Abstract
The ubiquitin ligase CBLL1 (also known as HAKAI) has been proposed to be a critical cellular factor exploited by West Nile virus (WNV) for productive infection. CBLL1 has emerged as a major hit in a recent RNA interference screen designed to identify cellular factors required for the early stages of the WNV life cycle. Follow-up experiments showed that HeLa cells knocked down for CBLL1 by a small interfering RNA (siRNA) failed to internalize WNV particles and resisted infection. Furthermore, depletion of a free-ubiquitin pool by the proteasome inhibitor MG132 abolished WNV endocytosis, suggesting that CBLL1 acts in concert with the ubiquitin proteasome system to mediate virus internalization. Here, we examined the effect of CBLL1 knockdown and proteasome inhibitors on infection by WNV and other flaviviruses. We identified new siRNAs that repress the CBLL1 protein and strongly inhibit the endocytosis of Listeria monocytogenes, a bacterial pathogen known to require CBLL1 to invade host cells. Strikingly, however, we detected efficient WNV, dengue virus, and yellow fever virus infection of human cells, despite potent downregulation of CBLL1 by RNA interference. In addition, we found that the proteasome inhibitors MG132 and lactacystin did not affect WNV internalization but strongly repressed flavivirus RNA translation and replication. Together, these data do not support a requirement for CBLL1 during flavivirus entry and rather suggest an essential role of the ubiquitin/proteasome pathway for flavivirus genome amplification.
West Nile virus (WNV), dengue virus (DV), and yellow fever virus (YFV) are mosquito-borne flaviviruses that cause neurological disease and hemorrhagic fever in humans and constitute major public health threats (11, 19). They are lipid-enveloped viruses that contain a single-stranded, positive-sense RNA genome (12, 16). To initiate their infectious life cycle, flaviviruses interact via their envelope glycoprotein (E protein) with still-uncharacterized cellular receptors (2), enter the host cell by clathrin-mediated endocytosis, and traffic through the endosomal pathway (5, 15, 27). Endosome acidification triggers major conformational changes in their E proteins that induce fusion of the viral and host cell membranes (4, 13, 20), resulting in the release of the viral capsid and genomic RNA into the cytosol. The viral genome is translated in the cytoplasm as a single polyprotein, which is co- and posttranslationally processed by both host cell signalases and the virus-encoded protease NS2B/NS3 into three structural and seven nonstructural (NS) proteins (12, 16). Because flaviviruses encode only 10 proteins, they need to subvert and/or counteract host cell functions for viral propagation (8). Understanding the interplay between flaviviruses and the host cell machinery could provide important insights into the molecular mechanisms of viral pathogenesis and could facilitate the development of a novel generation of antiviral drugs targeting host cell factors. A recent genome-wide RNA interference (RNAi) screen, designed to identify cellular factors associated with the early stages of the WNV life cycle (entry, viral-genome replication, and translation of the viral RNA [vRNA]), revealed that more than 300 host proteins affect the outcome of infection (14). These proteins are involved in crucial host cell processes, including intracellular protein trafficking; signal transduction; ion and molecular transport; and nucleic acid, protein, and lipid metabolism. In agreement with previous data (5, 15), this study confirmed the importance of both the clathrin-mediated pathway for endocytosis and host factors (e.g., viral ATPase [vATPase]) that regulate acidification of endosomes (14). In addition, it revealed an unexpected role for CBLL1 during WNV infection (14). CBLL1, also known as HAKAI, is a cbl-like ubiquitin ligase that has been recently identified as an E-cadherin binding protein (9). CBLL1 mediates ubiquitination of E-cadherin and regulates E-cadherin complex endocytosis (9), a mechanism that is coopted by the bacterium Listeria monocytogenes for internalization (3). Krishnan et al. found that silencing of the CBLL1 gene by RNA interference strongly impaired both WNV and DV serotype 2 (DV2) infection (14). Moreover, they proposed that CBLL1 acts in concert with the ubiquitin/proteasome system to mediate WNV endocytosis (14). Indeed, transfection of CBLL1 small interfering RNA (siRNA) in HeLa cells, as well as pretreatment of cells with the proteasome inhibitor MG132, impaired internalization of tetramethyl-rhodamine isocyanate-labeled WNV particles (14). In this report, we reevaluate the roles of both the ubiquitin ligase CBLL1 and the proteasome system in WNV infection and extend our study to other pathogenically relevant flaviviruses, such as DV and YFV.
MATERIALS AND METHODS
Cell culture and virus strains.
HeLa MZ (a gift of L. Pelkmans, ETH-Zurich), HeLa ATCC, A549 (a gift of A. Helenius, ETH-Zurich), and BHK NS3-Luc-NS3 (a gift of M. Jacobs, University College London Medical School) cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S). The cells were maintained in a 5% CO2 incubator at 37°C. C6/36 mosquito cells from Aedes albopictus adapted to grow at 28°C were cultured in Leibovitz's L-15 medium supplemented with 10% FBS, P/S, tryptose phosphate broth, and nonessential amino acids. WNV lineage I (Israeli IS-98-STI strain) and DV serotype 1 (FGA/NA d1d strain) were grown on C6/36 cells. DV serotype 2 (New Guinea C strain) and YFV (17D strain) were propagated on BHK-21 cells. Virus titers were determined by focus immunodetection assay (FIA) on C6/36 cells as previously described (18) and were expressed as focus-forming units (FFU). WNV-green fluorescent protein (WNII-GFP) was produced from a DNA-launched molecular clone of a lineage II strain of WNV (956 D117 3B) that encodes GFP (23) (a gift of T. Pierson, University of Pennsylvania) and was grown on BHK-21 cells. WNV reporter virus particles (RVPs) were produced by a complementation approach as described previously (1, 24). Briefly, HEK-293T cells were cotransfected with a DNA-launched WNV replicon and a plasmid encoding WNV C-prM-E polyprotein. Two days after transfection, RVPs were collected, filtered, and stored at −80°C until they were used. RVPs were titrated by fluorescence-activated cell sorter (FACS) on Vero cells.
Transfections and infections.
HeLa MZ, HeLa ATCC, and A549 cells were reverse transfected using the Lipofectamine RNAiMax protocol (Invitrogen) with 50 nM siRNAs. After 72 h, the cells were infected at the indicated multiplicity of infection (MOI), and the percentage of infected cells was quantified by flow cytometry 24 h postinfection. The CBLL1 siRNAs used in this study are the ON-TARGETplus SMARTpool (Dhar.OTP) (Dharmacon L-007069-00), the siGenome (GE) SMARTpool (Dhar.GE) (Dharmacon; M-007069-00; originally used by Krishnan et al. [14]), and a duplex siRNA from Qiagen (SI00137424). As a positive control, we used an siRNA targeting the ATP6V1B2 subunit (ON-TARGETplus SMARTpool L-011589-01) or the clathrin heavy chain (CHC) (Dharmacon; ON-TARGETplus SMARTpool L-004001-00). Negative-control siRNAs were the ON-TARGETplus nontargeting (NT) (Dharmacon; pool D-001810-10), the siGenome NT siRNA (Dharmacon; pool D-001206-05), and the All Stars Negative Control siRNA SI03650318 (purchased from Qiagen). For Listeria experiments, cells were transfected with pcDNA3-E-cadherin, or pcDNA empty vector as a control, 24 h before the experiments, using Gene Juice (Novagen) according to the manufacturer's instructions.
Flow cytometry assays (FACS).
Intracellular viral antigens were stained with anti-E specific monoclonal antibody (MAb). Briefly, infected cells were simultaneously fixed and permeabilized using Cytofix/Cytoperm buffer according to the manufacturer's instructions (Pharmingen). For labeling, cells were incubated with mouse MAb detecting WNV (4G2), DV (2H2), or YFV (2D12) E protein and then labeled with a polyclonal goat anti-mouse immunoglobulin/RPE (DakoCytomation; R0480). The cells were resuspended in phosphate-buffered saline (PBS) plus 4% paraformaldehyde (PFA) and subjected to flow cytometry analysis (FACSCalibur; Becton Dickinson). The data were analyzed by using CellQuest software (BD Biosciences).
Western blot analysis.
Cells were lysed in lysis buffer (50 mM Tris-HCl, 150 mM NaCl, and 1% Triton X-100) in the presence of complete protease inhibitors (Roche Diagnostics). Samples were adjusted for protein content (determined by the Bradford method) and subjected to sodium dodecyl sulfate-polyacrylamide gel (4 to 12%) electrophoresis (Bio-Rad, Hercules, CA), followed by transfer onto polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA). The membranes were probed using the following antibodies (Abcam): mouse MAb against CHC (1:200), rabbit polyclonal Ab against CBLL1 (1.25 μg/ml), and rabbit polyclonal Ab against ATP6V1B2 (1:500). To control for the amount of protein loaded per well, the membranes were probed using either a mouse MAb against GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Abcam; 1:50,000) or a rabbit polyclonal Ab against β-tubulin (Santa Cruz, CA; 1:1,000). Staining was revealed with horseradish peroxidase (HRP)-coupled secondary antibodies (Vector Laboratories) and developed using ECL Plus Western Blotting Detection reagents (Amersham) following the manufacturer's instructions. Image acquisition was performed by using an LAS-1000 charge-coupled-device (CCD) camera and Image Gauge software version 4.22 (Fuji Photo Film Co.).
Viral-RNA purification, reverse transcription, and real-time qPCR.
Total RNA was extracted from infected cells using an RNeasy Mini Kit (Qiagen), treated with DNase (RNase-free DNase set from Qiagen), and stored at −20°C until it was used. Total cDNA was produced by reverse transcription (RT) of equal amounts of purified RNA (0.5 μg) using random primers (Roche) and SuperScriptII Reverse Transcriptase (Invitrogen), according to the manufacturer's protocols. Samples were treated with RNase H (Biolabs). Real-time quantitative PCR (qPCR) was then used to quantify genomic viral RNA using an ABI Prism 7300 sequence detection system and the SYBR green PCR kit according to the manufacturer's instructions (Applied Biosystems). The cycling conditions were 10 min at 95°C and 40 cycles of 95°C for 15 s and 60°C for 1 min. The primers for the qPCR were designed with PrimerExpress 2.0 software (Applied Biosystems) within the conserved region of the capsid gene of WNV (5′ primer, CCGCGTGTTGTCCTTGATT; 3′ primer, GCCCCTTGCCGTCGAT), YFV17D (5′ primer, GCCGTTCCCATGATGTTCTG; 3′ primer, CACCCGTCATCAACAGCATT), and DV2NGC (5′ primer, CTGGTGGCGTTCCTTCGTT; 3′ primer, TGTTCCCCATCTCTTCAGTATCC). For GAPDH, the QuantiTect Primer Assay GAPDH primers (Qiagen) were used. The results were normalized to GAPDH gene copy numbers by using the 2−ΔΔCT method (17). To quantify the differences in vRNA production, the relative level of vRNA in control samples was assigned a value of 100%.
Drug treatments.
Experimental cells were pretreated before infection with the indicated amounts of the drug MG132 (Sigma-Aldrich C2211-5MG), lactacystin (Calbiochem 426100), chlorpromazine (Sigma-Aldrich C8138-5G), bafilomycin A1 (Calbiochem 196000), mycophelonic acid (MPA) (Sigma-Aldrich M3536-50MG), or heparin (Sigma-Aldrich H1027). The inhibitors were present throughout the assay unless otherwise stated. Mock-treated cells were incubated with an equivalent volume of drug solvent: dimethyl sulfoxide (DMSO) (for proteasome inhibitors) or methanol (for MPA).
Cell viability.
Cell viability was measured by using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, which is based on the conversion of MTT into formazan crystals by mitochondrial dehydrogenases. The test was performed after a 24-h incubation of HeLa MZ cells with MG132 or lactacystin according to the instructions of the manufacturer (Sigma). Absorbance was measured on an enzyme-linked immunosorbent assay (ELISA) reader using a test wavelength of 570 nm.
Gentamicin invasion assays and immunofluorescence analysis.
L. monocytogenes ΔinlB (7) was grown at 37°C to an optical density at 600 nm (OD600) of 0.8. Before infection, the bacteria were washed twice in PBS and diluted in DMEM to an MOI of 50. HeLa cells were washed twice with DMEM and incubated with bacteria for 1 h at 37°C. The cells were then washed three times with complete culture medium and incubated for 2 h at 37°C in complete culture medium containing gentamicin (25 μg·ml−1). The cells were then washed three times with PBS and lysed in 0.2% Triton X-100. The number of viable bacteria released from the cells was assessed by titering on agar plates. For immunofluorescence analysis, after the gentamicin treatment, the cells were washed three times with PBS and fixed in 4% paraformaldehyde. The cells were then incubated with the anti-L. monocytogenes antiserum R11 before permeabilization to specifically label extracellular bacteria (25), washed, and incubated with a Cy5-coupled polyclonal secondary antibody. The cells were then permeabilized and incubated with the same anti-L. monocytogenes antiserum to label total bacteria (extracellular and intracellular) and with a fluorescein isothiocyanate (FITC)-coupled polyclonal secondary antibody. In addition, the cells were labeled with the anti-E-cadherin monoclonal antibody HECD1 (Zymed), followed by a Cy3-coupled monoclonal secondary antibody. Comparing extracellular versus total bacteria assessed the efficiency of bacterial internalization.
Luciferase assay.
Luciferase activity (in relative light units [RLU]) was measured in cell lysates 12 h after cell treatment using the Promega Luciferase Kit according to the manufacturer's instructions. Luciferase activity was measured with a microplate reader (Mithras LB940; Berthold Technologies, Bad Wildbad, Germany) for 10 s.
Statistical analyses.
GraphPad Prism software was used for graphical representations and statistical analyses. All results are expressed as means and standard deviations. The error bars in the graphs represent standard deviations. The results were tested for significance using Student's t test. Results that had P values of <0.05 were considered significant and are indicated with asterisks.
RESULTS AND DISCUSSION
Potent CBLL1 silencing in HeLa cells using RNA interference.
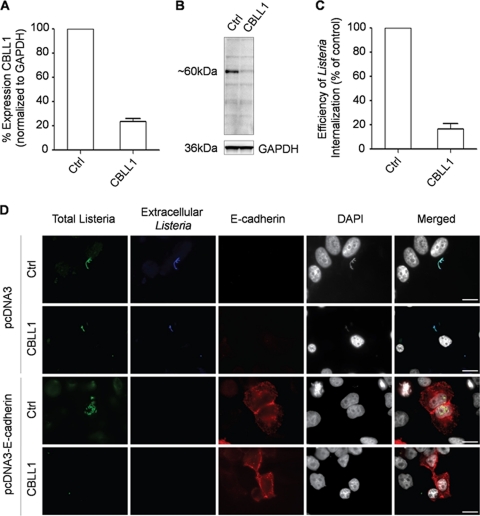
A recent RNAi screen showed that HeLa cells knocked down for CBLL1 expression failed to internalize WNV particles and resisted infection (14). This conclusion was mainly based on experiments done with a single siRNA targeting the CBLL1 gene, and only limited validation analyses are presented in this study. One recommended approach to prevent problems of false positives when performing RNAi screening is to confirm the observed phenotype using several siRNAs targeting the respective mRNAs at different sites (21). To strengthen the conclusion that CBLL1 is a critical host factor exploited by WNV to accomplish the early steps of infection, we first characterized a new siRNA pool directed against CBLL1 obtained via the Dharmacon ON-TARGETplus SMARTpool technology. ON-TARGETplus SMARTpool comprises chemically modified siRNAs designed to reduced off targets and enhance siRNA effectiveness. Our initial studies focused on HeLa MZ cells (a HeLa-derived cell line provided by Lucas Pelkmans, ETH-Zurich) that can be efficiently transfected with siRNA (26) and infected by WNV (this study) or other flaviviruses (26). siRNA-transfected cells were viable over the experimental procedure and presented no obvious defect in either morphology or growth (data not shown). Real-time PCR (qPCR) experiments indicated that HeLa MZ cells transfected with siRNA against CBLL1 displayed a strong reduction in steady-state CBLL1 mRNA levels compared to cells that received control siRNA (Fig. 1 A). The efficient knockdown of CBLL1 was also verified by Western blot analysis. As shown in Fig. 1B, a strong repression of CBLL1 protein expression was observed only in lysates of CBLL1 siRNA-transfected cells, but not in controls. The blots were also probed for GAPDH as a loading control, and similar amounts of GAPDH were detected in both CBLL1-depleted and control samples (Fig. 1B). To confirm the functionality of our CBLL1-targeting siRNA sequences, we followed the internalization of the bacterial pathogen L. monocytogenes, which invades epithelial cells by two pathways, depending on the bacterial surface proteins InlA and InlB. The receptor for InlA is E-cadherin, and it has been shown that InlA-mediated invasion depends exclusively on CBLL1 (3, 25). As HeLa cells do not express E-cadherin, control and CBLL1-depleted cells were transfected with either a vector encoding human E-cadherin or the corresponding empty vector as a control. After transfections, cells were infected with L. monocytogenes ΔinlB to specifically focus on the InlA internalization pathway, and the efficiency of bacterial internalization was evaluated by a gentamicin survival assay (Fig. 1C) and by immunofluorescence (Fig. 1D). As expected, bacteria could not invade cells not expressing E-cadherin regardless of CBLL1 depletion. On the other hand, bacteria efficiently infected E-cadherin-expressing cells, and such efficiency was reduced by 80% when CBLL1 was depleted, as assessed by titering bacteria on agar plates (Fig. 1C).
FIG. 1.
Efficient siRNA-mediated knockdown of CBLL1 expression in HeLa cells. HeLa MZ cells were reverse transfected for 72 h with the Dhar.OTP siRNA targeting CBLL1 or a nonspecific sequence (Ctrl). (A) CBLL1 mRNA levels in transfected cells were quantified by qPCR as described in Materials and Methods. These experiments were performed in triplicate samples, and the mean average results are shown, along with the standard deviation of the data, indicated by error bars. (B) Immunoblot analysis of CBLL1 protein levels in siRNA-transfected cells using polyclonal anti-CBLL1 antibodies. GAPDH was measured as a loading control. Control and CBLL1-depleted HeLa MZ cells were transfected with either a pcDNA3 empty vector or pcDNA3-E-cadherin and incubated with L. monocytogenes ΔinlB. (C) The efficiency of bacterial internalization was quantified by a gentamicin survival assay. These experiments were performed in triplicate samples, and the mean average results are shown, along with the standard deviations of the data, indicated by error bars. (D) Extracellular and total (extracellular and intracellular) bacteria were detected with the anti-Listeria antibody on nonpermeabilized cells (blue) and permeabilized cells (green), respectively. HeLa cells were labeled with DAPI and the anti-E-cadherin antibody HECD1 to detect E-cadherin-expressing cells (red). Scale bars, 10 μm. The results are representative of three independent experiments.
HeLa MZ cells knocked down for CBLL1 support normal levels of WNV and other flavivirus infectivity.
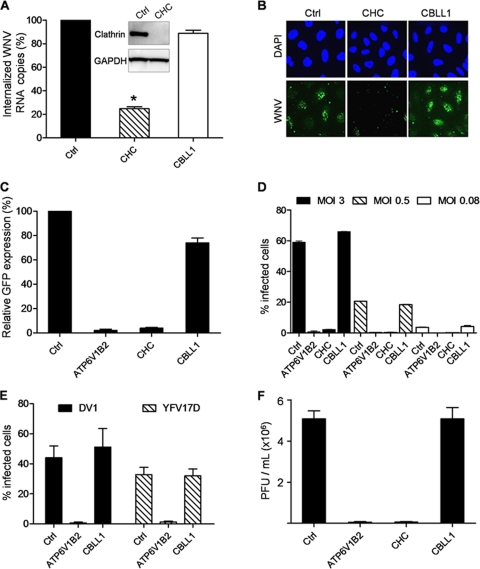
As CBLL1 has been proposed to be necessary for internalization of WNV (14), we used different assays to test whether CBLL1 knockdown would prevent virus entry into target cells. First, we transfected HeLa MZ cells with control or CBLL1 siRNA and then incubated them with mosquito-grown WNV particles (Israel 1998 strain; MOI, 10) for 1 h at 4°C. After being washed, the cells were incubated for 20 min at 37°C to allow virus endocytosis and incubated with proteinase K to remove virus remaining at the cell surface. Proteinase K treatment eliminated more than 90% of noninternalized cell surface-bound WNV particles, as assessed by staining cells with the anti-E MAb 4G2 (data not shown). Total RNA was extracted from infected cells, and viral-RNA penetration was quantified by qPCR experiments. As a control, the CHC, a cellular protein regulating clathrin-coated-pit formation, was also targeted for knockdown. Compared to control siRNA, CHC siRNA substantially reduced the level of CHC protein expression in HeLa MZ cells (Fig. 2 A), leading to a strong inhibition of clathrin-mediated uptake of transferrin (data not shown). CHC knockdown potently inhibited WNV RNA entry onto HeLa cells (Fig. 2A), confirming previous findings that WNV endocytosis is mediated by a clathrin-dependent mechanism (5, 15). In contrast, we observed that CBLL1 depletion did not affect viral-RNA uptake. qPCR analysis revealed that the amount of viral RNA internalized in cells transfected with CBLL1 siRNA was similar to the amount internalized in cells that received control siRNA (Fig. 2A). In a second approach, siRNA-transfected cells were incubated with WNV particles (MOI, 50) for 1 h at 4°C and shifted to 37°C for 30 min, and virus uptake was monitored by fluorescence microscopy using the anti-WNV E protein MAb E16 (22). As anticipated, treatment of HeLa MZ cells with CHC siRNA resulted in strong reduction of WNV E protein intracellular accumulation (Fig. 2B). Consistent with Fig. 2A, CBLL1 knockdown did not affect WNV E protein uptake in HeLa MZ cells (Fig. 2B). On the basis of these experiments, we concluded that WNV endocytosis could be significantly reduced in HeLa MZ cells upon potent knockdown of CHC. In contrast, despite potent knockdown of CBLL1 expression, HeLa MZ cells still efficiently internalized WNV particles. As the recent study by Krishnan et al. indicated that CBLL1 silencing resulted in a marked reduction of WNV-infected cells (14), additional efforts were made to investigate the role played by the ubiquitin ligase CBLL1 during the WNV life cycle. To test whether CBLL1 could affect an early step of infection (fusion, uncoating, or genome amplification), we tested the ability of our CBLL1 siRNA to suppress GFP encoded by WNV RVPs upon infection. For this purpose, we generated WNV RVPs by complementation of a subgenomic WNV replicon encoding GFP with a plasmid encoding WNV structural (C, PrM, and E) proteins provided in trans (24). RVPs are capable of only a single round of infection and are therefore particularly useful to study WNV entry and genome amplification (24). As a positive control, we transfected HeLa MZ cells with siRNA targeting CHC or the gene for ATP6V1B2, a subunit of a vacuolar ATPase that mediates endosomal acidification and is required for flavivirus pH-dependent fusion. As expected, both CHC and ATP6V1B2 knockdown strongly impaired WNV infection in HeLa cells, as indicated by the potent inhibition of GFP expression upon infection (Fig. 2C). In contrast, depletion of CBLL1 did not have a major impact on WNV RVP infection. Indeed, HeLa cells transfected with CBLL1 or control siRNA showed analogous levels of GFP expression, indicating similar susceptibility to WNV infection (Fig. 2C). To validate these findings, siRNA-transfected cells were challenged with mosquito-grown WNV particles at different MOIs (3, 0.5, and 0.08), and infection was evaluated 24 h later by flow cytometry analysis (FACS) using the 4G2 MAb that recognizes WNV E protein. Consistent with the results presented in Fig. 2A to C, WNV particles efficiently infected HeLa MZ cells transfected with CBLL1 siRNA (Fig. 2D). In contrast, WNV infection could be significantly reduced in HeLa MZ cells upon knockdown of CHC or ATPV1B2 (Fig. 2D). To further test whether CBLL1 plays a role during the replication cycles of other flaviviruses, siRNA-transfected cells were infected with DV1 or YFV-7D, and infection was monitored by FACS using the anti-E protein MAbs 2H2 and 2D12, respectively. As found for WNV, both DV1 and YFV 17D productively infected cells lacking CBLL1, while viral infection was strongly inhibited in cells transfected with siRNA targeting the ATP6V1V2 gene (Fig. 2E). To assess whether CBLL1 could regulate a late step in the WNV life cycle, titration experiments were performed on supernatants harvested from siRNA-transfected cells. As shown in Fig. 2F, CBLL1 silencing did not impair the production of infectious WNV particles. However, cells knocked down for CHC or ATPV1B62 expression showed a massive reduction in secretion of infectious particles compared to control cells. Altogether, these studies demonstrate that knocking down CBLL1 has no obvious effect on infection of HeLa MZ cells by WNV and other flaviviruses.
FIG. 2.
WNV enters into and productively infects HeLa MZ cells knocked down for CBLL1. (A) Cells were reverse transfected with Dhar.OTP siRNA targeting CBLL1, the CHC, or a nonspecific sequence (Ctrl). Transfected HeLa MZ cells were incubated with WNV (MOI, 10) for 2 h at 4°C, shifted to 37°C for 30 min, and treated with proteinase K. Total RNA was extracted from infected cells, and WNV genome copy numbers were quantified by qPCR. The inset shows a Western blot of clathrin (CHC) and GAPDH protein contents in siRNA-treated cells. The asterisk indicates a P value of <0.05 (B) SiRNA-transfected cells were incubated with WNV particles at an MOI of 50 for 2 h at 4°C, shifted to 37°C for 30 min, and fixed with paraformaldehyde. Virus internalization was analyzed by immunofluorescence using the anti-WNV E protein E16. (C) Cells were reverse transfected with Dhar.OTP siRNA targeting CBLL1, the CHC, APT6V1B2, or a nonspecific sequence (Ctrl). The cells were then challenged with WNV RVP, and infection was measured for 48 h as a function of GFP expression using flow cytometry. The results are expressed relative to the percentage of GFP-positive cells detected in cells transfected with control siRNA. The error bars indicate the standard errors of three independent experiments. (D) Cells transfected as indicated in panel C were challenged with WNV at MOIs of 3, 0.6, and 0.08 for 2 h, washed, and incubated for 24 h at 37°C. Viral infection was quantified by FACS after 24 h using the MAb 4G2. (E) Cells were reverse transfected as indicated in panel C and infected with DV1 or YFV17D (both at an MOI of 2). Viral infection was quantified by FACS 24 h later using the MAbs 2H2 and 2D12, respectively. (F) Titration of cell-free supernatants collected from siRNA-transfected cells and challenged with WNV. The viral titers in supernatants were expressed as PFU per ml in C6/36 cells. The values are means and standard deviations of three independent experiments done in triplicate.
Effects of different CBLL1 siRNAs on WNV infection of human cells.
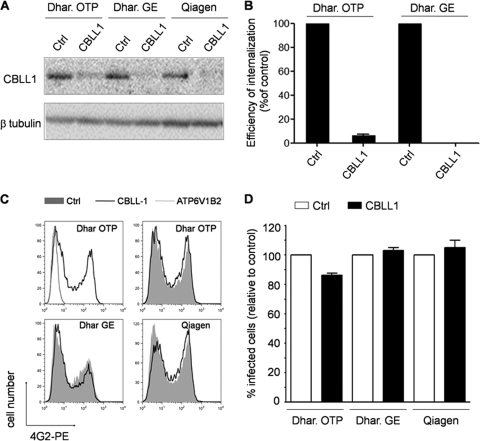
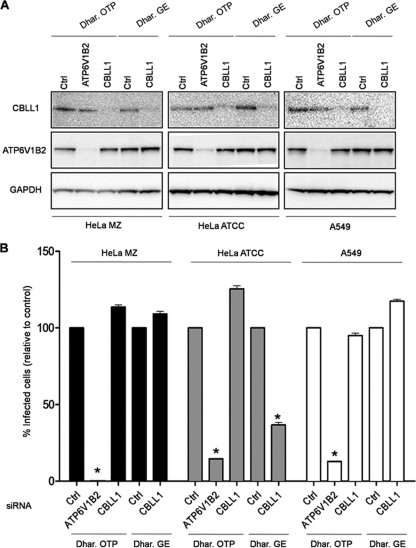
In light of the discrepancy between our results and those previously reported by Krishnan et al. (14), we set out to evaluate in more depth the potential involvement of CBLL1 during the early steps of WNV infection. First, we tested different siRNAs targeting different regions within the CBLL1 gene for the ability to inhibit WNV infection. To this end, we purchased, in addition to the On-Target plus Smart pool, the siGenome SMARTpool originally used by Krishnan et al. (14) and a duplex siRNA obtained from Qiagen. Upon transfection of HeLa MZ cells with these different siRNAs, CBLL1 protein levels were strongly repressed in comparison with their respective controls (Fig. 3 A). We also observed that the siRNA pool Dhar.GE strongly inhibits L. monocytogenes entry into HeLa MZ cells expressing E-cadherin, as efficiently as the siRNA pool Dhar.OTP (Fig. 3B). Strikingly, however, the Dhar.GE siRNA did not have a drastic effect on WNV infection of HeLa MZ cells (Fig. 3C). Similar results were obtained when cells were transfected with the CBLL1 Qiagen siRNA and challenged with WNV particles (Fig. 3C). To rule out the possibility that the absence of effect of CBLL1 depletion on WNV infection is dependent on the viral strain used in our study, similar experiments were conducted with a molecular clone of a lineage II WNV strain that expresses a GFP reporter gene (WNII-GFP; strain 956 D117 3B) (23). Infectious WNII-GFP particles were produced in HEK 239T cells according to the experimental procedure described by Pierson et al. (23) and were used to infect siRNA-transfected-HeLa MZ cells. Depletion of CBLL1 using three independent siRNA oligonucleotides did not impair WNII-GFP infection, as indicated by the accumulation of GFP in infected cells (Fig. 3D). Collectively, these data indicate that HeLa MZ cells with CBLL1 expression potently downregulated support normal WNV and other flavivirus infectivities. Because CBLL1 emerged as a hit in an RNAi screen performed in a HeLa cell derivative (HeLa ATCC cells) different from the one used in our study (HeLa MZ cells), we finally assessed whether discrepancies between our results and those previously reported by Krishnan et al. (14) could be explained by a cell-type-specific role of CBLL1. To explore this possibility, we reverse transfected three different human cell lines (HeLa MZ, HeLa ATCC, and A549) for 72 h with CBLL1 Dhar.OTP, CBLL1 Dhar.GE, or their cognate nontargeting siRNA controls and compared their abilities to support WNV infection. As indicated in Fig. 4 A, the siRNAs significantly reduced CBLL1 protein levels, with similar efficiencies in all the cell types tested. For positive-control purposes, HeLa MZ, HeLa ATCC, and A549 cells were also transfected with siRNA directed against ATPV1B62, which resulted in a strong reduction of ATPV1B62 protein levels and a potent inhibition of WNV infection (Fig. 4B). This study revealed that HeLa MZ, HeLa ATCC, and A549 cells transfected with Dhar.OTP siRNA against CBLL1 were susceptible to WNV infection with efficiencies similar to those of cells transfected with control nontargeting siRNA. Consistent with the findings of Krishnan et al. (14), we observed that the Dhar.GE CBLL1 siRNA pool significantly inhibits WNV infection when transfected in HeLa ATCC cells. However, we found that this oligonucleotide pool has virtually no effect on WNV infectivity when transfected in HeLa MZ or A549 cells, despite its ability to promote potent CBLL1 protein knockdown. Similar results were obtained when siRNA-transfected cells were infected with DV1 strain FGA and stained with the anti-E protein MAb 2H2 (data not shown). These data indicate that CBLL1 siRNA previously used in the work of Krishnan et al. (14) inhibits WNV infection only in a specific HeLa cell derivative but has no influence on WNV infection when transfected in other, related human cells. We therefore suggest that the apparent role of CBLL1 in WNV infection proposed by Krishnan et al. (14) could reflect an off-target effect produced by the Dhar.GE CBLL1 siRNA on HeLa ATCC cells. Collectively, the data obtained in this report strongly suggest that the ubiquitin ligase CBLL1 is dispensable for flavivirus infection.
FIG. 3.
Effects of different CBLL1 siRNAs on WNV infection. (A) HeLa MZ cells were transfected with three different siRNAs (Dhar.OTP, Dhar.GE, and Qiagen) targeting different regions within the CBLL1 gene. Control (Ctrl) siRNAs used in this study are the Dhar.OTP NT sequence, the Dhar.GE NT sequence, and a Qiagen NT sequence. Shown is immunoblot analysis of CBLL1 protein levels in siRNA-transfected cells using polyclonal anti-CBLL1 antibodies. β-Tubulin was measured as a loading control. (B) The efficiency of internalization of L. monocytogenes ΔinlB was evaluated by a gentamicin survival assay in HeLa MZ cells transfected with the indicated siRNAs and with pcDNA3-E-cadherin. (C and D) siRNA-transfected HeLa MZ cells were infected with mosquito-grown WNV particles or WNII-GFP, and viral infection was quantified by FACS analysis by measuring intracellular E protein (C) or GFP production (D). In panel C, a representative experiment out of four is shown. The filled histograms represent infection levels detected in cells receiving the control siRNA. The black lines indicate WNV infection in CBLL1 knockdown cells. In panel D, the values are means of three independent experiments with standard deviations.
FIG. 4.
WNV infection of different human cells knocked down for CBLL1 expression. (A) HeLa MZ, HeLa ATCC, and A549 cells were reverse transfected with the siRNA Dhar.OTP CBLL1 or Dhar.GE CBLL1 or their corresponding controls (Ctrl). The siRNA Dhar.OTP targeting the ATP6V1B2 gene was used as an internal control. Knockdown efficiency was assessed by Western blotting on cellular lysates and probed with anti-CBLL1, ATP6V1B2, and GAPDH antibodies. (B) Parallel siRNA-transfected samples were infected with WNV for 24 h, and E protein antigen was detected by flow cytometry. The results are expressed relative to the percentage of infected cells detected in cells transfected with control siRNA. The error bars represent standard deviations from the mean between three independent experiments, and the asterisks represent statistical significance compared to Ctrl siRNA transfection (P < 0.05).
Effect of proteasome inhibitors on flavivirus infection.
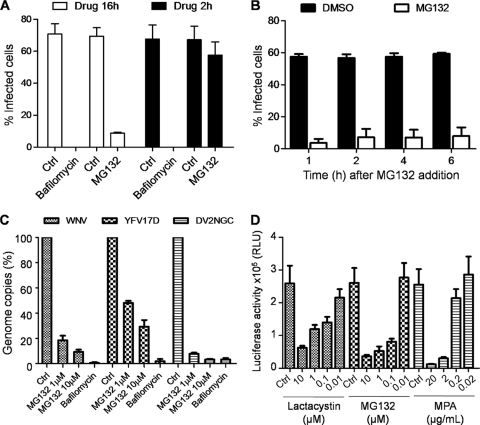
Based on the ability of CBLL1 to transfer a ubiquitin chain to cellular receptors and to promote their endocytosis and degradation by the 26S proteasome (9), it has been proposed that CBLL1 acts in concert with the ubiquitin-proteasome system to regulate WNV internalization (14). To investigate whether WNV endocytosis requires a functional proteasome activity, we performed internalization assays using either the irreversible (lactacystin) or reversible (MG132) specific proteasome inhibitor. In all the experiments described below, the no-drug controls received the same amount of DMSO solvent as the vehicle control. HeLa MZ cells were preincubated for 1 h with proteasome inhibitors (10 μM) or with heparin (50 μg/ml) or chlorpromazine (10 μg/ml) to block virus attachment and virus clathrin-mediated endocytosis, respectively. The cells were then incubated for 1 h at 4°C with WNV particles (MOI, 10), washed twice with cold PBS, and then incubated with drug-supplemented medium for 30 min at 37°C to allow virus endocytosis. Afterwards, the cells were treated with proteinase K, and genomic viral RNA was quantified by qPCR. The housekeeping GAPDH gene was used as an internal-control gene. As expected, we observed a strong inhibition of WNV internalization by heparin and chlorpromazine treatment (Fig. 5 A). However, neither MG132 nor lactacystin inhibited virus RNA entry, suggesting that the ubiquitin/proteasome system is not required for WNV internalization (Fig. 5A). Proteasome subunits were identified in a small-scale siRNA-based study (encompassing ∼5,500 genes) using WNV replicons containing a luciferase reporter, and proteasome was proposed to be required for a postinternalization step in WNV infection (10). This observation prompted us to decipher which step of the WNV life cycle would require a functional proteasome and whether exploitation of the ubiquitin/proteasome system is a hallmark of flavivirus infection. In a first set of experiments, we preincubated HeLa MZ cells for 30 min with different concentrations of either lactacystin or MG132 and then infected them with mosquito-grown WNV particles for 16 h in the continuous presence of the drugs. Viral infection was evaluated by FACS analysis as shown in Fig. 2. Compared to the DMSO-treated cells, MG132 and lactacystin reduced WNV E protein expression in a concentration-dependent manner (Fig. 5B). Neither of the drugs caused cytotoxicity, as measured by a standard MTT assay (Fig. 5C). Interestingly, under identical experimental conditions, similar results were obtained with the DV1 FGA strain (Fig. 5D) or YFV-17D (Fig. 5E). Furthermore, we observed that MG132 inhibits, in a dose-dependent manner, WNV infection of C6/36 mosquito cells (Fig. 5F), suggesting that the ubiquitin-proteasome system is also required for flavivirus infection in insect vectors. We then took advantage of the reversible property of MG132 to determine whether removing proteasome inhibitor treatment would restore WNV E protein production in infected cells. For this purpose, HeLa MZ cells were preincubated for 30 min with MG132 (10 μM) or bafilomycin A1 (Calbiochem; 25 nM), a specific inhibitor of vacuolar ATPase that blocks flavivirus fusion, and thereafter incubated the cells with WNV for 2 h. The cells were then washed twice in PBS and cultured for 16 h in the presence or absence of drugs. As shown in Fig. 6 A, MG132 had little or no effect on E protein expression when the drug was removed 2 h postinfection. In contrast, removal of bafilomycin A1 did not restore E protein accumulation in infected cells. To investigate whether a functional ubiquitin/proteasome system is required for a postentry step in WNV infection, we carried out time-of-addition experiments in which MG132 was added 1 h, 2 h, 4 h, or 6 h after WNV incubation. We observed that E protein expression measured at 16 h postinfection was severely reduced even at the 6-h time point (Fig. 6B), strongly suggesting that MG132 inhibits a postentry step during the flavivirus life cycle. To ascertain whether proteasome activity was required for flavivirus RNA amplification, HeLa cells were preincubated for 30 min with either MG132 (10 and 1 μM) or bafilomycin A1 and infected independently with WNV, DV2 (New Guinea C), or YFV 17D at an MOI of 1 for 1 h. After washes in PBS, the cells were cultured for 8 h in the presence of inhibitors. Total RNAs from infected cells were extracted, and viral-RNA amplification was quantified by qPCR using specific primers located in the capsid gene. As shown in Fig. 6C, WNV RNA amplification was strongly repressed in a dose-dependent manner in cells treated with MG132 or bafilomycin A1 compared to control cells. Interestingly, we observed that both DV2 and YFV-17D RNA amplifications are dramatically inhibited by MG132 (Fig. 6C), indicating that the proteasome pathway is coopted by several flaviviruses to promote viral replication. To selectively investigate whether the ubiquitin/proteasome system was required for flavivirus RNA translation and replication, we examined the effects of either lactacystin or MG132 treatment of BHK cells stably transfected with a firefly luciferase DV replicon (BHK-luc). Replicons are subgenomic viral RNAs in which the virus structural genes (region C-prM-E) are deleted. When introduced into host cells, replicons are translated by the cellular machinery and express the viral nonstructural proteins required for replication. MPA, a drug previously shown to inhibit flavivirus replication (6), was used as a positive control in the experiments. Twelve hours following incubation with different concentrations of drugs, luciferase activity in BHK lysates was quantified using a Mithras LB940 (Berthold). A dose-dependent reduction of firefly luciferase activity in BHK cells treated with the proteasome inhibitors was observed (Fig. 6D). These data strongly suggest that a functional ubiquitin/proteasome system is required for flavivirus RNA amplification.
FIG. 5.
Proteasome inhibitors prevent a postentry step during flavivirus infection. (A) HeLa MZ cells were incubated for 45 min with either MG132 (30 μM), lactacystin (30 μM), heparin (50 μg/ml), chlorpromazine (CPZ) (10 μg/ml), or equivalent mock control. The cells were incubated with WNV for 1 h at 4°C (MOI, 10), washed, and shifted to 37°C for 30 min in the presence of drugs. Total RNA was extracted from infected cells, and genomic viral RNA was quantified by qPCR as described in Materials and Methods. The results were expressed relative to the percentage of WNV genome copies in control cells treated with no drug. The values are means of three independent experiments with standard deviations. The asterisks represent statistical significance compared to mock-treated cells (P < 0.05). (B) HeLa MZ cells were preincubated for 30 min with different concentrations of MG132, lactacystin, or DMSO (Ctrl). The cells were infected with WNV (MOI, 2) in the continued presence of drugs or corresponding control. Viral infection was evaluated as described in the legend to Fig. 4. (C) Cell viability was measured by using the MTT assay, as described in Materials and Methods. Values are shown as percentages of mock-treated cells, and the error bars represent standard deviations from the mean of three independent experiments. (D) HeLa MZ cells were preincubated for 30 min with MG132 (10 μM), lactacystin (10 μM), or DMSO (Ctrl). The cells were infected with mosquito-grown DV1 (MOI, 1) in the continued presence of drugs. Viral infection was evaluated as described in the legend to Fig. 4 using the 2H2 MAb. One representative experiment out of four is shown. (E) HeLa MZ cells were preincubated for 30 min with MG132 (10 μM) or DMSO (Ctrl) and infected with YFV-17D (MOI, 1) in the continued presence of drugs. Viral infection was evaluated as described in the legend to Fig. 4 using the anti-YFV E protein MAb 2D12. One representative experiment out of three is shown. (F) C6/36 cells were preincubated with the indicated amounts of MG132 for 45 min and then infected with WNV (MOI, 3). E protein expression was detected in infected cells by flow cytometry 24 h postinfection. The values are means and standard deviations of three independent experiments.
FIG. 6.
A proteasome system is required for flavivirus RNA translation and replication. (A) HeLa cells were preincubated for 1 h with MG132 (10 μM), bafilomycin (25 nM), or DMSO and then infected with WNV (MOI, 3) for 2 h in the presence of drugs. The cells were then grown in medium supplemented (white bars) or not (black bars) with the drugs. The cells were stained for intracellular E protein production 16 h postinfection and analyzed by FACS. (B) HeLa MZ cells were infected with WNV for 2 h and washed extensively, and MG132 was added to the cell culture at different time points after infection. Viral infection was evaluated by FACS 16 h postinfection using the 4G2 MAb. (C) HeLa cells pretreated for 1 h with MG132 (1 or 10 μM) and bafilomycin A1 (25 nM) were infected with WNV (MOI, 3), YFV vaccine strain 17D (MOI, 3), or DV2 New Guinea C strain (MOI, 3) for 8 h. Total RNA was extracted from infected cells, and WNV, YFV17D, and DV2 NGC genome copy numbers were quantified by qPCR as described in Materials and Methods. The results were expressed as percentages of entry in control cells (Ctrl) treated with no drug. (D) BHK NS3-Luc-NS3 cells expressing a firefly DV2 replicon were incubated with different concentrations of proteasome inhibitors or MPA as a positive control. Cell lysates were prepared 12 h after drug addition and analyzed for luciferase activity. The values are means and standard deviations of three independent experiments done in triplicate.
Conclusion.
The work presented here indicates that CBLL1 is dispensable for WNV entry. Using several RNA duplexes, we found that different human cells efficiently knocked down for CBLL1 expression are still able to internalize WNV particles and are readily infected by several flaviviruses. We also presented data indicating that the previously reported CBLL1 siRNA inhibits WNV infection independently of its ability to repress CBLL1 protein but has no influence on WNV infection when transfected in human cells other than those used by Krishnan et al. (14). Furthermore, our results do not support direct involvement of the proteasome system during WNV endocytosis. In agreement with previous findings (10), we rather suggest that the ubiquitin/proteasome system plays a critical role during a postentry step of the WNV life cycle. Indeed, our findings revealed that the proteasome activity is required for amplification of several flavivirus genomes. The precise mechanism and molecular targets of this regulation are currently unknown and await further investigation.
Acknowledgments
We thank T. Pierson and Mike Diamond for generously supplying us with WNII-GFP constructs and the monoclonal anti-WNV E protein E16, respectively. We thank Pascale Lesage, Alessia Zamborlini, and Xavier Carnec for critical reading of the manuscript.
A.A. is supported by grants from the Fondation pour la Recherche Médicale, the Institut Universitaire d'Hématologie, the Institut National de la Recherche Médicale (INSERM), and the Marie Curie FP6 framework (INTRAPATH Program: early-stage training in infectious diseases). L.M is funded by Fondation pour la Recherche Médicale. M.-D.F.-G. is supported by a Marie Curie and “La Caixa” fellowship.
Footnotes
Published ahead of print on 29 December 2010.
REFERENCES
- 1.Ansarah-Sobrinho, C., S. Nelson, C. A. Jost, S. S. Whitehead, and T. C. Pierson. 2008. Temperature-dependent production of pseudoinfectious dengue reporter virus particles by complementation. Virology 381:67-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansarah-Sobrinho, C., S. Nelson, and T. C. Pierson. 2007. Flaviviruses, p. 1-14. In S. Pöhlmann and G. Simmons (ed.), Viral entry into host cells. Landes Bioscience, Austin, TX.
- 3.Bonazzi, M., E. Veiga, J. Pizarro-Cerda, and P. Cossart. 2008. Successive post-translational modifications of E-cadherin are required for InlA-mediated internalization of Listeria monocytogenes. Cell Microbiol. 10:2208-2222. [DOI] [PubMed] [Google Scholar]
- 4.Bressanelli, S., et al. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu, J. J., and M. L. Ng. 2004. Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J. Virol. 78:10543-10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diamond, M. S., M. Zachariah, and E. Harris. 2002. Mycophenolic acid inhibits dengue virus infection by preventing replication of viral RNA. Virology 304:211-221. [DOI] [PubMed] [Google Scholar]
- 7.Dramsi, S., et al. 1995. Entry of Listeria monocytogenes into hepatocytes requires expression of inIB, a surface protein of the internalin multigene family. Mol. Microbiol. 16:251-261. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez-Garcia, M. D., M. Mazzon, M. Jacobs, and A. Amara. 2009. Pathogenesis of flavivirus infections: using and abusing the host cell. Cell Host Microbe 5:318-328. [DOI] [PubMed] [Google Scholar]
- 9.Fujita, Y., et al. 2002. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat. Cell Biol. 4:222-231. [DOI] [PubMed] [Google Scholar]
- 10.Gilfoy, F., R. Fayzulin, and P. W. Mason. 2009. West Nile virus genome amplification requires the functional activities of the proteasome. Virology 385:74-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gould, E. A., and T. Solomon. 2008. Pathogenic flaviviruses. Lancet 371:500-509. [DOI] [PubMed] [Google Scholar]
- 12.Gubler, D. J., G. Kuno, and L. Markoff. 2007. Flaviviruses, p. 1153-1252. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 13.Heinz, F. X., and S. L. Allison. 2001. The machinery for flavivirus fusion with host cell membranes. Curr. Opin. Microbiol. 4:450-455. [DOI] [PubMed] [Google Scholar]
- 14.Krishnan, M. N., et al. 2008. RNA interference screen for human genes associated with West Nile virus infection. Nature 455:242-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnan, M. N., et al. 2007. Rab 5 is required for the cellular entry of dengue and West Nile viruses. J. Virol. 81:4881-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindenbach, B. D., H. J. Thiel, and C. M. Rice. 2007. Flaviviridae: the viruses and their replication, p. 1101-1152. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 17.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 18.Lozach, P. Y., et al. 2005. Dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN)-mediated enhancement of dengue virus infection is independent of DC-SIGN internalization signals. J. Biol. Chem. 280:23698-23708. [DOI] [PubMed] [Google Scholar]
- 19.Mackenzie, J. S., D. J. Gubler, and L. R. Petersen. 2004. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 10:S98-S109. [DOI] [PubMed] [Google Scholar]
- 20.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313-319. [DOI] [PubMed] [Google Scholar]
- 21.Mohr, S., C. Bakal, and N. Perrimon. 2010. Genomic screening with RNAi: results and challenges. Annu. Rev. Biochem. 79:37-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliphant, T., et al. 2005. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 11:522-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pierson, T. C., et al. 2005. An infectious West Nile virus that expresses a GFP reporter gene. Virology 334:28-40. [DOI] [PubMed] [Google Scholar]
- 24.Pierson, T. C., et al. 2006. A rapid and quantitative assay for measuring antibody-mediated neutralization of West Nile virus infection. Virology 346:53-65. [DOI] [PubMed] [Google Scholar]
- 25.Pizarro-Cerdá, J., et al. 2002. Distinct protein patterns associated with Listeria monocytogenes InlA- or InlB-phagosomes. Cell Microbiol. 4:101-115. [DOI] [PubMed] [Google Scholar]
- 26.Snijder, B., et al. 2009. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature 461:520-523. [DOI] [PubMed] [Google Scholar]
- 27.van der Schaar, H. M., et al. 2008. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 4:e1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]