Abstract
Rotavirus replication and virulence are strongly influenced by virus strain and host species. The rotavirus proteins VP3, VP4, VP7, NSP1, and NSP4 have all been implicated in strain and species restriction of replication; however, the mechanisms have not been fully determined. Simian (RRV) and bovine (UK) rotaviruses have distinctive replication capacities in mouse extraintestinal organs such as the biliary tract. Using reassortants between UK and RRV, we previously demonstrated that the differential replication of these viruses in mouse embryonic fibroblasts is determined by the respective NSP1 proteins, which differ substantially in their abilities to degrade interferon (IFN) regulatory factor 3 (IRF3) and suppress the type I IFN response. In this study, we used an in vivo model of rotavirus infection of mouse gallbladder with UK × RRV reassortants to study the genetic and mechanistic basis of systemic rotavirus replication. We found that the low-replication phenotype of UK in biliary tissues was conferred by UK VP4 and that the high-replication phenotype of RRV was conferred by RRV VP4 and NSP1. Viruses with RRV VP4 entered cultured mouse cholangiocytes more efficiently than did those with UK VP4. Reassortants with RRV VP4 and UK NSP1 genes induced high levels of expression of IRF3-dependent p54 in biliary tissues, and their replication was increased 3-fold in IFN-α/β and -γ receptor or STAT1 knockout (KO) mice compared to wild-type mice. Our data indicate that systemic rotavirus strain-specific replication in the murine biliary tract is determined by both viral entry mediated by VP4 and viral antagonism of the host innate immune response mediated by NSP1.
Group A rotaviruses (RVs) are segmented double-stranded RNA viruses that replicate primarily in mature epithelial cells on the tips of small intestinal villi (7). Rotavirus is the most common cause of severe dehydrating diarrhea in infants and young children worldwide; these infections result in more than 600,000 deaths annually, mostly in developing countries, and over 2 million hospitalizations each year (20). Rotavirus infection is ubiquitous among mammals; however, virulent viral strains isolated from one animal species generally have a diminished replication capacity and diminished virulence in heterologous hosts. This phenomenon of rotavirus host range restriction was employed for the development of two rotavirus vaccines in which animal rotaviruses that are naturally attenuated in humans were used to produce the genetic backbone of human vaccines. The pentavalent rotavirus vaccine Rotateq (Merck), for example, is derived from a bovine rotavirus strain, WC, with incorporated human rotavirus genes that encode VP7 of serotypes 1, 2, 3, and 4 and VP4 of serotype P1A to induce neutralizing antibody (Ab) responses to the most common human RV serotypes.
The viral factors underlying rotavirus host range restriction are not fully understood but are likely to be multigenic. Early studies using reassortants between simian and bovine strains demonstrated that the viral surface protein VP4 plays a critical role in determining virulence in a heterologous suckling mouse model (19). A role for the viral surface proteins VP4 and VP7 in modulating virulence was reported for mouse, rabbit, and gnotobiotic pig models using various homologous or heterologous RVs (6, 13, 18). Other viral proteins such as VP3 and NSP4 have also been associated with virulence in various model systems (13). Broome et al. previously examined reassortant RVs generated from the cross of a virulent, highly replication-competent, homologous murine rotavirus (EW) and a heterologous, less virulent, and poorly replicating simian RV, RRV, in a mouse model and demonstrated that the infection dose required to induce diarrhea in 50% of suckling mice (diarrhea dose 50) and the ability to transmit infection to noninoculated litter mates were associated with viral gene 5, which encodes nonstructural protein 1 (NSP1) (5).
The function of NSP1, the most sequence-diverse gene in the RV genome, was initially unclear, as it was found to be dispensable for rotavirus replication in some cell culture systems (25). Recently, it was shown that NSP1 is an antagonist of the host innate immune response, especially the type I interferon (IFN) response. NSP1 interacts with several cellular interferon regulatory factors (IRFs), including IRF3, IRF5, and IRF7, and the interaction induces the degradation of these IRFs, resulting in the suppression of the type I IFN response (2, 3). In addition, in at least some circumstances, NSP1 inhibits interferon signaling by degrading βTrCP, a host factor responsible for the activation of the transcription factor NF-κB (12).
Previously, we showed that simian rotavirus RRV replicates efficiently in mouse embryonic fibroblasts (MEFs), whereas the replication of a bovine RV strain (UK) is severely restricted. Using a genetic analysis of reassortants generated between UK and RRV, we linked the growth differences between these two viruses in MEFs to the differential abilities of the UK and RRV NSP1 proteins to degrade IRF3 and suppress the cellular type I IFN response. These genetic data indicated that rotavirus replication in vitro is regulated by the NSP1 antagonism of cellular innate immunity, which is highly dependent on the virus strains and species and/or tissue origins of the host cells (9, 24).
It is less clear whether or how the virus-mediated antagonism of innate immunity affects rotavirus infection in vivo. We previously reported that in RRV-infected 5-day-old mice, the virus replicates in both the intestine and extraintestinal organs such as liver, bile duct, and mesenteric lymph nodes although without apparent systemic symptoms. In IFN-αβ/γ signaling-deficient IFN receptor (IFNR) or STAT1 knockout (KO) mice, RRV replication is significantly enhanced relative to infection of wild-type mice, especially in the liver, pancreas, and biliary tracts. This infection often results in lethal hepatitis, pancreatitis, and biliary atresia, supporting the notion that IFN regulates RV replication in vivo (8). In contrast, the systemic replication of bovine and porcine rotaviruses was not detectable in infected suckling mice, and the systemic replication of murine rotavirus was not significantly enhanced in the IFNR KO mice relative to wild-type mice (8). These results suggest that the murine, simian, and other heterologous (bovine and porcine) rotaviruses have very distinct enteric and systemic replication phenotypes in mice.
The mouse biliary tract is highly susceptible to RRV but not bovine rotavirus infection (8). Several studies have shown that the intraperitoneal (i.p.) inoculation of RRV in newborn mice results in high levels of viral replication in biliary epithelia and lethal biliary atresia, which can be prevented by the administration of type I IFN (22). These results combined with our previous observations of interferon signaling-deficient mice indicate that rotavirus infection of the biliary tract is virus strain specific and is regulated, at least in part, by the host IFN response. In this study, we sought to identify the genetic and mechanistic basis for these distinct in vivo phenotypes by the direct inoculation of mouse gallbladders with selected UK × RRV reassortants to determine the genetic basis for strain-specific replication in the biliary tract. We found that the high-growth phenotype of RRV in mouse biliary tissue in vivo was determined by the RRV VP4 and NSP1 genes. The restricted-growth phenotype of the bovine UK virus in biliary tissue was conferred mainly by UK VP4, since reassortants with UK VP4 replicated at low levels regardless of the origins of the NSP1 genes. In additional studies of cultured murine biliary epithelial cells, we found that reassortant virus with UK VP4 had diminished viral entry, providing a possible mechanistic basis for the role of VP4 in the biliary-growth phenotype. We further demonstrated that the expression of the IRF3-dependent ISG54 gene in biliary tissues was significantly upregulated after infection with reassortant viruses carrying RRV VP4 and UK NSP1, whereas p54 expression was low in biliary tissues from mice infected with RRV or viruses carrying RRV VP4 and NSP1. These findings are consistent with the hypothesis that RRV NSP1 enhanced replication due to its ability to block interferon production. Our study indicates that rotavirus species-specific replication in the biliary tract is regulated by both VP4, which mediates the viral entry of target cells, and NSP1, which suppresses the host innate immune response. The latter conclusion was also supported by the partial restoration of infectivity of reassortants carrying RRV VP4 and UK NSP1 in the biliary tract of IFN signaling-deficient mice.
MATERIALS AND METHODS
Viruses and cells.
Simian rotavirus RRV, bovine rotavirus UK, and their reassortant viruses were propagated in the green monkey kidney MA104 cell line. Viruses used to inoculate mouse gallbladder were concentrated from infected cell lysates. Briefly, MA104 cells were infected with the indicated RV strains or reassortants at a multiplicity of infection (MOI) of 0.1. At 3 to 4 days postinfection or when cytopathic effects (CPEs) were extensive, the cell lysates were collected and frozen and thawed two times. Cell lysates were first centrifuged at 1,000 × g with a GH-3.8 rotor in an Allegra 6R centrifuge (Beckman Coulter, Brea, CA) for 20 min. Pellets from the centrifugation were resuspended in medium 199 (M199) from Gibco (Grand Island, NY) supplemented with 1,000 U/ml penicillin and 1,000 μg/ml of streptomycin without fetal calf serum (FCS), extracted with Genetron, and added to supernatants. The supernatants were further centrifuged at 10,000 × g with a JLA-16.250 rotor in an Avanti J-26 XP centrifuge (Beckman Coulter) for 30 min to remove cell debris. Supernatants were collected, and viruses were pelleted at 83,000 × g with an SW-28 rotor in a L8-60m ultracentrifuge (Beckman Coulter) for 2 h using a 40% sucrose cushion. The pellets were collected and resuspended in M199. The virus titers in the concentrated viral stocks were determined by a plaque assay with MA104 cells and expressed as PFU per ml as described previously (14).
The MA104 cells were maintained in M199 supplemented with 1,000 U/ml penicillin, 1,000 μg/ml of streptomycin, and 10% FCS as described previously (14). The cultured mouse cholangiocyte line was a kind gift from Cara Mack (1). These cells are simian virus 40 (SV40) large-antigen-transformed primary mouse cholangiocytes originally generated by Yoshiyuki Ueno (27). The cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 1,000 U/ml penicillin, 1,000 μg/ml of streptomycin, and 10% FCS.
Mice.
IFN-α/β and -γ receptor double-knockout mice (IFN-αγR KO) on an 129sv background were a generous gift from H. W. Virgin IV (15). Wild-type 129sv mice and STAT1 knockout (STAT1 KO) mice, also on a 129sv background, were purchased from Taconic (Germantown, NY). All mice were maintained at the Veterinary Medical Unit of the Palo Alto VA Health Care System. All animal studies were approved by the Stanford Institutional Animal Care Committee.
Rotavirus intra-gallbladder infection and biliary tissue virus detection.
Twenty-day-old mice were anesthetized with ketamine (40 mg/kg of body weight) and xylazine (4 mg/kg). Hair on the abdominal area of the mice was shaved. A 1-cm-long incision was made in the upper mid-abdominal area. The gallbladder was isolated with a forceps, and 2 × 106 PFU of the indicated virus in a 5-μl volume was injected directly into the gallbladder. Wounds were closed with Autoclip wound clips (Mik Ron, Sparks, MD), and mice were given buprenorphine (0.05 mg/kg) to relieve pain postsurgery. At 16 h postinoculation, mice were sacrificed, and extrahepatic bile ducts, gallbladders, and a small amount of attached liver tissues were collected and stored at −80°C for subsequent virus detection. Each sample was weighed, and M199 without FCS was added to each sample to make a 10% (wt/vol) suspension that was then homogenized with a pestle. Samples were clarified by centrifugation at 20,000 × g in an Eppendorf microcentrifuge for 3 min.
Before virus titration, tissue suspensions were treated with trypsin (5 μg/ml) for 30 min at 37°C. The virus titration plaque assay was performed as described previously, and the titers of virus in tissues are presented as PFU per gram of tissue (8).
Histology and immunohistology.
Bile ducts from infected mice were collected at 16 h postinfection, frozen in optimal cutting temperature compound (OCT), and kept at −80°C until sectioning and staining. Sections 6 μm thick were cut, air dried, and fixed in a cold acetone-methanol mixture (1:1) for 10 min at −20°C. Tissues sections were stained with Texas Red-labeled monoclonal antibody (MAb) to VP6 (clone 1E11, which detects all rotavirus strains used in this study) and unlabeled rat MAb anti-K19 (clone Troma III, which specifically stains biliary epithelial cells; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA); Alexa 488-labeled goat anti-rat IgG was used as a secondary Ab (Invitrogen, Carlsbad, CA). The cell nucleus was stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Invitrogen). After staining, slides were mounted with Aqua Poly/Mount (Polysciences, Warrington, PA) and observed by using a Nikon Eclipse TE300 inverted fluorescent microscope equipped with a QImaging Retica 200R charge-coupled-device (CCD) camera (QImaging, Surrey, BC, Canada), and images were acquired and analyzed with the QCapture Pro program (QImaging, Surrey, BC, Canada).
Immunofluorescence, confocal imaging, and quantification of RV in cell cultures.
Cultured mouse cholangiocytes were seeded on 12-mm polycarbonate tissue culture inserts with 0.4-μm pores (Costar transwell filters; Corning, Corning, NY) at a density of 105 cells/cm2 and supplemented with fresh medium every day for 5 days. Cells were infected with the indicated trypsin-activated virus at an MOI of 200 for 1 h, washed three times with phosphate-buffered saline (PBS), and fixed with 3% paraformaldehyde in 100 mM phosphate buffer (pH 7.4) for 10 min as recently described (28). Infected cells were immunostained with anti-VP6 MAb 1E11 and Alexa Fluor (AF) 488-conjugated anti-mouse IgG secondary antibody (Invitrogen) under permeabilizing conditions (PBS, 1% saponin, 1% Triton X-100, 3% bovine serum albumin) and AF 594 phalloidin (Invitrogen). Each sample was then mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, CA) and imaged with a confocal microscope (LSM510; Zeiss, Jena, Germany). z stacks were reconstructed into three dimensions with Volocity software (Improvision, Coventry, England). Using Volocity software, VP6-immunostained dots representative of input virus were identified and quantified based on intensity and size (>1 μm or <20 μm). Dots were counted in at least three separate experiments, and each experiment consisted of the quantification of three representative fields under each condition.
Detection of p54 expression in biliary tissues using immunoblotting.
Biliary tissues from infected or control mice were collected and snap-frozen as described above. Frozen tissues were weighed and lysed in cold radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris [pH 7.5], 1% NP-40, 0.5% sodium deoxycholate, 0.2% SDS). Tissues were homogenized by using a pestle. An equal volume of 2× Laemmli buffer containing 2% SDS was added to the homogenates before boiling for 5 min. Samples were separated on 12% SDS-PAGE gels and immunoblotted as described previously (24). Blots were probed for p54 and tubulin levels using commercially available antibodies from Thermo Scientific (Waltham, MA) and Sigma (St. Louis, MO), respectively.
Statistical analysis.
Multiple-regression analysis was used to assess the association between the UK or RRV origin of each rotavirus gene and the levels of viral replication in biliary tissues with the SAS program (SAS Institute Inc., Cary, NC). Because the experimental design was fractional, collinearity may exist among gene effects. Collinearity can inflate standard errors, reduce statistical power, and, if severe enough, create numerical instabilities that cause regression calculations to fail. To address this possibility, regression modeling employed orthogonal coding (−1/2 for UK and +1/2 for RRV) of an individual gene's effects, and the Gentleman-Givens transformation was used to decompose the design matrix (11). To compare the levels of biliary viral replication among different VP4 and NSP1 genotype groups, virus titers of every sample from each of the group were log transformed and analyzed by using analysis of variance (ANOVA) with the PASW Statistics 12 program (SPSS Inc., Chicago, IL). A post hoc Scheffe test was used for pairwise comparisons between each two genotype groups. Levels of viral entry into cultured biliary cholangiocytes after infection with UK, RRV, or selected reassortants were analyzed by using the same methods.
RESULTS
Replications of UK × RRV reassortants in mouse biliary tissues.
Bovine rotavirus UK or simian rotavirus RRV was inoculated directly into the gallbladders of 20-day-old wild-type 129sv mice. At 16 h postinfection, gallbladders and extrahepatic bile ducts were collected for virus titration. As summarized in Table 1 and Fig. 1, UK RV replication was very restricted, and titers in biliary tissues were about 500 times lower than those of RRV (P < 0.001).
TABLE 1.
Viral titers in biliary tract of 20-day-old 129sv mice 16 h after intra-gallbladder inoculation with RRV, UK, or the indicated UK × RRV reassortant viruses
| Virus | Rotavirus genea |
Titer (PFU/g)b | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||
| 25-1-1 | R | U | U | R | R | U | R | U | R | R | U | 16,708,333 |
| 24-1-1 | R | U | U | R | R | U | R | U | R | U | U | 14,333,333 |
| 7-1-1 | R | R | R | R | R | R | R | R | R | R | U | 1,300,000 |
| 14-1-1 | U | R | U | R | R | R | R | R | R | R | R | 70,555,556 |
| 11-2-1-1 | R | U | R | R | R | U | R | R | R | R | U | 3,341,667 |
| 27-3-1 | U | R | R | R | R | R | R | R | R | R | R | 23,888,889 |
| 25-2-1 | R | R | R | R | R | U | R | R | R | U | R | 35,888,889 |
| 13-1-1 | R | R | R | R | R | U | R | R | R | R | R | 15,666,667 |
| 22-1-1 | R | R | R | R | R | R | R | U | R | R | R | 565,556 |
| 8-1-1 | R | R | U | R | R | R | R | R | R | R | U | 4,436,667 |
| 6-1-1 | R | R | R | R | U | U | R | U | R | R | R | 88,889 |
| 27-2-1 | R | R | R | R | U | U | R | R | R | R | R | 158,889 |
| 4-1-1 | R | R | R | R | U | U | R | R | R | R | U | 194,286 |
| 36-1-1 | R | U | R | R | U | R | R | U | U | R | R | 35,095 |
| 9-8-2 | U | U | U | R | U | U | R | U | U | U | U | 53,944 |
| 19-1-1 | R | R | R | U | R | R | R | U | R | R | R | 7,500 |
| 3-2-1 | U | U | U | U | R | U | U | U | U | U | U | 467 |
| 36-2-1 | U | U | U | U | U | U | U | U | R | U | R | 1,778 |
| 21-1-1 | U | U | U | U | U | R | U | U | U | R | R | 667 |
| 27-1-1-1 | U | U | R | U | U | R | U | R | U | U | U | 1,889 |
| 32-2-1-1 | U | U | R | U | U | U | R | U | U | U | R | 4,111 |
| 20-1-1 | U | R | U | U | U | R | U | U | U | R | R | 1,667 |
| UK | U | U | U | U | U | U | U | U | U | U | U | 8,467 |
| RRV | R | R | R | R | R | R | R | R | R | R | R | 4,266,667 |
U, gene from UK; R, gene from RRV. Viral VP4, NSP1, and VP7 are encoded by genes 4, 5, and 7, respectively.
Mean RV titer per gram of biliary tissue in mice infected with 2 × 106 PFU of RRV, UK, or UK × RRV reassortant virus for 16 h. The numbers of mice in each reassortant infected group varied from 4 to 10.
FIG. 1.
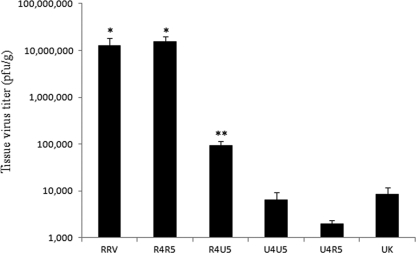
Mean RV titers in biliary tissue of mice infected with different gene 4 and 5 constellations of UK × RRV reassortant viruses. RRV, UK, or UK × RRV reassortant viruses (2 × 106 PFU) were inoculated directly into gallbladders of 20-day-old 129sv mice. Extrahepatic biliary ducts were collected, and virus titers were determined by plaque assay 16 h later. Mean tissue titers were calculated by grouping reassortants based on four different gene 4/5 constellations: RRV VP4 and RRV NSP1 (R4R5), RRV VP4 and UK NSP1 (R4U5), UK VP4 and UK NSP1 (U4U5), and UK VP4 and RRV NSP1 (U4R5). See Table 1 for viral titers. *, not significantly different from each other (P > 0.05) but significantly different from other groups (P < 0.05); **, significantly different from all other groups (P < 0.05).
To study the genetic basis of the differential replication between RRV and UK in the mouse biliary tract, we analyzed the replication of a series of UK × RRV reassortant viruses. The parental gene assignments for each reassortant virus and viral titers in biliary tissues 16 h postinoculation are presented in Table 1. Using multiple-regression analysis, we correlated the origin of each gene with the biliary tract titers. We found that the origins of the VP4, NSP1, and VP7 genes (genes 4, 5, and 7, respectively) were significantly correlated with the titers in biliary tissues. The regression coefficients were 4.55, 2.67, and 3.08 for VP4, NSP1, and VP7, respectively (P < 0.05), indicating that tissue titers increased significantly if these genes were derived from RRV. Further examination revealed that the biliary titers for reassortant viruses that had both VP4 and NSP1 derived from RRV (25-1-1, 24-1-1, 7-1-1, 14-1-1, 11-2-1-1, 27-3-1, 25-2-1, 13-1-1, 22-1-1, and 8-1-1) replicated to titers above 500,000 PFU/g (most were over 1,000,000 PFU/g). The mean virus titer for this reassortant group was 15.6 × 106 PFU/g (±4.2 × 106 PFU/g [standard error of the mean {SEM}]) and was not statistically different from that of RRV (12.7 × 106 PFU/g ± 5.7 × 106 PFU/g; P > 0.05), suggesting that RRV VP4 and NSP1 are strongly associated with the RRV biliary tract growth phenotype in mouse biliary tract (Fig. 1). The mean virus titer of reassortants with VP4 derived from UK but NSP1 from RRV (19-1-1 and 3-2-1) was 6.4 × 103 PFU/g ± 3.0 × 103 PFU/g, similar to that of the parental UK strain (8.5 × 103 PFU/g ± 3.4 × 103 PFU/g). Reassortants with both UK VP4 and NSP1 (36-2-1, 21-1-1, 27-1-1-1, 32-2-1-1, and 20-1-1) had a mean titer of 2.0 × 103 PFU/g ± 0.4 × 103 PFU/g (P > 0.05), indicating that the UK VP4 gene cosegregated with the UK growth phenotype in the mouse biliary tissues (Fig. 1). On the other hand, reassortants with RRV VP4 and UK NSP1 (1-2-1, 6-1-1, 9-8-2, 27-2-1, 4-1-1, and 36-1-1) had an intermediate replication phenotype (9.2 × 104 PFU ± 2.0 × 104 PFU) that was significantly lower than that of RRV or reassortants with RRV VP4 and NSP1 (P < 0.001) but significantly higher than that of UK or reassortants with UK VP4 (P < 0.001), as shown in Fig. 1. These data indicate that UK NSP1 plays a role in limiting rotavirus replication in biliary tissues.
RRV gene 7 encodes the viral surface glycoprotein VP7 and was associated with most high-yield reassortants, but it was also present in reassortants of the intermediate-yield group (6-1-1), which has RRV VP4 and UK NSP1, and the low-yield group (19-1-1 and 32-2-1-1), which have UK VP4 and RRV or UK NSP1 (Table 1). Although RRV VP7 may play a role in enhancing rotavirus replication in the biliary tract, it does not appear to be an independent determinant for the RRV growth phenotype in this tissue.
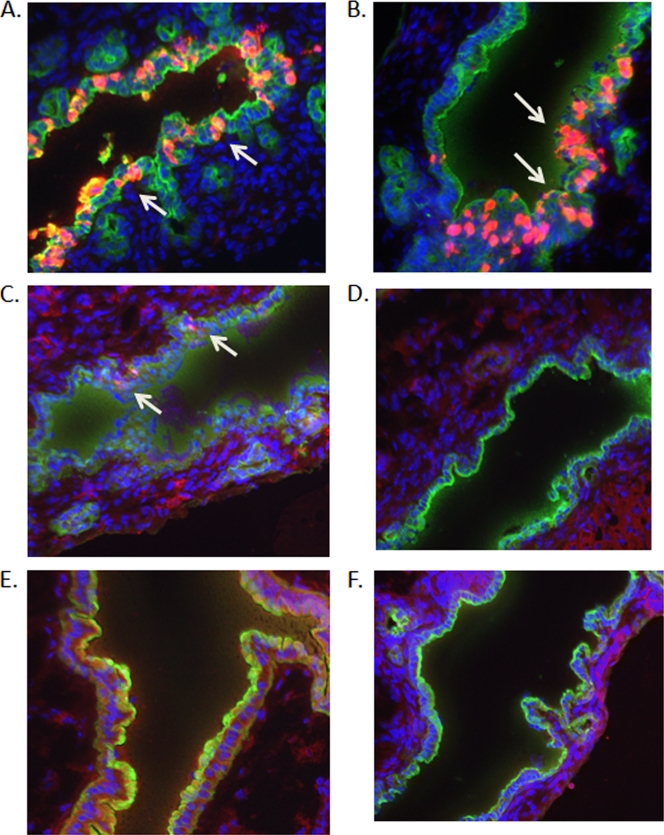
We then used immunofluorescent staining of biliary tissue sections to evaluate levels of RV antigens at 16 h after direct gallbladder inoculation with UK, RRV, and selected UK × RRV reassortants (Fig. 2). VP6 was detected primarily in cytokeratin K-19-positive cells from animals infected with RRV or reassortant 24-1-1 (RRV VP4 and NSP1), indicating that these virus strains replicated specifically in biliary epithelial cells in bile ducts (Fig. 2A and B). Viral antigen was not detected in bile ducts of animals infected with UK or reassortants with UK VP4 (19-1-1 and 27-1-1) (Fig. 2D to F). Infection with reassortant 27-2-1 (RRV VP4 and UK NSP1) resulted in greatly reduced levels of viral antigen in biliary epithelial cells compared to levels in RRV-infected animals (Fig. 2C). These results are consistent with the viral yield data presented in Table 1 and indicate that most replication occurred in biliary epithelial cells.
FIG. 2.
Detection of viral antigens in extrahepatic bile ducts 16 h after intra-gallbladder inoculation of RRV, UK, or selected UK × RRV reassortant viruses. Gallbladders of 20-day-old 129sv mice were inoculated directly with 2 × 106 PFU of RRV (A), 24-1-1 (R4R5) (B), 27-2-1 (R4U5) (C), UK (D), 19-1-1 (U4R5) (E), and 27-1-1 (U4U5) (F). Extrahepatic bile ducts were collected, frozen, and sectioned. Tissue sections were stained with anti-VP6 (red) and anti-cytokeratin K-19 (green) MAbs and DAPI (blue) and observed by using fluorescent microscopy. Arrows indicate infected cells.
Entry of UK, RRV, and selected UK × RRV reassortants into cultured biliary epithelial cells.
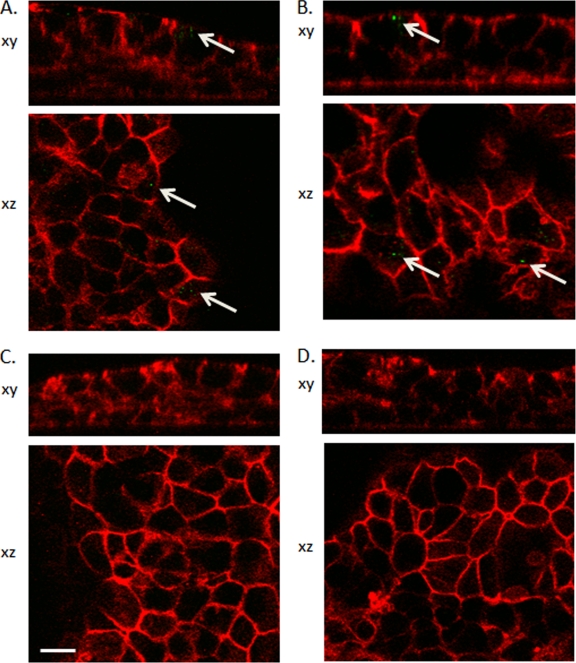
To investigate the mechanistic role of VP4 in the regulation of the differential infectivity of RRV and UK in biliary epithelial cells, we infected a cultured mouse cholangiocyte monolayer with UK, RRV, and selected reassortant viruses for 1 h. Immunofluorescent staining and confocal microscopy were used to visually examine and quantify entry into the cells, as described previously (28). Levels of RRV and reassortant virus with RRV VP4 and UK NSP1 (27-2-1) were significantly higher than levels of UK or reassortant virus with UK VP4 and RRV NSP1 (19-1-1), suggesting that viruses with UK VP4 had a diminished capacity for viral entry into murine biliary epithelial cells (Fig. 3 and Table 2). In addition, we also measured the levels of replication of these viruses in a cell line of murine cholangiocytes at 16 h (viral antigen staining) and 2 days (virus titer determination) postinfection and found that, similarly to in vivo studies, UK VP4 correlated with low levels of viral replication, and levels of replication of viruses with RRV VP4 were higher but were determined by the origin of the NSP1 proteins (data not shown).
FIG. 3.
Examination of virus entry into cultured mouse cholangiocytes. Mouse cholangiocytes were cultured in transwell plates for 5 days and inoculated with RRV (A), UK × RRV reassortant 27-2-1 (R4U5) (B), UK (C), or UK × RRV reassortant 19-1-1 (U4R5) (D) for 1 h at an MOI of 200. Cells were washed and fixed for staining. Optical sections were taken at a 0.2-μm resolution through the cells, and z stacks were reconstructed into three dimensions. RV antigens were visualized in green (anti-VP6), and red staining corresponds to the F-actin network visualized using labeled phalloidin. Arrows indicate viral antigens. These micrographs are representative of at least three separate experiments, and in each experiment, at least three fields were quantified. Scale bar, 10 μm.
TABLE 2.
Quantitative analysis of viral entry into cultured mouse cholangiocytes
The quantification of immunostained dots was based on the fluorescent intensity and size of each dot (see Materials and Methods). Results are mean numbers of immunostained dots (±SEM) quantified from four separate experiments.
Significantly different from UK or 19-1-1 (P < 0.05) but not different from each other (P > 0.05).
Expression of p54 in biliary tissues after infection with UK × RRV reassortant viruses.
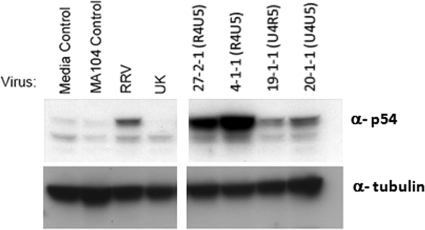
We previously showed that NSP1 proteins from UK and RRV degrade IRF3 with very different efficiencies in MEFs, and this results in significant differences in the activation of the type I IFN response and viral replication (9, 24). In order to quantify the ability of different reassortants to induce an interferon response in vivo, we examined the levels of p54 expression in biliary tissue following infection; the levels of the ISG54 gene product correlate with IRF3 activation (26). In biliary tissues of mice inoculated with either RRV or UK, low or undetectable levels of p54 expression were observed (Fig. 4). In contrast, the expression of p54 was strongly induced in the biliary tissues of mice infected with reassortants 27-2-1 and 4-1-1, which carry VP4 derived from RRV and NSP1 from UK, but not in mice infected with reassortant 20-1-1, which has both VP4 and NSP1 derived from UK (Fig. 4). These data suggest that the UK and RRV NSP1 proteins have significantly different effects on the IRF3-mediated innate responses in vivo; however, this effect is apparent only for viruses that possess RRV VP4 and are entry competent.
FIG. 4.
Detection of p54 expression in biliary tissues after rotavirus infection. Gallbladders of 20-day-old mice were inoculated with RRV, UK, or the indicated UK × RRV reassortants. Extrahepatic bile ducts were collected at 16 h postinfection. P54 and α-tubulin expressions in biliary tissues were detected by Western blotting with the indicated antisera.
To further examine the role of the innate immune response on rotavirus replication in biliary tissues, we inoculated gallbladders of IFN-αβ/γ receptor KO (IFNR KO) mice directly with UK, RRV, or selected UK × RRV reassortants. The virus titers in biliary tissues of IFNR KO mice infected with UK or RRV were 2.2 × 103 PFU/g ± 619 PFU/g and 1.7 × 106 PFU/g ± 0.3 × 106 PFU/g, respectively, and were not statistically different from the titers in wild-type mice (P > 0.05). The virus titers in biliary tissue of IFNR KO mice infected by reassortants with UK VP4 and UK NSP1, UK VP4 and RRV NSP1, or RRV VP4 and RRV NSP1 were 6.3 × 103 PFU/g ± 2.9 × 103 PFU/g, 10.0 × 103 PFU/g ± 3.4 × 103 PFU/g, and 11.5 × 106 PFU/g ± 2.4 × 106 PFU/g, respectively, and differences between IFNR KO and wild-type mice were not significant (P > 0.05). In contrast, the mean virus yield after infection of the gallbladders with RRV VP4 and UK NSP1 reassortants in IFNR KO mice (2.5 × 105 PFU/g ± 0.5 × 105 PFU/g) was 2.7 times higher than that of wild-type mice (P < 0.001). This significant difference in replication was also observed for STAT1 KO mice (data not shown).
DISCUSSION
The genetic basis of rotavirus virulence and host restriction has been studied with several animal model systems using various strains of virus originating from different homologous and heterologous host species. It is clear that rotavirus host or species restriction and virulence are both likely determined by multiple viral genes, and the relative importance of specific viral genes is highly dependent on the choice of virus strain and animal host studied. Based on our previous results with cell cultures and in vivo, we chose to study reassortants derived from the coinfection of cells with bovine UK and simian RRV strains. We chose these two strains for several reasons. First, both strains have been used as heterologous backbone strains for human RV vaccines (10, 17). Second, the RRV strain has a relatively unique phenotype vis-à-vis its ability to replicate efficiently in the murine biliary tree and cause a systemic liver and biliary tract disease in newborn or IFNR KO mice (8, 21, 23). We and others have shown previously that bovine and simian rotaviruses have significantly different replication efficiencies in intestine and some extraintestinal organs, including biliary tracts, after oral infection (8). In mouse embryonic fibroblasts, the difference in replication between these viral strains was mapped to the viral gene encoding NSP1 (9). The differential replication of UK and RRV in MEFs is determined by differences in cellular IRF3 degradation and type I IFN suppression by UK or RRV NSP1 (9). These prior observations provide an excellent rationale to study the genetic basis of strain-specific differences in systemic replication in the biliary tree in vivo using a classic genetic approach.
In this study of differences between the two heterologous nonmurine RV strains RRV and UK, we found that the rotavirus VP4 gene plays a pivotal role in regulating RV replication in the mouse biliary tract (Table 1 and Fig. 1). The expression of UK VP4 conferred a low-growth phenotype in mouse biliary tissues, whereas RRV VP4 conferred the high-growth phenotype but only if NSP1 was also derived from RRV. Rotavirus VP4 is the spike protein on the viral surface that is primarily responsible for the initial stages of viral attachment and entry into susceptible cells (16). We observed that viruses with VP4 derived from UK had a significantly reduced replication capacity and entered cultured biliary epithelial cells less efficiently than viruses with an RRV-derived VP4 gene (Table 2 and Fig. 3). These data suggest that VP4's critical role in regulating replication in the murine biliary tree takes place, at least in part, at the level of viral entry into target cells.
Earlier studies demonstrated that VP4 is one of the viral proteins responsible for the differential infectivity and virulence of several strains of rotavirus in cell culture and intestine (13, 19). Of note, a recent genetic study with a mouse model of rotavirus-induced biliary atresia (G. Tiao, personal communication) demonstrated that VP4 also determines this disease phenotype when RRV was compared to a non-disease-inducing simian RV (TOUCH). In this analysis of differences between TOUCH and RRV, the genetic origin of NSP1 was not significant. VP4 was not a primary determinant of enteric virulence and host range restriction when the homologous wild-type murine RV strain (EW) was compared to the significantly less virulent RRV strain (5). It should be noted that in these studies, the EW parental strain was not adapted to cell culture, so modest changes in the replication capacity between EW and reassortants containing RRV VP4 could not be accurately assessed because none of the cell culture-adapted reassortants contained a murine VP4 gene.
Results from our current studies of UK and RRV indicated that viral entry mediated by VP4 and/or VP7 is crucial but is not the sole determinant regulating systemic replication in the murine biliary tree. Interestingly, the role of UK VP4 in restricting viral replication in the mouse biliary tree was not observed for MEFs. In these cells, RRV × UK reassortant viruses replicated equally well irrespective of the genetic origin of VP4 (9). In MEFs, only UK NSP1 was clearly associated with the reduction of rotavirus replication, suggesting that the restrictive effects of VP4 are likely to be cell type specific.
We found that, along with VP4, the efficient biliary tree replication phenotype associated with RRV was also dependent on the genetic origin of NSP1. Only reassortants with both VP4 and NSP1 derived from RRV had phenotypes resembling that of the parental RRV strain and replicated to high titers in the biliary tree (Table 1 and Fig. 1). The replication of reassortants with RRV VP4 but UK-derived NSP1 grew less well but still to higher titers than parental UK or reassortants with UK VP4. These results suggest that after viral entry, which is enhanced substantially by the presence of RRV VP4, the efficiency of viral replication in biliary tissue is strongly influenced by the genetic origin of NSP1. The role of the NSP1 protein in modulating rotavirus host range restriction was first reported by Broome et al. for a suckling mouse model using genetic reassortment between RRV and a virulent homologous murine rotavirus, EW (5). In contrast, subsequent genetic studies using diverse animal models and different parental virus strains showed no association between NSP1 and rotavirus virulence (4, 6, 13). Although the reasons for these apparently conflicting results are not clear, disparate results are likely due to the fact that in some of the studies, virus shedding and diarrhea rather than direct measurements of viral loads in vivo were studied, viral replication differences were not examined, or the parental NSP1 genes studied may not have differed significantly. For example, in a recent comparison of biliary tract disease between the two simian strains TOUCH and RRV, the NSP1 sequences of the two strains shared 91% amino acid homology, diminishing the likelihood that in this comparison, NSP1 would be identified as a key determinant in replication restriction (G. Tiao, personal communication).
Rotavirus NSP1 induces the degradation of several interferon regulatory factors, including IRF3, IRF7, and IRF5, and the NF-κB regulatory factor βTrCP and suppresses the cellular IFN response in vitro (2, 3, 12). Of note, the efficiency of NSP1 antagonism of the host innate immune response is highly dependent on the virus strain and cell origin (24). The RV strain- and species-specific effects of NSP1 on the innate immune response in vivo have not yet been well studied. In this study we were not able to measure the NSP1-mediated degradation of IRF3 directly in infected tissues due to the constitutive expression of IRF3 in many cell types in the biliary tract. However, we demonstrated that viruses with RRV VP4 (which appears to facilitate biliary cell entry) and UK NSP1 (which does not efficiently degrade IRF3 and does efficiently activate IRF3 in MEFs) stimulated strong p54 expression in biliary tissues, whereas viruses with RRV VP4 and NSP1 induced a very weak p54 response. p54 expression is dependent on the activation of the IRF3 pathway by virus infection in a tissue-specific manner (26). Hence, we propose that the reduced biliary replication of reassortants with RRV VP4 and UK NSP1 is due to the inefficient suppression of IRF3 activation by UK NSP1. This conclusion is supported by the observation that the replication of viruses with RRV VP4 and UK NSP1 in biliary tissues was increased significantly (approximately 3-fold) in IFN signaling-deficient IFN receptor KO and STAT1 KO mice compared to wild-type hosts.
Previously, we found that the replication of UK in MEFs isolated from IFN receptor KO or STAT1 KO mice was enhanced more than 100 times compared to the replication in wild-type MEFs (9), suggesting that the effects of the IFN response on rotavirus replication are much stronger in vitro than in vivo. It is possible that other innate antiviral mechanisms that are differentially affected by UK or RRV NSP1 gene products are also involved in regulating rotavirus replication in the biliary tract. Graff et al. have shown previously that NSP1 mediates the proteasome-dependent degradation of β-TrCP and affects NFκB activation (12). In this study we did not directly examine the role of rotavirus NSP1 on other innate pathways activated during rotavirus infection in the biliary tract, but this topic is currently under investigation.
The role of VP7 in rotavirus species restriction was reported by several early studies (6, 13, 18), and the parental origin of VP7 was statistically correlated with viral titers in biliary tissues in this study. We found that all the reassortants with a high viral yield in the biliary tree had VP7 genes derived from RRV, whereas most (but not all) of the low-yield reassortants had UK VP7, suggesting that VP7 may also play an important role in regulating rotavirus replication in the biliary tract. However, the VP7 origin did not perfectly cosegregate with growth phenotypes, since RRV VP7 was harbored by several low-replicating reassortants, including 27-1-1-1 and 19-1-1. Interestingly, reassortant 19-1-1, which has UK VP4 but RRV VP7 and NSP1, displayed a reduced-viral-entry phenotype in cultured biliary epithelial cells, similarly to the parental UK virus. This indicated that RRV VP7 alone was not sufficient to mediate effective viral entry. Therefore, our results suggest that VP7 may function in collaboration with VP4 and NSP1 to enhance rotavirus infection in biliary tissues, but it does not independently determine rotavirus infectivity in vivo.
Our findings clearly demonstrate that the host restriction in biliary tract replication of the heterologous RV strain UK can be rescued in vivo by gene segments encoding VP4 and NSP1 from RRV, another heterologous RV strain that resembles murine viruses in its ability to efficiently suppress the mouse interferon response in vitro in MEFs (9, 24). Of note, RRV was previously shown to be uniquely capable of infecting the biliary tree and causing a lethal biliary tract disease in newborn and INFR KO mice (8, 21, 23). The data presented here reveal that VP4 determines whether a virus can enter biliary epithelial cells, providing a mechanistic explanation for the VP4-dependent phenotype observed in vivo. Furthermore, we found that among reassortant viruses carrying entry-competent VP4, the ability to replicate in biliary epithelial cells was associated with the ability to regulate IRF3-mediated interferon responses (as determined by levels of p54 expression in biliary tissue), supporting our previously reported observations in vitro (9, 24). Furthermore, this study provides direct evidence of the role of NSP1 in interferon-mediated RV restriction in vivo. Future studies will be needed to extend this type of analysis to the host- and strain-specific regulation of RV replication in the small intestine.
Acknowledgments
We sincerely thank Herbert W. Virgin IV for the kind gift of IFN-αβ/γ receptor KO mice and Cara Mack for the kind gift of cultured mouse cholangiocytes. We also thank Tyson H. Holmes for his assistance with the statistical analysis.
This study was supported in part by a VA Merit Award and NIH grants R01 AI021362 and P30DK56339.
Footnotes
Published ahead of print on 29 December 2010.
REFERENCES
- 1.Barnes, B. H., et al. 2009. Cholangiocytes as immune modulators in rotavirus-induced murine biliary atresia. Liver Int. 29:1253-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barro, M., and J. T. Patton. 2005. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc. Natl. Acad. Sci. U. S. A. 102:4114-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barro, M., and J. T. Patton. 2007. Rotavirus NSP1 inhibits expression of type I interferon by antagonizing the function of interferon regulatory factors IRF3, IRF5, and IRF7. J. Virol. 81:4473-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridger, J. C., W. Dhaliwal, M. J. Adamson, and C. R. Howard. 1998. Determinants of rotavirus host range restriction—a heterologous bovine NSP1 gene does not affect replication kinetics in the pig. Virology 245:47-52. [DOI] [PubMed] [Google Scholar]
- 5.Broome, R. L., P. T. Vo, R. L. Ward, H. F. Clark, and H. B. Greenberg. 1993. Murine rotavirus genes encoding outer capsid proteins VP4 and VP7 are not major determinants of host range restriction and virulence. J. Virol. 67:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciarlet, M., M. K. Estes, C. Barone, R. F. Ramig, and M. E. Conner. 1998. Analysis of host range restriction determinants in the rabbit model: comparison of homologous and heterologous rotavirus infections. J. Virol. 72:2341-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estes, M., and A. Z. Kapikian. 2007. Rotaviruses, p. 1917-1974. In D. M. Knipe et al. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 8.Feng, N., et al. 2008. Role of interferon in homologous and heterologous rotavirus infection in the intestines and extraintestinal organs of suckling mice. J. Virol. 82:7578-7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng, N., et al. 2009. Variation in antagonism of the interferon response to rotavirus NSP1 results in differential infectivity in mouse embryonic fibroblasts. J. Virol. 83:6987-6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flores, J., et al. 1993. Reactogenicity and immunogenicity of a high-titer rhesus rotavirus-based quadrivalent rotavirus vaccine. J. Clin. Microbiol. 31:2439-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentleman, W. M. 1997. Least squares computations by Givens transformations with square roots. J. Inst. Math. Appl. 12:329-336. [Google Scholar]
- 12.Graff, J. W., K. Ettayebi, and M. E. Hardy. 2009. Rotavirus NSP1 inhibits NFkappaB activation by inducing proteasome-dependent degradation of beta-TrCP: a novel mechanism of IFN antagonism. PLoS Pathog. 5:e1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoshino, Y., et al. 1995. Identification of group A rotavirus genes associated with virulence of a porcine rotavirus and host range restriction of a human rotavirus in the gnotobiotic piglet model. Virology 209:274-280. [DOI] [PubMed] [Google Scholar]
- 14.Hoshino, Y., R. G. Wyatt, H. B. Greenberg, J. Flores, and A. Z. Kapikian. 1984. Serotypic similarity and diversity of rotaviruses of mammalian and avian origin as studied by plaque-reduction neutralization. J. Infect. Dis. 149:694-702. [DOI] [PubMed] [Google Scholar]
- 15.Leib, D. A., et al. 1999. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J. Exp. Med. 189:663-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez, S., and C. F. Arias. 2006. Early steps in rotavirus cell entry. Curr. Top. Microbiol. Immunol. 309:39-66. [DOI] [PubMed] [Google Scholar]
- 17.Midthun, K., et al. 1985. Reassortant rotaviruses as potential live rotavirus vaccine candidates. J. Virol. 53:949-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mori, Y., et al. 2003. Roles of outer capsid proteins as determinants of pathogenicity and host range restriction of avian rotaviruses in a suckling mouse model. Virology 316:126-134. [DOI] [PubMed] [Google Scholar]
- 19.Offit, P. A., G. Blavat, H. B. Greenberg, and H. F. Clark. 1986. Molecular basis of rotavirus virulence: role of gene segment 4. J. Virol. 57:46-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parashar, U. D., C. J. Gibson, J. S. Bresse, and R. I. Glass. 2006. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 12:304-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersen, C., et al. 1997. New aspects in a murine model for extrahepatic biliary atresia. J. Pediatr. Surg. 32:1190-1195. [DOI] [PubMed] [Google Scholar]
- 22.Petersen, C., E. Bruns, M. Kuske, and P. von Wussow. 1997. Treatment of extrahepatic biliary atresia with interferon-alpha in a murine infectious model. Pediatr. Res. 42:623-628. [DOI] [PubMed] [Google Scholar]
- 23.Riepenhoff-Talty, M., et al. 1993. Group A rotaviruses produce extrahepatic biliary obstruction in orally inoculated newborn mice. Pediatr. Res. 33:394-399. [DOI] [PubMed] [Google Scholar]
- 24.Sen, A., N. Feng, K. Ettayebi, M. E. Hardy, and H. B. Greenberg. 2009. IRF3 inhibition by rotavirus NSP1 is host cell and virus strain dependent but independent of NSP1 proteasomal degradation. J. Virol. 83:10322-10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taniguchi, K., K. Kojima, and S. Urasawa. 1996. Nondefective rotavirus mutants with an NSP1 gene which has a deletion of 500 nucleotides, including a cysteine-rich zinc finger motif-encoding region (nucleotides 156 to 248), or which has a nonsense codon at nucleotides 153-155. J. Virol. 70:4125-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terenzi, F., C. White, S. Pal, B. R. Williams, and G. C. Sen. 2007. Tissue-specific and inducer-specific differential induction of ISG56 and ISG54 in mice. J. Virol. 81:8656-8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueno, Y., et al. 2003. Evaluation of differential gene expression by microarray analysis in small and large cholangiocytes isolated from normal mice. Liver Int. 23:449-459. [DOI] [PubMed] [Google Scholar]
- 28.Wolf, M., P. Vo, and H. B. Greenberg. Rhesus rotavirus entry into a polarized epithelium is endocytosis dependent and involves sequential VP4 conformational changes. J. Virol., in press. [DOI] [PMC free article] [PubMed]