a black line, and key amino acid residues in the motif are marked with asterisks. Amino acid residues included in this alignment are the following: KSHV K3, 1 to 89; KSHV K5, 1 to 95; γHV68 K3, 1 to 88; HVS ORF 12, 1 to 88; and RHVP R11, 33 to 127. (F) Alignment of RHVP R16 with mouse Bcl-2 and γHV68 M11. The BH1 domain is indicated by a black line. Amino acid residues included in this alignment are as follows: Bcl-2, 84 to 179; M11, 28 to 123; and R16, 24 to119.
The R6 ORF.
Although R6 did not have significant sequence similarity to known viral or cellular proteins in the initial BLAST analysis, further analysis using the HHpred program (68) to compare the R6 sequence to the Pfam database (18) in a combined sequence similarity and secondary-structure prediction search detected weak similarity to the interleukin-4 (IL-4)/IL-13 short-chain, left-handed, 4-helical-bundle family of cytokines. Similarly, analysis of the R6 sequence using Phyre (36) revealed a weak alignment to the structure of human IL-13. The IL-13 type II receptor is composed of two chains, the IL-4 receptor α chain (IL-4Rα) and the IL-13 receptor α1 chain (IL-13Rα1), and the structure of IL-13 bound to this receptor has been reported (40). Alignment of the R6 sequence with those of human, mouse, rat, and cotton rat IL-13 (Fig. 3B) shows similarity between R6 and IL-13 in some of the key contact residues in the IL-13/IL-4Rα interface (site I; marked with “1” on the figure) and the IL-13/IL-13Rα1 interface (sites 2 and 3; marked on the figure with “2” or “3,” respectively), suggesting that R6 may have retained receptor binding function.
The R8 ORF.
R8 was found to have strong similarity to members of mouse C-type lectin family 2 (CLEC2) by BLAST analysis (E values in the e−38 range), and computational translation of R8 predicted a type II transmembrane protein containing a C-type lectin-like domain (CTLD). R8 also has significant sequence similarity to a rat cytomegalovirus (RCMV) C-type lectin-like protein (RCTL) (86) that likely serves as a viral decoy for the natural killer (NK) cell inhibitory receptor, NKR-P1B, resulting in the inhibition of NK cell-mediated cytotoxicity (85). Multiple-sequence alignment of R8 with a mouse C-type lectin, Clr-g, and RCTL (Fig. 3C) shows conservation in R8 of 5 of the 6 cysteine residues that define the consensus lectin-like domain, as well as an arginine residue that is uniquely conserved among CLEC2 family members (72). The fifth cysteine (C5), which is consistently absent from members of the Clr family (60), is also absent in RHVP R8.
The R11 ORF.
R11 is similar to mouse mammary tumor virus superantigen (MMTV sAg; E value of 6e−6) and also has detectable sequence similarity to the ORF 14 sAg-like molecules of both HVS and the closely related herpesvirus ateles when queried specifically against herpesvirus sequences (E values of 2e−5 and 9e−6, respectively). Neither KSHV nor γHV68 appears to carry ORF 14 homologs, and since the overall amino acid identity between R11 and HVS ORF 14 is only 18% (Table 1), we designated R11 a unique ORF rather than a conserved gammaherpesvirus ORF. An alignment of R11 with the MMTV sAg and ORF 14 of HVS and HVA (Fig. 3D) shows the greatest degree of sequence conservation in the C-terminal portion of the protein, which in the MMTV sAg mediates interactions with the MHC class II I-A molecule (92).
The R12 ORF.
R12 is similar to the γHV68 K3 protein (E value of 3e−8), which is a relative of the KSHV K3 protein that functions as an E3 ubiquitin ligase. Both the γHV68 and KSHV K3 proteins can downregulate MHC class I expression by ubiquitinating class I molecules and targeting them for degradation (9, 26, 27, 88). KSHV encodes a second E3 ubiquitin ligase, K5, which targets MHC class I as well as other molecules (10, 30, 31, 50) but does not appear to be present in γHV68. HVS encodes ORF 12, which has sequence similarity to the K3 family of proteins (20), but the function of HVS ORF 12 has not been characterized. Although the amino acid identity between R12 and either γHV68 or KSHV K3 is low (15% and 10%, respectively; [Table 1]), an alignment of R12 with KSHV K3 and K5 and γHV68 K3 and HVS ORF 12 (Fig. 3E) shows conservation of residues forming the consensus sequence of the RING-CH (C4HC3) domain (12), which is important for mediating ubiquitination of target molecules (20).
The R14 ORF.
R14 has similarity to a γHV68 ORF, M7 (E value of 3e−5), which encodes a membrane glycoprotein, gp150. M7 is located in the same genomic position, between ORFs 50 and 52, as ORF 51 in HVS but does not have significant similarity to ORF 51 by BLASTP and was therefore designated an M ORF (84). Similarly, RHVP R14 is located between ORFs 50 and 52 but has similarity only to γHV68 M7 and not to HVS ORF 51. The corresponding genomic position in KSHV is occupied by K8, which has no significant similarity to either HVS ORF 51 or γHV68 M7. The gp150 molecule encoded by γHV68 M7 has been suggested to function as an immunogenic decoy by inducing an antibody response that does not promote virion neutralization (23). Whether RHVP R14 may serve a similar function remains to be determined.
The R16 ORF.
Like other gammaherpesviruses, RHVP appears to encode a Bcl-2-like molecule in ORF R16 that has sequence similarity to the γHV68 viral Bcl-2 (vBcl-2) homolog encoded by the M11 ORF (E value of 2e−6). Although both KSHV and HVS encode vBcl-2 molecules in ORF 16, they are in different genomic locations from either RHVP R16 or γHV68 M11, and R16 does not have significant sequence similarity with either KSHV or HVS ORF 16. Thus, RHVP R16 may be more closely related to γHV68 M11 than to KSHV and HVS ORF 16. An alignment of R16 with mouse Bcl-2 and γHV68 M11 (Fig. 3F) indicates that most of the sequence similarity between these molecules lies in the Bcl-2 homology 1 (BH1) domain, which is critical for Bcl-2 antiapoptotic activity (94). Similarly, mutation of conserved residues in the BH1 domain of γHV68 M11 abrogates binding to peptides from proapoptotic BH3-only proteins and to the autophagy protein, Beclin-1, and results in deficient viral reactivation from latency and persistent viral replication (48, 67). Thus, although the overall sequence similarity between RHVP R16 and either Bcl-2 or γHV68 M11 is low, R16 appears to have conserved a domain that is functionally important in both Bcl-2 and M11.
The R5, R7, and R18 ORFs.
Finally, RHVP contains 3 ORFs, R5, R7, and R18, all of which have sequence similarity to the tree shrew adenovirus 105R (E values of 2e−17, 2e−21, and 6e−38, respectively) (11). The function of tree shrew adenovirus 105R is unknown, but all three ORFs were predicted by SignalP to have signal peptides (16) and to contain Ig domains when searched against the Pfam database (18) and analyzed using Phyre (36). This suggests that R5, R7, and R18 may share structural features with 105R rather than being true homologs of that protein.
ORFs with limited or no similarity to known proteins.
The remaining R ORFs, which are shown as red arrows in Fig. 1, did not have any matches in the database with significant E values by either BLASTP or TBLASTX. However, further analysis comparing every translated reading frame of greater than 20 amino acids to the Pfam database identified the conserved US22 domain in R9. The US22 domain is found in members of the US22 multigene family of proteins from various betaherpesviruses, including human cytomegalovirus (HCMV) and murine cytomegalovirus (MCMV). Several US22 proteins have been shown to confer efficient replication of MCMV in macrophages (25, 51), suggesting a role for these proteins in viral replication and/or cell tropism. R13 and R15 are in the same genomic positions as the HVS and KSHV ORFs 28 and 65, respectively (2, 65), although they do not have detectable sequence similarity to these ORFs. ORFs 28 and 65 also do not appear to be conserved in γHV68, although γHV68 carries the M9 ORF in the position corresponding to that of ORF 65 (84). As an alignment of RHVP R15 and γHV68 M9 reveals only very weak sequence similarity (data not shown), it is unclear whether R15 is related to M9. In summary, these data show that RHVP carries unique ORFs with potential immunomodulatory functions that are not shared with γHV68, suggesting that RHVP may have evolved immune evasion mechanisms that are distinct from those of γHV68.
RHVP is most closely related to rodent rhadinoviruses.
Recent studies have identified numerous new rodent gammaherpesviruses, including the first reported gammaherpesvirus of Mus musculus (15). To assess the relationship between RHVP and other known gammaherpesviruses, we performed phylogenetic analysis using concatenated amino acid sequences representing segments from each virus encoding glycoprotein B and DNA polymerase, as previously described (15). We found that RHVP grouped with several rodent rhadinoviruses that were previously found to form a distinct clade (Fig. 4) (15). These viruses include γHV68 and rhadinoviruses from various mouse, rat, and vole species. However, RHVP formed a separate branch from these other viruses and is clearly distinct from γHV68, confirming that RHVP is a novel rodent rhadinovirus.
FIG. 4.
RHVP is most closely related to rodent rhadinoviruses. Concatenated amino acid sequences corresponding to glycoprotein B and DNA polymerase from various gammaherpesviruses were aligned, and phylogenetic analysis was performed using the neighbor-joining method with 1,000 bootstrap replicates. The corresponding sequence from the betaherpesvirus murine cytomegalovirus (MCMV) was included for comparison. Subclassifications of the gammaherpesviruses are indicated, and bootstrap values over 65% are shown. Abbreviations: BoHV-4, bovine herpesvirus 4; EHV-2, equine herpesvirus 2; BsavRHV1, Bandicota savilei rhadinovirus 1; MmusRHV1, Mus musculus rhadinovirus 1; McerRHV1, Mus cervicolor rhadinovirus 1; AlHV-1, alcelaphine herpesvirus 1; PLHV-1, porcine lymphotropic herpesvirus 1; EBV, Epstein-Barr virus; CalHV-3, callitrichine herpesvirus 3; RHVP, rat herpesvirus Peru; BindRHV4, Bandicota indica rhadinovirus 4; γHV68, murine gammaherpesvirus 68; AflaRHV1, Apodemus flavicollis rhadinovirus 1; AsylRHV1, Apodemus sylvaticus rhadinovirus 1; MglaRHV1, Myodes glareolus rhadinovirus 1; MagrRHV1, Microtus agrestis rhadinovirus 1; KSHV, Kaposi's sarcoma-associated herpesvirus; RRV, rhesus monkey rhadinovirus; HVS, herpesvirus saimiri; HVA, herpesvirus ateles; MCMV, murine cytomegalovirus.
Interferons and adaptive immunity are both required for protection against lethal RHVP infection.
To determine if RHVP infection can cause disease in laboratory mice, we injected wild-type and various immunodeficient mice on either the B6 or 129 background with 106 PFU of virus and monitored them for pathogenesis and lethality. As expected for a gammaherpesvirus capable of establishing long-term infection and similar to observations after infection with γHV68 (90), wild-type B6 and 129 mice survived for over 100 dpi and did not appear ill during the course of infection (Fig. 5A and data not shown). Interestingly, whereas mice deficient in STAT1 (STAT−/−), a key signal transducer in both the type I and type II interferon pathways, were resistant to RHVP infection, all mice deficient in both the IFN-αβ and IFN-γ receptors (IFNαβγR−/−) died by 12 dpi (Fig. 5A). Infection of IFNαβγR−/− mice with 104 PFU of RHVP still resulted in significant lethality, but the approximately 35% of mice that did not die during the first 20 days of infection survived for the duration of the experiment. Together, these data demonstrate an important role for type I and type II interferons in protection against RHVP infection. These results are consistent with the existence of a STAT1-independent antiviral effect of interferons, which has previously been observed for γHV68, MCMV, and Sindbis virus (4, 22). We found that RAG1-deficient (RAG−/−) mice, which lack B and T cells, exhibit complete but delayed lethality following RHVP infection. Infected RAG−/− mice appeared grossly normal early after infection (data not shown) but began to die at 56 dpi, and all succumbed by 91 dpi (Fig. 5A). Mice deficient in both RAG1 and STAT1 (RAGXSTAT−/−) were highly susceptible to infection with RHVP and died, with kinetics similar to that of infected IFNαβγR−/− mice. These data suggest that while interferons are critical for early control of RHVP infection, they are insufficient for clearance of infection and that the adaptive immune response is required for long-term protection from RHVP lethality. Thus, RHVP can infect laboratory mice, and protection against in vivo RHVP infection requires both the interferon response and adaptive immunity.
FIG. 5.
RHVP infection is lethal in mice lacking interferon responses and/or B and T cells. (A) Various strains of mice on the 129 (left) or B6 (right) background were infected with RHVP and monitored for lethality. 106 PFU of virus per mouse was used for all mice except IFNαβγR−/− mice, which were also tested using 104 PFU of virus per mouse, as indicated. These data represent at least 2 independent experiments. The total number of mice examined for each strain on the B6 background is as follows: B6, 14; STAT−/−, 10; RAG−/−, 21; and RAGXSTAT−/−, 14. The total number of mice examined for each strain on the 129 background is as follows: 129, 9; STAT−/−, 10; IFNαβγR−/− (106 PFU), 9; and IFNαβγR−/− (104 PFU), 49. (B) RAGXSTAT−/− (left) or IFNαβγR−/− (right) mice were infected with 106 PFU of either RHVP or RHVP-L per mouse and monitored for lethality. These data represent 2 independent experiments. The total numbers of RAGXSTAT−/− mice examined are as follows: RHVP, 9, and RHVP-L, 13. The total numbers of IFNαβγR−/− mice examined are as follows: RHVP, 8, and RHVP-L, 13. (C) IFNαβγR−/− mice were infected with RHVP, RHVP-L, or RHVP-E and monitored for lethality. 106 PFU of virus per mouse was used for all virus isolates except RHVP-E, which was also examined using 8 × 106 PFU per mouse, as indicated. These data represent 2 independent experiments. The total number of mice examined for each virus isolate is as follows: RHVP, 8; RHVP-L, 13; RHVP-E (106 PFU), 6; and RHVP-E (8 × 106 PFU), 9.
Generation of a cloned RHVP isolate with pathogenic potential.
Since the original RHVP virus stock we studied was grown from lung homogenate from a wild rat, it was possible that the in vivo phenotype observed in infected mice was due to the presence of viruses other than RHVP in the original virus stock. We therefore generated purified isolates of RHVP by two sequential rounds of limiting dilution cloning and sequenced the genome of the isolate RHVP-L. The assembled genome of RHVP-L was slightly shorter than that of RHVP, which has an additional 121 bp of sequence at the left end and 302 bp of sequence at the right end that are not in RHVP-L (data not shown). These additional sequences are not predicted to carry any ORFs, however, and the RHVP and RHVP-L genomes differ at only one nucleotide position over the 123,913-bp overlap in sequence. Infection of RAGXSTAT−/− or IFNαβγR−/− mice with RHVP-L resulted in lethality with kinetics similar to that seen with RHVP infection (Fig. 5B). RHVP-L thus appears to be virtually identical to the parental RHVP in both sequence and virulence in mice.
We also used BLASTN to analyze sequence reads that were generated by 454 mass sequencing of RHVP but were not assembled into the RHVP genome. We found that the RHVP nucleic acid sample yielded 11 reads with sequence similarity to betaretrovirus sequences (E values ranging from e−67 to 0) and 7 reads with similarity to human adenovirus sequences (E values ranging from e−66 to 0). Of the 11 betaretrovirus-like sequences, 8 were highly similar to sequences from an endogenous simian retrovirus found in Vero cells (E values ranging from e−157 to 0) (83), in which the RHVP viruses were grown. We performed PCR for all 11 betaretrovirus-like sequences and were able to detect 9 of them in the RHVP and RHVP-L virus stocks, as well as in a “mock” stock generated from uninfected Vero cells (data not shown). This confirms the presence of endogenous retroviral sequences in Vero cells and indicates that the betaretrovirus-like sequences detected in the RHVP sequencing sample are likely from the host cells. We were unable to detect any of the adenovirus-like sequences in the virus stocks by PCR (data not shown). However, the 454 sequencing data set from both RHVP-L and a second purified RHVP isolate, designated RHVP-E (data not shown), also contained similar adenovirus-like sequence reads even though the RHVP-E isolate was nonlethal after inoculation into IFNαβγR−/− mice at a dose at which RHVP and RHVP-L are 100% lethal (Fig. 5C). RHVP-E was able to kill IFNαβγR−/− mice when given at an 8-fold-higher dose. Thus, the presence of adenovirus-like sequences does not appear to correlate with virulence in mice, indicating that the lethality observed in infected mice is most likely due to RHVP rather than to a contaminating adenovirus-like agent. We are unsure of the source of these adenovirus-like sequences.
RHVP replicates in multiple organs of mice deficient in interferon responses.
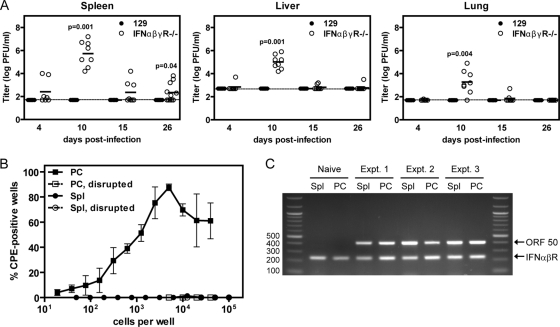
Although infection of wild-type mice with RHVP did not result in lethality or detectable disease, it is possible that RHVP replicated in wild-type mice but that the infection was asymptomatic. We therefore tested whether RHVP could be detected in organs from infected wild-type 129 mice by plaque assay on Vero cells. We also determined viral titers in organs from infected IFNαβγR−/− mice, in which RHVP is virulent, to assess the tissue tropism and kinetics of RHVP acute replication in vivo. As 106 PFU of virus killed all IFNαβγR−/− mice by 12 dpi, this set of infections was done using 104 PFU of virus, a dose which kills approximately 65% of IFNαβγR−/− mice (Fig. 4). We were unable to detect infectious virus in spleen, liver, or lung from infected wild-type mice at various times postinfection with the limits of detection for the plaque assay set at 1.7-log PFU/ml for spleen and lung samples and 2.7-log PFU/ml for liver samples. By contrast, we detected virus in all three organs of IFNαβγR−/− mice, with peak viral titers observed at 10 dpi (Fig. 6A). By 26 dpi, the virus was largely undetectable in liver and lung, although 6 of 11 mice tested still had splenic viral titers above the limit of detection. These results are consistent with lethality data showing that some IFNαβγR−/− mice survive infection with this dose of virus.
FIG. 6.
RHVP replicates acutely in IFNαβγR−/− mice and establishes latent infection in wild-type mice. (A) 129 and IFNαβγR−/− mice were infected with 104 PFU of RHVP per mouse, tissues were collected at the time points indicated, and the viral titers in each tissue were determined by plaque assay. P values are given for statistically significant differences, and the limit of detection for the assay is indicated by dotted lines. These data represent at least 2 independent experiments. The total number of 129 mice examined per day is as follows: day 4, 7; day 10, 7; day 15, 6; and day 26, 7. The total number of IFNαβγR−/− mice examined per day is as follows: day 4, 7; day 10, 8; day 15, 8; and day 26, 11. (B) B6 mice were infected with 106 PFU of RHVP per mouse. At 28 dpi, peritoneal cells (PC; filled squares) and splenocytes (Spl; filled circles) were collected and plated in limiting 2-fold dilutions to assess their capacities to reactivate latent virus. Peritoneal cells and splenocytes that were mechanically disrupted (open squares and open circles, respectively) were analyzed in parallel. These data represent the means ± standard errors of the means (SEM) of the results of 3 independent experiments with cells pooled from 2 to 4 mice per experiment. (C) Total nucleic acid was prepared from the same splenocyte and peritoneal cell samples analyzed for Fig. 6B, as well as from splenocytes and peritoneal cells from uninfected mice. PCR was performed using primers to RHVP ORF 50 and to the IFNαβR gene, and their respective products are indicated with arrows. The sizes of relevant bands in the DNA ladder are shown.
RHVP establishes latent infection in peritoneal cells and splenocytes of wild-type mice.
The observation that wild-type mice did not develop apparent disease or have detectable viral titers in organs following infection with RHVP suggested that the virus may replicate poorly in wild-type mice or may even be unable to infect them. Since establishment of latent infection is a hallmark of herpesvirus biology and γHV68 mutant viruses that are replication defective can establish latent infection in vivo (43, 54, 79), we tested whether RHVP can establish latent infection in wild-type B6 mice. We found that peritoneal cells harvested from infected B6 mice at 28 dpi could reactivate lytic RHVP virus in ex vivo cultures (Fig. 6B), as detected by the development of CPE in the Vero cell indicator monolayers. To determine whether this lytic viral replication was due to reactivation from latency or to the presence of preformed lytic virus in the cell samples, we also tested in parallel peritoneal cells that had been disrupted and were therefore unable to reactivate latent virus. Cell disruption was performed using conditions that killed 99% of cells, as assessed by trypan blue exclusion, but decreased the titers of preformed lytic virus in the samples by less than 5% (data not shown). We did not detect any CPE in ex vivo cultures of disrupted peritoneal cells (Fig. 6B), confirming that the lytic viral replication observed in wells containing nondisrupted peritoneal cells was the result of viral reactivation from latency. By contrast, no CPE was observed when splenocytes from infected mice were cultured ex vivo. Similar results were obtained when ex vivo reactivation was assessed in infected B6 or 129 mice at 42 dpi (not shown).
Since the frequency of ex vivo reactivation of γHV68 from infected splenocytes has been shown to decrease dramatically between 16 and 42 dpi (78), it is possible that the frequency of RHVP reactivation from infected splenocytes at 28 dpi is too low to detect. To further investigate whether RHVP can establish latent infection in splenocytes, we performed PCR using primers to RHVP ORF 50 to detect the presence of the viral genome in the same splenocyte and peritoneal cell samples analyzed in the ex vivo reactivation assays above. We were able to detect PCR products of the correct size in both infected splenocytes and peritoneal cells from all three experiments but not in splenocytes and peritoneal cells from uninfected mice (Fig. 6C). The identities of these ORF 50 PCR products were confirmed by sequencing (data not shown). These data demonstrate that splenocytes from infected mice are positive for the RHVP viral genome and suggest that RHVP can also establish latent infection in splenocytes.
DISCUSSION
We present here the annotated genome of a novel rodent gammaherpesvirus that establishes acute and latent infection in laboratory mice and contains multiple novel ORFs that may encode immunomodulatory factors. The RHVP genome contains a core group of conserved gammaherpesvirus genes that includes those encoding essential structural and replication proteins (Fig. 1 and Table 1) and is very similar to those of other gammaherpesviruses sequenced to date. RHVP also carries several unique ORFs, some of which have sequence similarity to other viral or cellular genes but are not present in γHV68. Phylogenetic analysis confirmed that RHVP is a member of the rhadinovirus genus that is distinct from γHV68 (Fig. 4). Thus, RHVP may provide a new small animal model for studying in vivo gammaherpesvirus infection and pathogenesis.
Annotation of ORFs in the RHVP genome was based on the criteria that the ORFs be predicted by both of the gene prediction algorithms used and that they contain an initiating methionine and a stop codon. As removal of either requirement resulted in the prediction of many more ORFs, this annotation may represent an underestimate of the coding potential of the RHVP genome. However, analysis of every open reading frame in the genome of at least 20 amino acids by BLASTP or comparison to the Pfam database yielded no additional conserved protein domains that did not correlate to annotated ORFs. This is consistent with the observation that none of the additional ORFs predicted using less-stringent criteria had significant matches to known genes by BLAST and suggests that all potential RHVP ORFs with significant similarity to known genes have been included in this annotation.
Overall, the RHVP genome is very similar to that of γHV68 (Fig. 1), and RHVP carries all the conserved gammaherpesvirus ORFs that have been found in γHV68 (84). RHVP and γHV68 also have similar numbers of predicted ORFs that are not conserved gammaherpesvirus ORFs, with 17 M ORFs in γHV68 and 18 R ORFs in RHVP. However, as ORFs in the RHVP and γHV68 genomes were not predicted using identical algorithms and criteria, the coding potential of the two viruses cannot be directly compared based on their annotated genomes. Importantly, ORF-based annotation of the γHV68 genome failed to reveal expressed genomic regions that were subsequently detected by tiled array (33). Moreover, although the RHVP genome does not appear to have tRNA-like molecules like those in γHV68 that encode the viral miRNAs, other gammaherpesviruses, including EBV and KSHV, have been shown to encode miRNAs that are not part of a tRNA-miRNA structure (8, 58, 59). Thus, it is possible that further examination may reveal viral miRNAs in the RHVP genome as well. We therefore predict that the ORF-based annotation presented here does not represent all of the genetic elements in the RHVP genome.
Like γHV68, RHVP encodes ORFs that may modulate host cellular pathways or the host immune response, including molecules similar to the KSHV K3 protein, Bcl-2, cyclin D, and G protein-coupled receptors (Table 1). As observed with γHV68, sequence conservation of the K3- and Bcl-2-like molecules appears to be concentrated in the RING-CH and BH1 domains, respectively (Fig. 3E and F), which suggests specific preservation of functional domains. One notable difference between RHVP and other gammaherpesviruses is the presence of two copies of the ORF 4 complement regulatory protein (ORFs 4a and 4b) in RHVP; γHV68, KSHV, and HVS each encodes only one copy (2, 65, 84). Similar to the ORF 4 homologs from γHV68, KSHV, and HVS, which have been shown to function as bona fide complement regulatory proteins (19, 35, 70, 71), both copies of the RHVP ORF 4 appear to contain key conserved residues that define the SCR motifs and to have four SCRs (Fig. 2). Interestingly, HVS also encodes a second complement regulatory protein, ORF 15, although it is most similar to CD59, an inhibitor of the complement membrane attack complex, and has been shown to block complement-mediated lysis at a terminal step in the pathway (1, 2, 64). Further study will be required to determine whether ORFs 4a and 4b provide distinct mechanisms for complement regulation.
Several RHVP ORFs that are not present in γHV68 may also function as immunomodulatory factors, including a superantigen-like molecule, three ORFs similar to MHC class I, and a C-type lectin-like protein. RHVP R11 is similar to the MMTV superantigen as well as to the ORF 14 superantigens in HVS and HVA (Fig. 3D). The function of HVA ORF 14 has not been extensively studied, but HVS ORF 14 has been shown to bind to the human MHC class II molecule, HLA-DR, and to stimulate T cell proliferation (93). Studies examining whether ORF 14 is required for T cell transformation and oncogenesis have, however, yielded conflicting results (14, 38, 39). We have found no evidence to date that RHVP infection is tumorigenic in laboratory mice (data not shown), and thus, the significance of the encoding of a superantigen by RHVP is unknown. Neither KSHV nor γHV68 has homologs of ORF 14, but the γHV68 M1 protein was recently shown to stimulate Vβ4+ CD8+ T cells in a superantigen-like manner, resulting in suppression of viral reactivation via long-term production of IFN-γ (17). Additionally, EBV has been shown to transactivate the env gene of the human endogenous retrovirus K18 (HERV-K18), which encodes a superantigen (74). The biological significance of this transactivation is unclear, but these observations suggest that association with superantigen activity is a feature that is common to multiple gammaherpesviruses.
Each of the three class I-like ORFs, R1, R2, and R3, has sequence similarity to human and mouse class I molecules, but interestingly, each is similar to a different region of the host class I molecules (Fig. 3A). This suggests the possibility that these three ORFs may be spliced to express a larger class I-like protein. An example of a gammaherpesvirus homolog of a cellular gene that has an exon-intron structure like that of the host gene is the ovine herpesvirus 2 (OvHV-2) IL-10 homolog, which retains the exact exon structure of the host IL-10 (32). As many of the key residues which contribute to peptide binding in the A or F pockets appear to be conserved in these three RHVP ORFs (Fig. 3A), it is possible that a class I-like molecule incorporating all three ORFs may be able to bind peptide. The presence of viral class I-like molecules in RHVP is, to our knowledge, unique among gammaherpesviruses. Unlike cytomegaloviruses, which encode multiple class I-like molecules that are thought to counter susceptibility to NK cell cytotoxicity resulting from viral downregulation of host class I (reviewed in reference 55), the gammaherpesviruses, such as KSHV and γHV68, that encode class I-downregulating proteins have not been reported to express viral class I homologs as decoy ligands. As RHVP encodes an ORF similar to K3, it is interesting to speculate that the RHVP class I-like ORFs may function to protect infected cells from NK killing following downregulation of host class I by RHVP K3.
Additionally, the RHVP R8 ORF encodes a C-type lectin-like molecule (Fig. 3C) similar to the murine CLEC2 family of proteins, which are ligands for the NK cell NK cell receptor protein 1 (NKR-P1) receptors (29). R8 also has sequence similarity to the RCMV RCTL molecule that functions as a decoy ligand for the NK inhibitory receptor, NKR-P1B (85). Although C-type lectin-like molecules are also encoded by several poxviruses (86), this is the first example of a C-type lectin-like molecule in a gammaherpesvirus. The similarity of RHVP R8 to RCTL and a member of the Clr family, including the absence of the C5 residue that is conserved in lectin-like domains but not in Clr family members (60), suggests that R8 may similarly function to engage NK cell receptors. Although further studies are required to determine the functions of R8 and the MHC class I-like ORFs, their presence in the RHVP genome suggests that the virus may encode molecules aimed at inhibiting NK cytotoxicity, which in turn suggests that NK cells may be involved in the control of RHVP infection.
The role of NK cells in the host immune response to RHVP infection was not examined in this study. However, data from in vivo RHVP infection demonstrate that intact interferon responses, as well as T and/or B cells, are important for protection from lethal infection (Fig. 5A). In mice lacking an interferon response, RHVP can replicate in multiple organs during acute infection, although the waning of viral titers by 26 dpi and the survival of some infected mice suggest that viral infection can be controlled even in the absence of an interferon response (Fig. 6A). The observation that RAG−/− mice survive for 60 days before universally succumbing to lethal infection (Fig. 5A) suggests that the innate immune response can control viral infection for a time but that T and/or B cells are required for long-term survival. The lack of detectable disease or viral titers in infected wild-type mice indicates that RHVP may replicate poorly in immunocompetent mice, although the mice can clearly be infected, as evidenced by the capacity of the virus to reactivate from latency following ex vivo culture of peritoneal cells from infected wild-type mice (Fig. 6B). It is possible that a higher infecting dose would result in detectable RHVP titers in organs and in the development of disease.
Like γHV68, RHVP can establish latent infection in peritoneal cells with the capacity to reactivate ex vivo (Fig. 6B). Although the specific cell type in which RHVP establishes latency has not been determined, it is possible that macrophages are the latently infected cells, since they are the major latent reservoir for γHV68 in the peritoneum (91). In contrast, we did not observe ex vivo reactivation of RHVP from splenocytes collected from infected mice at either 28 or 42 dpi (Fig. 6B and data not shown), although we were able to detect the presence of the viral genome by PCR in splenocytes from infected mice at 28 dpi (Fig. 6C). This suggests that RHVP is capable of establishing latency in splenocytes but may not reactivate at a high enough frequency at 28 dpi to be detected in our ex vivo reactivation assay. Future studies examining ex vivo reactivation at earlier time points will be helpful for assessing the ability of RHVP to reactivate from infected splenocytes. In summary, RHVP may provide a new and interesting mouse model for gammaherpesvirus infection in addition to γHV68, particularly for the study of potential immune evasion genes that are not carried by γHV68.
Acknowledgments
We acknowledge support from the Washington University/Pfizer Biomedical Agreement and the Center for Structural Genomics of Infectious Disease. This work was also supported by grants U54 AI057160 Project 15 to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (for initial virus identification), U54 AI057160-06S1 ARRA supplement (for sequencing and genomic analysis), and R01 CA096511 (for analysis of interferons in gammaherpesvirus latency). R.B.T. and H.G. were supported in part by NIH contract NO1-AI30027.
We thank the staff of the U.S. Naval Medical Research Institute Detachment, Lima, Peru, for their logistical and technical help in collecting rodent samples and facilitating their transport.
Footnotes
Published ahead of print on 5 January 2011.
REFERENCES
- 1.Albrecht, J. C., et al. 1992. Herpesvirus saimiri has a gene specifying a homologue of the cellular membrane glycoprotein CD59. Virology 190:527-530. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht, J.-C., et al. 1992. Primary structure of the herpesvirus saimiri genome. J. Virol. 66:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton, E. S., M. L. Lutzke, R. Rochford, and H. W. Virgin. 2005. Alpha/beta interferons regulate murine gammaherpesvirus latent gene expression and reactivation from latency. J. Virol. 79:14149-14160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Besemer, J., A. Lomsadze, and M. Borodovsky. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjorkman, P. J., et al. 1987. Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329:506-512. [DOI] [PubMed] [Google Scholar]
- 8.Bogerd, H. P., et al. 2010. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral microRNAs. Mol. Cell 37:135-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boname, J. M., and P. G. Stevenson. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15:627-636. [DOI] [PubMed] [Google Scholar]
- 10.Coscoy, L., and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. U. S. A. 97:8051-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davison, A. J., M. Benko, and B. Harrach. 2003. Genetic content and evolution of adenoviruses. J. Gen. Virol. 84:2895-2908. [DOI] [PubMed] [Google Scholar]
- 12.Dodd, R. B., et al. 2004. Solution structure of the Kaposi's sarcoma-associated herpesvirus K3 N-terminal domain reveals a novel E2-binding C4HC3-type RING domain. J. Biol. Chem. 279:53840-53847. [DOI] [PubMed] [Google Scholar]
- 13.Dong, B. Q., et al. 2007. Detection of a novel and highly divergent coronavirus from Asian leopard cats and Chinese ferret badgers in Southern China. J. Virol. 81:6920-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duboise, M., et al. 1998. A role for herpesvirus saimiri orf14 in transformation and persistent infection. J. Virol. 72:6770-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehlers, B., et al. 2007. Identification of novel rodent herpesviruses, including the first gammaherpesvirus of Mus musculus. J. Virol. 81:8091-8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emanuelsson, O., S. Brunak, G. von Heijne, and H. Nielsen. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2:953-971. [DOI] [PubMed] [Google Scholar]
- 17.Evans, A. G., et al. 2008. A gammaherpesvirus-secreted activator of Vβ4+ CD8+ T cells regulates chronic infection and immunopathology. J. Exp. Med. 205:669-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finn, R. D., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211-D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fodor, W. L., et al. 1995. The complement control protein homolog of herpesvirus saimiri regulates serum complement by inhibiting C3 convertase activity. J. Virol. 69:3889-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Früh, K., E. Bartee, K. Gouveia, and M. Mansouri. 2002. Immune evasion by a novel family of viral PHD/LAP-finger proteins of gamma-2 herpesviruses and poxviruses. Virus Res. 88:55-69. [DOI] [PubMed] [Google Scholar]
- 21.Gangappa, S., L. F. Van Dyk, T. J. Jewett, S. H. Speck, and H. W. Virgin. 2002. Identification of the in vivo role of a viral bcl-2. J. Exp. Med. 195:931-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gil, M. P., et al. 2001. Biologic consequences of Stat1-independent IFN signaling. Proc. Natl. Acad. Sci. U. S. A. 98:6680-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillet, L., J. S. May, S. Colaco, and P. G. Stevenson. 2007. The murine gammaherpesvirus-68 gp150 acts as an immunogenic decoy to limit virion neutralization. PLoS One 2:e705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen, T. H., S. Huang, P. L. Arnold, and D. H. Fremont. 2007. Patterns of nonclassical MHC antigen presentation. Nat. Immunol. 8:563-568. [DOI] [PubMed] [Google Scholar]
- 25.Hanson, L. K., J. S. Slater, Z. Karabekian, G. Ciocco-Schmitt, and A. E. Campbell. 2001. Products of US22 genes M140 and M141 confer efficient replication of murine cytomegalovirus in macrophages and spleen. J. Virol. 75:6292-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herr, R. A., J. Harris, S. Fang, X. Wang, and T. H. Hansen. 2009. Role of the RING-CH domain of viral ligase mK3 in ubiquitination of non-lysine and lysine MHC I residues. Traffic 10:1301-1317. [DOI] [PubMed] [Google Scholar]
- 27.Hewitt, E. W., et al. 2002. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J. 21:2418-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang, S., et al. 2009. Conserved herpesviral kinase promotes viral persistence by inhibiting the IRF-3-mediated type I interferon response. Cell Host Microbe 5:166-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iizuka, K., O. V. Naidenko, B. F. Plougastel, D. H. Fremont, and W. M. Yokoyama. 2003. Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat. Immunol. 4:801-807. [DOI] [PubMed] [Google Scholar]
- 30.Ishido, S., et al. 2000. Inhibition of natural killer cell-mediated cytotoxicity by Kaposi's sarcoma-associated herpesvirus K5 protein. Immunity 13:365-374. [DOI] [PubMed] [Google Scholar]
- 31.Ishido, S., C. Wang, B. S. Lee, G. B. Cohen, and J. U. Jung. 2000. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 74:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jayawardane, G., et al. 2008. A captured viral interleukin 10 gene with cellular exon structure. J. Gen. Virol. 89:2447-2455. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, L. S., E. K. Willert, and H. W. Virgin. 2010. Redefining the genetics of murine gammaherpesvirus 68 via transcriptome-based annotation. Cell Host Microbe 7:516-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapadia, S. B., B. Levine, S. H. Speck, and H. W. Virgin. 2002. Critical role of complement and viral evasion of complement in acute, persistent, and latent gamma-herpesvirus infection. Immunity 17:143-155. [DOI] [PubMed] [Google Scholar]
- 35.Kapadia, S. B., H. Molina, V. van Berkel, S. H. Speck, and H. W. Virgin IV. 1999. Murine gammaherpesvirus 68 encodes a functional regulator of complement activation. J. Virol. 73:7658-7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelley, L. A., and M. J. Sternberg. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363-371. [DOI] [PubMed] [Google Scholar]
- 37.Kieff, E., and A. Rickinson. 2007. Epstein-Barr virus and its replication, p. 2605-2654. In D. M. Knipe et al. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 38.Knappe, A., et al. 1997. The superantigen-homologous viral immediate-early gene ie14/vsag in herpesvirus saimiri-transformed human T cells. J. Virol. 71:9124-9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knappe, A., et al. 1998. T-cell lymphoma caused by herpesvirus saimiri C488 independently of ie14/vsag, a viral gene with superantigen homology. J. Virol. 72:3469-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LaPorte, S. L., et al. 2008. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell 132:259-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larkin, M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 42.Lee, B. J., U. H. Koszinowski, S. R. Sarawar, and H. Adler. 2003. A gammaherpesvirus G protein-coupled receptor homologue is required for increased viral replication in response to chemokines and efficient reactivation from latency. J. Immunol. 170:243-251. [DOI] [PubMed] [Google Scholar]
- 43.Li, H., K. Ikuta, J. W. Sixbey, and S. A. Tibbetts. 2008. A replication-defective gammaherpesvirus efficiently establishes long-term latency in macrophages but not B cells in vivo. J. Virol. 82:8500-8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang, C., X. E, and J. U. Jung. 2008. Downregulation of autophagy by herpesvirus Bcl-2 homologs. Autophagy 4:268-272. [DOI] [PubMed] [Google Scholar]
- 45.Liang, X., et al. 2006. Deregulation of DNA damage signal transduction by herpesvirus latency-associated M2. J. Virol. 80:5862-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang, X. Z., Y. C. Shin, R. E. Means, and J. U. Jung. 2004. Inhibition of interferon-mediated antiviral activity by murine gammaherpesvirus 68 latency-associated M2 protein. J. Virol. 78:12416-12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liszewski, M. K., T. C. Farries, D. M. Lublin, I. Rooney, and J. P. Atkinson. 1996. Control of the complement system. Adv. Immunol. 61:201-283. [DOI] [PubMed] [Google Scholar]
- 48.Loh, J., et al. 2005. A surface groove essential for viral bcl-2 function during chronic infection in vivo. PLoS Pathog. 1:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mansouri, M., et al. 2006. Kaposi sarcoma herpesvirus K5 removes CD31/PECAM from endothelial cells. Blood 108:1932-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ménard, C., et al. 2003. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J. Virol. 77:5557-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore, P. S., and Y. Chang. 2003. Kaposi's sarcoma-associated herpesvirus immunoevasion and tumorigenesis: two sides of the same coin? Annu. Rev. Microbiol. 57:609-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moorman, N. J., H. W. Virgin, and S. H. Speck. 2003. Disruption of the gene encoding the γHV68 v-GPCR leads to decreased efficiency of reactivation from latency. Virology 307:179-190. [DOI] [PubMed] [Google Scholar]
- 54.Moser, J. A., M. L. Farrell, L. T. Krug, J. W. Upton, and S. H. Speck. 2006. A gammaherpesvirus 68 gene 50 null mutant establishes long-term latency in the lung but fails to vaccinate against a wild-type virus challenge. J. Virol. 80:1592-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orange, J. S., M. S. Fassett, L. A. Koopman, J. E. Boyson, and J. L. Strominger. 2002. Viral evasion of natural killer cells. Nat. Immunol. 3:1006-1012. [DOI] [PubMed] [Google Scholar]
- 56.Page, R. D. 2002. Visualizing phylogenetic trees using TreeView. Curr. Protoc. Bioinformatics 6.2.1-6.2.15. [DOI] [PubMed]
- 57.Parry, C. M., et al. 2000. A broad spectrum secreted chemokine binding protein encoded by a herpesvirus. J. Exp. Med. 191:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pfeffer, S., et al. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 59.Pfeffer, S., et al. 2004. Identification of virus-encoded microRNAs. Science 304:734-736. [DOI] [PubMed] [Google Scholar]
- 60.Plougastel, B., C. Dubbelde, and W. M. Yokoyama. 2001. Cloning of Clr, a new family of lectin-like genes localized between mouse Nkrp1a and Cd69. Immunogenetics 53:209-214. [DOI] [PubMed] [Google Scholar]
- 61.Powers, A. M., et al. 1999. Isolation and genetic characterization of a hantavirus (Bunyaviridae: Hantavirus) from a rodent, Oligoryzomys microtis (Muridae), collected in northeastern Peru. Am. J. Trop. Med. Hyg. 61:92-98. [DOI] [PubMed] [Google Scholar]
- 62.Rickinson, A., and E. Kieff. 2007. Epstein-Barr virus, p. 2655-2700. In D. M. Knipe et al. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 63.Rodrigues, L., M. Pires de Miranda, M. J. Caloca, X. R. Bustelo, and J. P. Simas. 2006. Activation of Vav by the gammaherpesvirus M2 protein contributes to the establishment of viral latency in B lymphocytes. J. Virol. 80:6123-6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rother, R. P., et al. 1994. Inhibition of complement-mediated cytolysis by the terminal complement inhibitor of herpesvirus saimiri. J. Virol. 68:730-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Russo, J. J., et al. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. U. S. A. 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siegel, A. M., J. H. Herskowitz, and S. H. Speck. 2008. The MHV68 M2 protein drives IL-10 dependent B cell proliferation and differentiation. PLoS Pathog. 4:e1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sinha, S., C. L. Colbert, N. Becker, Y. Wei, and B. Levine. 2008. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy 4:989-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Söding, J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951-960. [DOI] [PubMed] [Google Scholar]
- 69.Song, W. J., et al. 2004. Functional genomics analysis of Singapore grouper iridovirus: complete sequence determination and proteomic analysis. J. Virol. 78:12576-12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spiller, O. B., D. J. Blackbourn, L. Mark, D. G. Proctor, and A. M. Blom. 2003. Functional activity of the complement regulator encoded by Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 278:9283-9289. [DOI] [PubMed] [Google Scholar]
- 71.Spiller, O. B., et al. 2003. Complement regulation by Kaposi's sarcoma-associated herpesvirus ORF4 protein. J. Virol. 77:592-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spreu, J., E. C. Kienle, B. Schrage, and A. Steinle. 2007. CLEC2A: a novel, alternatively spliced and skin-associated member of the NKC-encoded AICL-CD69-LLT1 family. Immunogenetics 59:903-912. [DOI] [PubMed] [Google Scholar]
- 73.Stevenson, P. G., et al. 2002. K3-mediated evasion of CD8(+) T cells aids amplification of a latent gamma-herpesvirus. Nat. Immunol. 3:733-740. [DOI] [PubMed] [Google Scholar]
- 74.Sutkowski, N., B. Conrad, D. A. Thorley-Lawson, and B. T. Huber. 2001. Epstein-Barr virus transactivates the human endogenous retrovirus HERV-K18 that encodes a superantigen. Immunity 15:579-589. [DOI] [PubMed] [Google Scholar]
- 75.Tarakanova, V. L., F. Kreisel, D. W. White, and H. W. Virgin. 2008. Murine gammaherpesvirus 68 genes both induce and suppress lymphoproliferative disease. J. Virol. 82:1034-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tarakanova, V. L., et al. 2007. Gamma-herpesvirus kinase actively initiates a DNA damage response by inducing phosphorylation of H2AX to foster viral replication. Cell Host Microbe 1:275-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tibbetts, S. A., et al. 2003. Establishment and maintenance of gammaherpesvirus latency are independent of infective dose and route of infection. J. Virol. 77:7696-7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tibbetts, S. A., F. Suarez, A. L. Steed, J. A. Simmons, and H. W. Virgin. 2006. A gamma-herpesvirus deficient in replication establishes chronic infection in vivo and is impervious to restriction by adaptive immune cells. Virology 353:210-219. [DOI] [PubMed] [Google Scholar]
- 80.van Berkel, V., et al. 2000. Identification of a gammaherpesvirus selective chemokine binding protein that inhibits chemokine action. J. Virol. 74:6741-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Dyk, L. F., H. W. Virgin, and S. H. Speck. 2000. The murine gammaherpesvirus 68 v-cyclin is a critical regulator of reactivation from latency. J. Virol. 74:7451-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Dyk, L. F., H. W. Virgin, and S. H. Speck. 2003. Maintenance of gammaherpesvirus latency requires viral cyclin in the absence of B lymphocytes. J. Virol. 77:5118-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Victoria, J. G., et al. 2010. Viral nucleic acids in live-attenuated vaccines: detection of minority variants and an adventitious virus. J. Virol. 84:6033-6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Virgin, H. W., et al. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Voigt, S., et al. 2007. Cytomegalovirus evasion of innate immunity by subversion of the NKR-P1B:Clr-b missing-self axis. Immunity 26:617-627. [DOI] [PubMed] [Google Scholar]
- 86.Voigt, S., G. R. Sandford, L. Ding, and W. H. Burns. 2001. Identification and characterization of a spliced C-type lectin-like gene encoded by rat cytomegalovirus. J. Virol. 75:603-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vossen, M. T., E. M. Westerhout, C. Soderberg-Naucler, and E. J. Wiertz. 2002. Viral immune evasion: a masterpiece of evolution. Immunogenetics 54:527-542. [DOI] [PubMed] [Google Scholar]
- 88.Wang, X., et al. 2007. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol. 177:613-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weck, K. E., M. L. Barkon, L. I. Yoo, S. H. Speck, and H. W. Virgin. 1996. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J. Virol. 70:6775-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weck, K. E., et al. 1997. Murine gammaherpesvirus 68 causes severe large vessel arteritis in mice lacking interferon-gamma responsiveness: a new model for virus induced vascular disease. Nat. Med. 3:1346-1353. [DOI] [PubMed] [Google Scholar]
- 91.Weck, K. E., S. S. Kim, H. W. Virgin, and S. H. Speck. 1999. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J. Virol. 73:3273-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wirth, S., et al. 2002. Regions of mouse mammary tumor virus superantigen involved in interaction with the major histocompatibility complex class II I-A molecule. J. Virol. 76:11172-11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yao, Z., et al. 1996. Herpesvirus saimiri open reading frame 14, a protein encoded by T lymphotropic herpesvirus, binds to MHC class II molecules and stimulates T cell proliferation. J. Immunol. 156:3260-3266. [PubMed] [Google Scholar]
- 94.Yin, X. M., Z. N. Oltvai, and S. J. Korsmeyer. 1994. BH1 and BH2 domains of Bcl-2 are required for inhibition of apoptosis and heterodimerization with Bax. Nature 369:321-323. [DOI] [PubMed] [Google Scholar]