Abstract
The genetic elements of herpesvirus origins of lytic replication have been characterized in detail; however, much remains to be elucidated concerning their functional role in replication initiation. In the case of the Epstein-Barr virus (EBV), we have found that in addition to the two well-defined critical elements required for lytic replication (the upstream and downstream essential elements, UEE and DEE), the origin of lytic replication (OriLyt) also requires the presence of a GC-rich RNA in cis. The BHLF1 transcript is similar to the essential K5 transcript identified at the Kaposi's sarcoma-associated herpesvirus OriLyt. We have found that truncation of the BHLF1 transcript or deletion of the TATA box, but not the putative ATG initiation codon, reduce OriLyt function to background levels. By using an antibody specific for RNA-DNA hybrid molecules, we found the BHLF1 RNA stably annealed to its DNA template during the early steps of lytic reactivation. Furthermore, expression of human RNase H1, which degrades RNA in RNA-DNA hybrids, drastically reduces OriLyt-dependent DNA replication as well as recruitment of the viral single-stranded DNA binding protein BALF2 to OriLyt. These studies suggest that a GC-rich OriLyt transcript is an important component of gammaherpesvirus lytic origins and is required for initial strand separation and loading of core replication proteins.
Epstein-Barr virus (EBV) is a human gammaherpesvirus 1 (also known as human herpesvirus 4 [HHV4]) and the etiological agent responsible for infectious mononucleosis, oral hairy leukoplakia, AIDS immunoblastic lymphomas, posttransplant lymphoproliferative disease, 50% of Hodgkin's lymphomas, and the endemic forms of nasopharyngeal carcinoma and Burkitt's lymphoma (56, 79). Successful infection and viral spread, within and between individual hosts, are necessary prerequisites for EBV pathogenesis. Each of these requires productive lytic replication of the virus, which includes the duplication of its 170- to 175-kbp double-stranded DNA genome. To date, no antiviral drug has been approved, nor shown to be highly effective, in blocking EBV lytic replication, suggesting that mechanisms controlling viral DNA replication are sufficiently diverged from related members of the herpesvirus family (2).
Upon host cell infection and establishment of latency, herpesvirus genomes, including EBV, adopt a closed circular conformation (11, 64). During lytic reactivation from viral latency, DNA replication must initiate from this circular template. The viral basic leucine zipper (b-ZIP) protein Zta (encoded by the immediate-early BZLF1 gene, and also known as Z, ZEBRA, and EB1) governs this process (10, 13, 17, 57, 69). Zta has been described as both a transcription factor and an origin binding protein, activating both virus early gene transcription and the EBV origin of lytic replication (OriLyt) (18, 24, 36, 38, 65). Zta's ability to activate OriLyt is thought to be at least partially due to its ability to bind viral replication proteins, perhaps recruiting them to the origin (19, 21, 34, 35, 82). The functional equivalent of Zta in herpes simplex virus 1 (HSV1) is the origin binding protein (OBP) encoded by the HSV1 UL9 gene. The HSV1 OBP is an ATP-dependent DNA helicase that appears to work together with the single-stranded DNA (ssDNA) binding protein ICP8 to accomplish DNA strand separation at the HSV1 origin OriS (25, 32). The EBV genome contains an ICP8 orthologue, referred to as BALF2, and a processive helicase (encoded by the BBLF4 gene) but lacks a replication initiator helicase like UL9. It remains unclear how a transcription factor like Zta, with no known enzymatic activity, mediates initiation of DNA replication and, specifically, DNA strand unwinding at OriLyt (53).
EBV typically encodes two identical copies of OriLyt (although there are functional strains that contain only one copy), which include binding sites for Zta that are called Zta response elements (ZREs) (24, 38). Initial mapping studies by Hammerschmidt and Sugden identified two regions within OriLyt as necessary for replication (24). These regions, defined by testing overlapping EBV sequences in plasmid replication assays, mapped to nucleotides 52,632 to 52,944 (SstI-KpnI) and 53,207 to 53,581 (KpnI-NsiI) on the genome, respectively, and were later named the “upstream” and “downstream” essential elements (UEE and DEE). Finer mapping using deletions within the context of the larger 7.2-kbp BamHI-SalI fragment (nucleotides 48,848 to 56,084) narrowed the required regions to a 67-bp UEE (nucleotides 52,811 to 52,877) and an 87-bp DEE (nucleotides 53,342 to 53,428) (67). The UEE comprised the BHLF1/LF3 promoter (BHLF1p), including the TATA box, two ZREs (ZRE1 and -2), and a CCAAT box. All mutants tested that impaired BHLF1p function equally affected replication; however, not every mutation that impaired replication also impaired promoter function, suggesting that BHLF1p activity is required, though not sufficient, for replication. The DEE contains binding sites for the Sp1, Sp3, and ZBP-89 proteins, which interact with the core viral replication proteins (1, 23, 34, 65), including the EA-D processivity factor, which is able to activate the BHRF1 promoter via the downstream element (81, 82). The DEE also contains a homopurine-homopyrimidine sequence capable of forming a triple helix in vitro, and mutations that impair triple helix formation in vitro also disrupt DNA replication in vivo (48). These two essential core regions were thought to be flanked by nonessential auxiliary regions that influence the efficiency with which OriLyt-containing plasmids replicate in transient experiments (67).
The approximately 2.5-kb BHLF1 RNA, transcribed off what was initially referred to as the NotI repeat, or ntr gene, in the BamHI H fragment of the EBV genome and whose promoter (BHLF1p) overlaps the UEE of the “left” copy of OriLyt (OriLytL), was first mapped by Jeang and Hayward (28). They were able to show that BHLF1 RNA was produced in response to 12-O-tetradecanoylphorbol-13-acetate (TPA) treatment in both B95-8 and Raji EBV-positive cell lines. The RNA start site was mapped to the +29 position, separated from the TATA box by a partially S1 nuclease-resistant 21-bp palindrome, and it spanned the NotI repeat (also known as internal repeat 2 [IR2]) region of the virus ending at an AATAAA poly(A) signal sequence (28, 30). The nucleotide sequence of the 5′ region of the RNA, up to the NotI repeats, was shown to be 82% GC rich. Additionally, an equally GC-rich transcript produced by the EBV LF3 gene, the positional equivalent of BHLF1 found in the second EBV lytic origin (OriLytR) with an identical promoter, was also identified (20). Interestingly, this gene, which encompasses the PstI repeat region of the EBV genome (also known as internal repeat 4 [IR4]), shares only limited sequence identity with BHLF1. The BHLF1/LF3 promoter (referred to as BHLF1p throughout this paper) is the strongest known Zta-responsive promoter (37), and BHLF1 and LF3 RNAs are the most abundant transcripts found during lytic replication (20, 30, 40). In fact, due to their prevalence, assays that detect these RNAs have been frequently used as tools for diagnosing EBV lytic infection in the clinic (6, 7, 62).
In this work, we further investigated the mechanism of initiation of DNA replication at OriLytL. In an effort to define the minimal OriLyt, we found that one of the two divergent transcripts (BHLF1 or BHRF1) is essential for efficient DNA replication. We also found that the BHLF1 transcript provides a critical activity for OriLyt function in cis. The high guanine and cytosine (GC) content of the BHLF1 transcript, and its unknown function in lytic replication, prompted us to investigate its potential to form a stable RNA-DNA hybrid, or R-loop, structure similar to what has been observed at the mitochondrial DNA origin of replication (31, 76, 77) and at the immunoglobulin (Ig) locus during class switch recombination in B lymphocytes (22, 26). We used an antibody specific for RNA-DNA hybrid molecules to demonstrate the formation of such structures within the BHLF1 transcription unit. Furthermore, we show that human RNase H1, an enzyme specific for RNA-DNA duplex hybrids, is a potent inhibitor of OriLyt replication and, specifically, recruitment of the ssDNA binding protein BALF2 to OriLyt, suggesting that the RNA-DNA hybrid may generate ssDNA important for lytic replication initiation.
MATERIALS AND METHODS
Plasmids.
Full-length BZLF1 and BALF2 genes were cloned into the EcoRI-SalI and EcoRI-BamHI sites of the p3xFLAG-myc-CMV24 vector (Sigma), respectively, for mammalian cell expression. Full-length BZLF1 gene was also cloned into the EcoRI site of the pSRα vector (71) for mammalian cell expression. The plasmid encoding full-length RNase H1 protein fused to green fluorescent protein (GFP) was a gift from Robert J. Crouch (NICHD) (8, 9). Various-length OriLyt sequences were cloned into the NotI-PstI (or NotI-BamHI) sites of the pBluescript II KS(+) vector (Stratagene) using appropriate primers (data available upon request). The +60 to −400 regions of wild-type or mutant OriLyt BHLF1 promoters were amplified from the pBluescript-OriLyt plasmids and each inserted into the pHEBO-luciferase mammalian expression vector (3) using the primers 5′-CACGCTAGCCTGAATCCTACCTAGCTCCACCCA-3′ (BHLF1p fwd) and 5′-CACAAGCTTAGGCCGCCGCAAGGACGCCGGGCC-3′ (BHLF1p rev).
Cell lines.
ZKO-293 cells (a gift from H. J. Delecluse) are human 293 cells transformed with a hygromycin-resistant EBV bacmid containing a deletion of the BZLF1 gene. ZKO-293 and 293 cells were grown in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS), 20 mM GlutaMAX (Gibco), and 100 μg/ml hygromycin. The Mutu I and Raji cell lines are human Burkitt's lymphoma cell lines that harbor latent EBV. Both were cultured in RPMI 1640 medium with 10% FBS, 100 U/ml penicillin, 100 μl/ml streptomycin, and 20 mM GlutaMAX (Gibco).
Antibodies.
Anti-FLAG polyclonal rabbit antibody (Sigma), IgG nonspecific control rabbit antibody (Santa Cruz Biotechnology), anti-GFP polyclonal rabbit antibody (Santa Cruz Biotechnology), anti-poly(ADP-ribose) polymerase 1 (anti-PARP1) polyclonal rabbit antibody (Alexis Biochemicals), anti-β-actin monoclonal mouse antibody (Sigma), anti-RecQL1 rabbit polyclonal antibody (provided by Perry J. Blackshear, NIEHS), anti-RNA-DNA hybrid monoclonal antibody from the mouse hybridoma cell line HB-8730 (5) (a gift from Ramin Shiekhattar, The Wistar Institute), and anti-Zta rabbit and anti-BALF2 polyclonal antibodies (both custom-made by Pocono Rabbit Farm and Laboratory) were used.
OriLyt plasmid replication assay.
Transient plasmid transfection of ZKO-293 cells was carried out using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. A total of 5 × 106 ZKO-293 cells were transfected with 2 μg Zta and 2 μg OriLyt plasmid or vector control plasmids. At 48 h posttransfection, cells were washed once with phosphate-buffered saline (PBS) and lysed with 1 ml lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris [pH 8]) for 10 min on ice. Lysates were analyzed by Western blotting with antibody against BALF2, Zta, or actin. Alternatively, cells were lysed with Hirt lysis buffer (0.6% SDS, 10 mM EDTA, 10 mM Tris [pH 7.5]) for 5 min at 25°C. After lysis, sodium chloride was added to a final concentration of 750 mM and lysates were incubated at 4°C for at least 2 h. Supernatant was separated from debris by centrifugation and treated with 60 μg/ml RNase A for 1 h at 37°C followed by 120 μg/ml proteinase K for 2 h at 50°C. DNA was phenol-chloroform extracted and ethanol precipitated. Hirt DNA was then digested with or without HindIII (New England BioLabs) to linearize the pBluescript plasmids and DpnI (New England BioLabs), according to the manufacturer's instructions, to digest input plasmid. Following restriction digestion, DNA was phenol-chloroform extracted, ethanol precipitated, dissolved in DNA loading buffer (0.04% bromophenol blue, 0.04% xylene cyanol, 0.04% Orange G, 2.5% Ficoll 400), and separated by electrophoresis on a 0.7% agarose-Tris-borate-EDTA gel. Separated DNA was transferred to positively charged nylon membranes by using established methods for Southern blotting (68) and detected by hybridization with a digoxigenin (DIG)-labeled probe specific for pBluescript using digoxigenin EasyHyb reagents (Roche), according to the manufacturer's instructions. The probe was labeled using a DIG-High Prime DNA labeling kit (Roche), according to the manufacturer's instructions.
Northern blot assays.
Transient plasmid transfection of 293 cells was carried out using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. A total of 5 × 106 293 cells were transfected with 2 μg Zta and 2 μg of OriLyt or vector control plasmids. At 48 h posttransfection total RNA was isolated from transfected cells with TRIzol reagent (Invitrogen) and separated by electrophoresis in 1% agarose NorthernMax denaturing gel in 1× NorthernMax running buffer (Ambion). Each lane was loaded with RNA from 2 × 107 cells. The RNA was transferred to a nylon membrane and detected by hybridization with a DIG-labeled probe specific for the BHLF1 RNA (5′-GCAGCCGCGGCCCAGCGCGCCCCGTTCACG-3′), or the antisense control, using digoxigenin EasyHyb reagents (Roche) according to the manufacturer's instructions. The probe was labeled using a DIG oligonucleotide 3′-end-labeling kit (Roche) according to the manufacturer's instructions.
Luciferase assay.
Transient plasmid transfection of 293 cells was carried out using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. A total of 5 × 106 293 cells were transfected with 2 μg Zta and 2 μg of BHLF1p-luciferase or vector control plasmids. At 48 h posttransfection, cells were harvested and luciferase assays were performed by using the luciferase assay system (Promega). Zta expression levels were monitored by Western blot analysis.
Guanine density plots.
Guanine densities were determined and plotted using the online European Molecular Biology Open Software Suite (EMBOSS) Freak program (http://emboss.bioinformatics.nl/cgi-bin/emboss/freak) with a stepping value of 1 and an averaging window of 500.
RNA-DNA hybrid immunoprecipitation.
Mutu I or Raji cells were treated with 1 mM sodium butyrate and 50 ng/ml TPA to induce lytic replication or with a vehicle control. At various times postinduction cells were harvested and washed once in PBS, and 10 × 106 cells were resuspended in lysis buffer (20 mM Tris [pH 8], 4 mM EDTA, 20 mM NaCl, 1% SDS) with 0.2 U/ml RNase inhibitors (Roche) and 0.7 μg/μl proteinase K and incubated at 37°C for 18 h. The DNA was phenol-chloroform extracted, ethanol precipitated, resuspended in 300 μl of Tris-EDTA buffer and sonicated to between 500 and 800 bp. Eight micrograms of DNA was then resuspended in immunoprecipitation (IP) buffer (10 mM Na-phosphate [pH 7], 140 mM NaCl, 0.05% Triton X-100) and incubated with 5 μg of anti-RNA-DNA hybrid or 5 μg mouse IgG (Santa Cruz Biotechnology) for 18 h at 4°C. The immunocomplexes were precipitated by adding 50 μl of protein G-Dynabeads (Invitrogen) for 2 h at 4°C. The beads were collected by using a magnetic rack and washed twice for 10 min with 1 ml of IP buffer. The DNA was eluted by incubating the beads with 250 μl of proteinase digestion buffer (50 mM Tris [pH 8], 10 mM EDTA, 0.5% SDS, 0.3 μg/μl proteinase K) at 50°C for 3 h with shaking. The DNA was purified and analyzed by real-time quantitative PCR (qPCR).
Real-time qPCR.
Real-time qPCRs were run in a 7900 Applied Biosystems real-time qPCR machine using 6 ng DNA and 1.5 mM primers in 12 μl of 1× Sybr green master mix solution (Roche), according to the manufacturer's specified parameters. The oligonucleotide primer pairs used for quantitative real-time qPCR in this study were designed using the Primer Express software (Applied Biosystems) and included 5′-TCGCCTTCTTTTATCCTCTTTTTG-3′ (OriLyt-fwd), 5′-CCAACGGGCTAAAATGACA-3′ (OriLyt-rev), 5′-GCGGCCTTCACGAATGC-3′ (BNRF1-fwd), 5′-GGAACGTGTTGTCCCTAACCTC-3′ (BNRF1-rev), 5′-TCACCACCATGGAGAAGGCT-3′ (GAPDH-fwd), 5′-GCCATCCACAGTCTTCTGGG-3′ (GAPDH-rev), 5′-CTATGATGACACAAACCCCG-3′ (HSV-TKp-fwd), and 5′-GAGTTTCACGCCACCAAGAT-3′ (HSV-TKp-rev).
Dot blot assays.
Ten-microliter aliquots of DNA and/or RNA nucleotides oligomers (100 μM in 10 mM Tris-HCl [pH 8], 1 mM EDTA, and 100 mM NaCl) were incubated at 95°C for 5 min, then gradually cooled to 25°C to allow annealing, and then spotted onto a positively charged nylon membrane by using a 96-well Minifold Spot-Blot apparatus (Whatman). The membrane was air dried, cross-linked at 250 mJ in a UV Stratalinker 2400 (Stratagene), washed in 2× SSC (300 mM NaCl, 30 mM sodium citrate; pH 7.2), washed in TBST (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% Tween 20), blocked using TBST supplemented with 5% (wt/vol) milk powder, and probed using an anti-RNA-DNA hybrid antibody by standard Western blotting techniques. The oligonucleotide sequences used were as follows: DNA TTAGGGTTAGGGTTAGGGTTAGGGTTAGGG (oPL2070) with or without its complement, CCCTAACCCTAACCCTAACCCTAACCCTAA (oPL2071), or RNA UUAGGGUUAGGGUUAGGGUUAGGGUUAGGG (oPL2628), with or without its DNA complement (oPL2071).
EBV genome replication assay.
Transient plasmid transfection of ZKO-293 cells was carried out using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. A total of 5 × 106 ZKO-293 cells were transfected with 2 μg Zta or vector control and 0.5, 1, 2, or 4 μg of RNase H1-GFP or vector control plasmids. At 48 h posttransfection, cells were washed once with PBS and lysed with 1 ml lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris [pH 8]) for 10 min on ice. Lysates were sonicated on ice by using a Diagenode Bioruptor on high setting with 30-s on/off pulses for a total of 25 min until the DNA was reduced to an average length of 200 to 800 bp. Lysates were analyzed by Western blotting with antibodies against Zta and BALF2. Lysates were then diluted 1/10 in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris [pH 8], 167 mM NaCl) and treated with 120 μg/ml proteinase K for 2 h at 50°C, and DNA was phenol-chloroform extracted and ethanol precipitated. DNA quantity was measured by real-time qPCR.
Cell cycle analysis.
Transient plasmid transfection of ZKO-293 cells was carried out using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. A total of 5 × 106 ZKO-293 cells were transfected with 2 μg Zta or vector control and 0.5, 1, 2, or 4 μg of RNase H1-GFP or vector control plasmids. At 48 h posttransfection, cells were washed with PBS and resuspended in 3 × 106 cells/ml of cold PBS. Cells were fixed by drop-wise addition of 1 volume of 95% ethanol and incubation for 1 h at 4°C. Cells were then washed twice with cold PBS, treated with 1 μg/ml RNase A in PBS at 37°C for 15 min, and stained with 20 μg/ml propidium iodide (PI) in the dark at 25°C for 15 min, and the cell cycle was analyzed by PI staining using flow cytometry.
HSV1 genome replication assay.
Transient plasmid transfection of 293 cells was carried out using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. A total of 5 × 106 cells were transfected with 4 μg of RNase H1-GFP or vector control plasmid. At 6 h posttransfection, cells were seeded into a 24-well plate at 2 × 106 cell/well in 0.5 ml growth medium. At 24 h posttransfection RNase H1-GFP expression was confirmed by fluorescence microscopy (data not shown), and cells were treated with 1.4 × 103 PFU/μl HSV1 (KOS strain; a gift from Nigel W. Fraser, University of Pennsylvania) in 0.5 ml serum-free DMEM or mock infected with medium alone. Infection was permitted to proceed for 50 min at 25°C, after which virus-containing medium was removed and 0.5 ml of fresh growth medium was carefully added. Cells were incubated at 37°C for 10 h and then harvested. Cells were washed once with PBS and lysed with 1 ml lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris [pH 8]) for 10 min on ice. Lysates were sonicated on ice using a Diagenode Bioruptor on high setting with 30-s on/off pulses for a total of 25 min, until the DNA was reduced to an average length of 200 to 800 bp. Lysates were analyzed by Western blotting with antibody against GFP to confirm RNase H1-GFP expression levels (data not shown). Lysates were then diluted 1/10 in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris [pH 8], 167 mM NaCl) and treated with 120 μg/ml proteinase K for 2 h at 50°C, and DNA was phenol-chloroform extracted and ethanol precipitated. DNA quantity was measured by using a real-time qPCR with primers for the HSV1 genome (specifically the thymidine kinase promoter region [TKp]) or a the cellular genome (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) as a control.
Protein purification.
ZKO-293 or 293 cells were grown on 50 dishes (25 cm) with or without 200 μg/ml phosphonoacetic acid (PAA) and transfected with 500 μg (10 μg/plate) of expression plasmid for FLAG-Zta or control FLAG vector by using Lipofectamine 200 reagent (Invitrogen). Cells were harvested 48 h posttransfection by cell scraping, washed two times with ice-cold PBS, transferred to hypotonic buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], and protease inhibitor cocktail [Sigma]), and subjected to 10 strokes of a Dounce homogenizer to isolate nuclei. The nuclei were pelleted by centrifugation at 5,000 rpm for 10 min in a Sorvall SS34 rotor. Nuclei were then resuspended in buffer B (10 mM HEPES [pH 7.9], 400 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, and protease inhibitor cocktail [Sigma]) using a Dounce homogenizer (B pestle). Nuclei were stirred for 30 min in buffer B and then pelleted by centrifugation at 25,000 rpm for 30 min in a Sorvall SS34 rotor. The supernatant was designated the soluble nuclear fraction. The residual nuclear pellet was then resuspended in buffer E (10 mM Tris [pH 7.5], 10% glycerol, 400 mM NaCl, 0.05% Ipegal, 1 mM DTT, 1 mM PMSF, and protease inhibitor cocktail [Sigma]). The nuclear pellet was subjected to sonication for 10 min in a Diagenode Bioruptor until the bulk of the pellet was solubilized. The insoluble residual material was pelleted by centrifugation at 10,000 rpm for 10 min, and the remaining solubilized nuclear pellet was used for immunopurification using FLAG-agarose (Sigma). The extract was incubated with FLAG-agarose beads (10 μl/mg of extract) for 16 h at 4°C and then washed four times with buffer E and once with buffer C. The FLAG-agarose was then incubated with a 1-column volume of buffer C containing 1 mg/ml of FLAG peptide. Eluted material was either subjected to Western blot analysis or concentrated by trichloroacetic acid (TCA) precipitation and subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and colloidal blue staining for analysis by mass spectrometry.
Protein IP assays.
Cell lysates were generated by incubating cells in NET buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 0.05% NP-40, 1 mM PMSF, and protease inhibitor cocktail [Sigma]) at 4°C for 30 min with gentle agitation. The nuclei were pelleted at 15,000 rpm in a Microfuge centrifuge at 4°C. The supernatant was subjected to a preclearing with protein A- or protein G-Sepharose for 4 h and then incubated with primary antibody for 4 h and then with protein A-Sepharose (rabbit antibodies) or protein G-Sepharose (mouse antibodies) was added for 1 h. The immune complex was washed three times with NET buffer and one time with PBS and then was eluted by SDS-PAGE and heated at 95°C for 5 min.
ChIP assays.
For the chromatin IP (ChIP) assays we followed a modification of the protocol provided by Upstate Biotechnology, Inc., with minor modifications as previously described (15). DNAs were sonicated to between 200- and 800-bp fragments by using a Diagenode Bioruptor, and real-time qPCR was performed.
RESULTS
The BHLF1 or BHRF1 transcript regions are required for OriLyt function.
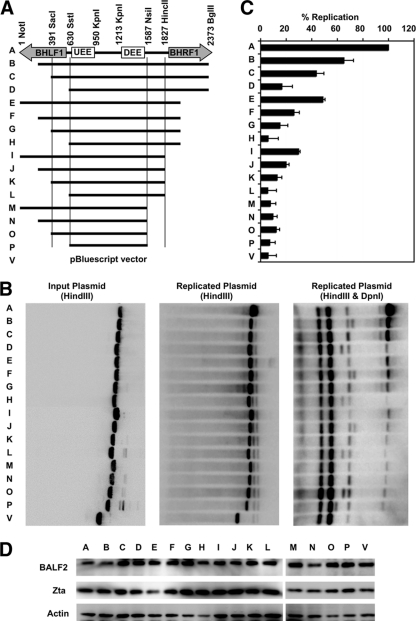
Previous studies have shown that the upstream and downstream essential elements of the EBV OriLyt are necessary for OriLyt function (66, 67). We tested whether these regions were sufficient to confer origin activity during lytic reactivation. To this end, we cloned sequences of various lengths from the BamHI H fragment of the Epstein-Barr virus, each containing the two essential OriLyt elements (Fig. 1A), into a pBluescript plasmid vector and assayed for their abilities to function as origins in the context of EBV lytic viral replication. pBluescript-OriLyt or vector control plasmids were transected into ZKO-293 cells along with the BZLF1 gene to induce lytic replication. Expression of Zta protein and the early lytic protein BALF2 (a transcriptional target of Zta) was monitored by Western blot analysis (Fig. 1D) to ensure successful induction of lytic replication in all transfected cultures. Plasmid DNA was analyzed by Southern blotting before transfection (Fig. 1B, left panel) or after recovery (Fig. 1B, center and right panels). DpnI enzyme was used to distinguish between input (DpnI-sensitive) and de novo, replicated (DpnI-resistant) plasmids (Fig. 1B, right panel). Surprisingly, the plasmid containing only the UEE, the DEE, and intervening sequence (plasmid P) was unable to replicate above background (empty vector, plasmid V) levels (Fig. 1B and C). Similarly, a plasmid incorporating all of the intervening sequence between the BHLF1 and BHRF1 genes (plasmid L) was also functionally dead. The largest plasmid tested, incorporating the 2,372-bp region spanning from the NotI repeats to the unique BglII site, including large portions of the BHLF1 and BHRF1 genes (plasmid A), replicated robustly. The intermediate-sized plasmids (plasmids B through K) gave intermediate phenotypes, suggesting that several regions within the BHLF1 and BHRF1 genes contributed to efficient OriLyt function.
FIG. 1.
The BHLF1 or BHRF1 transcript regions are required for OriLyt function. Sequences of various lengths were amplified from the BamHI H fragment of Epstein-Barr virus, cloned into a pBluescript plasmid vector, and tested for the ability to support lytic replication. (A) Diagram of the full-length (2,372-bp) region of OriLytL and each of the various truncations experimentally tested. (B) pBluescript-OriLyt or vector control plasmids were transected into ZKO-293 cells along with the BZLF1 gene. Plasmids were analyzed by Southern blotting before (left panel) or after (center and right panels) transfection and lytic induction. Plasmids recovered from cells were digested with or without DpnI enzyme (as indicated). One representative experiment is shown. (C) Relative plasmid amounts were quantified using quantitative Southern blotting. Results were calculated as the DpnI-digested signal divided by the input plasmid signal and normalized to the value obtained for full-length (2,372-bp) OriLyt. Data averages from three identical, independent experiments are shown, with error bars representing the standard deviations. (D) Western blotting was used to monitor Zta and BALF2 protein expression levels in all transfected cells.
It is important to note that these sequences are required in cis, as these experiments were performed in the context of full-length BHLF1 and BHRF1 genes present on the EBV bacmid genome harbored by the cells. Furthermore, the truncated plasmid sequences are not inhibiting replication of the full viral genome, as observed by Southern blot analysis and measured by real-time qPCR (data not shown). We also noted that there appeared to be some compensation between the BHLF1 and BHRF1 regions; a plasmid with truncated BHLF1 and BHRF1 regions (e.g., plasmid L) can be partially rescued by adding BHFL1 sequence (e.g., plasmid I) or BHRF1 sequence (e.g., plasmid D). These observations led us to the conclusion that the transcript regions of both of these genes are important.
BHLF1 transcription is required, in cis, for OriLyt function.
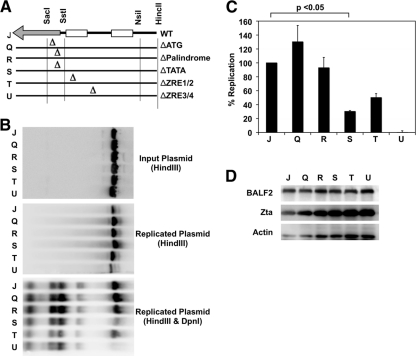
Given the potential compensatory roles of the BHLF1 and BHRF1 transcript regions, we decided to focus on one at a time. We chose the BHLF1 region for this study because BHLF1 RNA is the most abundant transcript found during lytic replication (30, 40), yet there is no known function for the transcript or any potential encoded protein. To determine whether the BHLF1 sequence requirement in cis is due to DNA contributions or due to the RNA transcript, we created several mutations in an OriLyt plasmid that disrupted either the putative BHLF1 ATG initiation codon, the TATA box, a 21-bp palindrome adjacent and immediately downstream of the TATA box, the first and second Zta response elements (ZRE1/2), or the third and fourth ZREs (ZRE3/4) (Fig. 2A). We then tested these mutant plasmids for their abilities to support replication during lytic infection in ZKO-293 cells (Fig. 2B). As before, expression of Zta and BALF2, as monitored by Western blotting (Fig. 2D), was normal, as was efficient replication of the EBV bacmid genome (data not shown). We found plasmid replication was significantly impaired in the case of the ΔTATA, ΔZRE1/2, and ΔZRE3/4 mutants, while the ΔATG and ΔPalindrome mutations had no significant effect (Fig. 2C). This suggests that BHLF1 RNA transcription is essential for OriLyt replication rather than DNA sequence along the BHLF1 gene.
FIG. 2.
BHLF1 RNA is required for OriLyt function in cis. (A) Insertional mutations were made to disrupt several regions of the OriLyt sequence cloned into the pBluescript vector. These mutant plasmids were then tested for their ability to support lytic replication. (B) Wild-type or mutant pBluescript-OriLyt or vector control plasmids were transected into ZKO-293 cells along with the BZLF1 gene. Plasmids were analyzed by Southern blotting before (top panel) or after (center and bottom panels) transfection and lytic induction. Plasmids recovered from cells were digested with or without DpnI enzyme (as indicated). One representative experiment is shown. (C) Relative plasmid amounts were quantified using quantitative Southern blotting. Results were calculated as the DpnI-digested signal divided by the input plasmid signal and normalized to the value obtained for wild-type OriLyt. Data averages from three independent experiments are shown, with error bars representing the standard deviations. Statistical significance was determined using a two-tailed, unpaired t test. (D) Western blot assays were used to monitor Zta and BALF2 protein expression levels in all transfected cells.
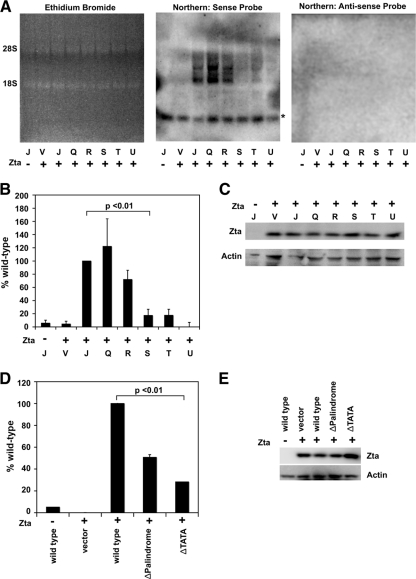
To directly measure transcriptional activity of the wild-type and mutant OriLyt plasmids, we used a Northern blot assay (Fig. 3A). OriLyt plasmids were cotransfected with or without Zta expression plasmids into 293 cells. Expression of Zta was monitored by Western blotting to ensure successful transfection (Fig. 3C). BHLF1 RNA was produced off wild-type OriLyt (Fig. 3A, center panel, lane 3) but not off a vector control (lane 2) or in the absence of Zta (lane 1). We did not detect a signal when we used an antisense probe (right panel). Ethidium bromide staining prior to Northern blot transfer (left panel) and a nonspecific band (noted by an asterisk in the center panel) were used as loading controls. As expected, the ΔTATA, ΔZRE1/2, and ΔZRE3/4 mutants were transcriptionally impaired, while the ΔATG and ΔPalindrome mutations produced near-wild-type levels of RNA (Fig. 3B). This indicates that BHLF1 RNA is expressed in the ΔATG and ΔPalindrome mutants, which also replicate to near-wild-type levels.
FIG. 3.
Effects of OriLyt mutations on BHLF1 transcription. (A to C) Wild-type or mutant pBluescript-OriLyt plasmids were transected into 293 cells along with the BZLF1 gene. (A) RNA was isolated and fractionated by agarose gel electrophoresis in the presence of ethidium bromide (left panel) and analyzed by Northern blotting with BHLF1 sense (center panel) or antisense (right panel) probes. One representative experiment is shown. A nonspecific band is indicated by the asterisk. (B) Relative RNA amounts were quantified using quantitative Northern blot analysis. Results were normalized to the value obtained for wild-type OriLyt. Data averages from three identical, independent experiments are shown, with error bars representing the standard deviations. Statistical significance was calculated using a one-tailed, paired t test. (C) Western blot assays were used to monitor Zta and BALF2 protein expression levels in all transfected cells. (D and E) Identical mutations were generated in the promoter of the pHEBO-BHLF1p-luciferase plasmid. These plasmids were cotransfected with the BZLF1 gene into 293 cells and assayed for the ability to express firefly luciferase. (D) Results were first normalized to an internal Renilla luciferase control and then to the wild-type signal. Data averages from three identical, independent experiments are shown, with error bars representing the standard deviations. Statistical significance was calculated using a two-tailed, unpaired t test. (E) Western blot assays were used to monitor Zta protein expression levels in all transfected cells.
To confirm that the BHLF1p ΔTATA box mutation was indeed reduced for transcription activity, we tested its activity in a luciferase reporter gene assay (Fig. 3D). Plasmids containing the wild-type or ΔTATA mutant promoter sequences cloned upstream of the luciferase gene were cotransfected with or without the BZLF1 gene into 293 cells and assayed for firefly luciferase compared to a Renilla luciferase internal negative control (Fig. 3D). Once again, Zta expression was monitored by Western blotting to ensure successful transfection (Fig. 3E). Firefly luciferase protein production was dependent on the presence of both the Zta protein and a BHLF1p-luciferase reporter plasmid. Insertional mutagenesis of the BHLF1p TATA box sequences (ΔTATA) led to an ∼70% decrease in reporter protein expression (Fig. 3D). The decrease in luciferase activity produced by the BHLF1p-luciferase-ΔTATA plasmid is consistent with the Northern blotting data (Fig. 3A, plasmid R), which indicate that a loss of BHLF1 transcription correlates with a loss of OriLyt replication. The mutations in the BHLF1 palindrome inhibited luciferase expression more significantly than they affected RNA expression measured in the Northern blot assay, suggesting that this mutation may also inhibit protein translation. These data suggest that production of the RNA transcript in cis, and not the DNA sequence, is required for efficient replication at OriLyt.
An RNA-DNA hybrid is formed at OriLyt during initiation of lytic replication.
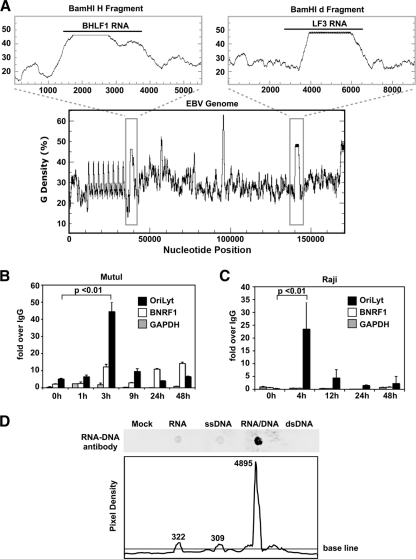
BHLF1 and its paralogue, LF3, in the second lytic origin (OriLytR), are >40% G-rich, including in their 5′ regions (Fig. 4A, upper panels). These genes are among the most enriched for guanine density compared to the rest of the EBV genome (lower panel). Since G-rich RNA transcripts have been shown to be critical for the formation of R-loops (58), we investigated the possibility that BHLF1 may persist as a stable RNA-DNA hybrid molecule at OriLyt. To determine whether or not a stable RNA-DNA hybrid molecule is formed at OriLyt during initiation of lytic replication, we employed an immunoprecipitation assay with an antibody specific for RNA-DNA hybrid molecules (5) (Fig. 4D). Latently infected B lymphocytes were treated with or without sodium butyrate (NaB) and TPA to induce viral lytic replication. Total DNA was isolated under RNase-free conditions and then immunoprecipitated with the RNA-DNA hybrid antibody or a nonspecific IgG control (Fig. 4B and C). Immunoprecipitated DNA was analyzed by real-time qPCR using primers for OriLyt (black boxes), the BNRF1 transcript region of the virus (white boxes), or GAPDH (gray boxes) as negative controls. A stable RNA-DNA hybrid was detected specifically at OriLyt within 3 h postinduction of lytic replication in Mutu I cells, and this structure disappeared over time (Fig. 4B). A similar finding was observed using Raji cells, in which RNA-DNA hybrids were also detected at OriLyt at early stages after treatment with lytic cycle-inducing agents (Fig. 4C). Since Raji cells cannot complete DNA replication due to their lack of the BALF2 ssDNA binding protein, our findings suggest that RNA-DNA hybrid formation occurs independently of, and prior to, DNA replication and BALF2 recruitment. This result supports the idea that the G-rich RNA within OriLyt is able to reanneal to its parental DNA strand and remain stably bound during the early initiating steps of DNA replication.
FIG. 4.
An RNA-DNA hybrid is formed at the G-rich BHLF1 region during initiation of lytic replication. (A) The guanine density of the entire EBV genome was plotted using the EMBOSS Freak program (lower plot). The BamHI H and BamHI d fragments, containing the BHLF1 and LF3 genes, respectively (gray boxes), have been enlarged (upper panels). (B and C) Mutu I (B) or Raji (C) cells were treated with or without Na-butyrate and TPA for times (in hours) indicated. DNA was then isolated and analyzed via immunoprecipitation using an antibody specific for RNA-DNA hybrid molecules or an IgG negative control, followed by real-time qPCR using primers for the OriLyt region (black boxes), the BNRF1 transcript region of the virus (white boxes), or the cellular region for GAPDH (gray boxes) as a negative control. Results are shown as the fold enrichment of the RNA-DNA signal over that of the IgG control. Error bars represent the standard deviations of at least three qPCR replicate reactions run side by side. Three identical, independent experiments were conducted; data from one representative experiment are shown. Statistical significance was calculated using a two-tailed, unpaired t test. (D) Nucleotides of various types (as indicated) were annealed, spotted onto a nylon membrane, and probed using the RNA-DNA hybrid antibody to demonstrate antibody specificity (top panel). Pixel density across a horizontal section of the top panel image was calculated using the ImageJ software (NIH). Numerical values indicate integrated densities for each peak.
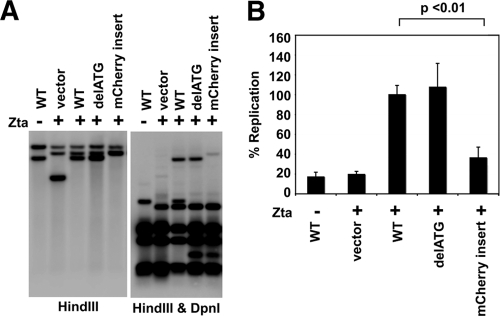
Insertion of a non-G-rich transcript impairs OriLyt-dependent replication.
Our data suggest that transcription of a specifically G-rich RNA in cis is important for OriLyt activation due to its ability to form an RNA-DNA hybrid molecule at OriLyt. To test this hypothesis, we inserted the non-G-rich mCherry gene (which has a guanine frequency of only 31%) into OriLyt at the site of the putative BHLF1 ATG initiation codon and tested the ability of this mCherry OriLyt construct to initiate lytic replication. Genetic disruption of the ATG codon had no significant effect on OriLyt function (Fig. 2 and 5); however, insertion of the non-G-rich mCherry gene drastically impaired lytic replication, as predicted (Fig. 5). These data suggest that transcription in and of itself is not sufficient to promote OriLyt activation. Instead, production of a specifically G-rich RNA (e.g., BHLF1) is required.
FIG. 5.
Insertion of a non-G-rich transcript disrupts OriLyt function. (A) Wild-type or mutant pBluescript-OriLyt or vector control plasmids were transected into ZKO-293 cells with or without the BZLF1 gene. Plasmids were analyzed by Southern blotting before (data not shown) or after (as indicated) transfection and lytic induction. Plasmids recovered from cells were digested with or without DpnI enzyme (as indicated). One representative experiment is shown. (B) Relative plasmid amounts were quantified using quantitative Southern blotting. Results were calculated as the DpnI-digested signal divided by the input plasmid signal and normalized to the value obtained for wild-type OriLyt. Data averages from three independent experiments are shown, with error bars representing the standard deviations. Statistical significance was determined using a two-tailed, unpaired t test.
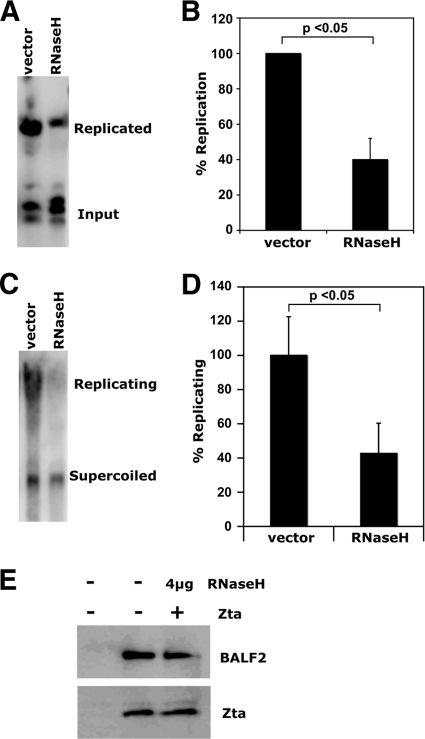
Human RNase H1 impairs OriLyt-dependent plasmid replication.
To determine whether an RNA-DNA hybrid is required for OriLyt-dependent DNA replication, we repeated our OriLyt plasmid replication assay in the presence of overexpressed human RNase H1, which specifically degrades the RNA in RNA-DNA hybrid molecules. ZKO-293 cells were cotransfected with full-length (NotI-BamHI), wild-type pBluescript OriLyt, the BZLF1 expression plasmid, and an expression plasmid encoding the RNase H1 fused to GFP (a kind gift from Robert J. Crouch, NICHD), or to a vector control (Fig. 6). Transfection efficiencies were monitored by examination of RNase H1-GFP expression by microscopy (data not shown), and Zta and BALF2 expression was monitored by Western blot analysis (Fig. 6E). Importantly, RNase H1-GFP expression did not significantly affect transcription of viral proteins. Plasmids were recovered from cells 48 h posttransfection, digested with HindIII and DpnI enzymes, and analyzed by quantitative Southern blotting (Fig. 6A and B). In the cells cotransfected with RNase H1-GFP, OriLyt-dependent plasmid replication was significantly impaired. To see if RNase H1 decreased the levels of replicating versus supercoiled plasmid in the cells, we also analyzed uncut plasmids by quantitative Southern blotting (Fig. 6C and D). As expected, the percentage of higher-molecular-weight (replicating) forms of OriLyt plasmid was much greater in cells transfected with the vector control than with RNase H1-GFP.
FIG. 6.
RNase H1 impairs OriLyt-dependent plasmid replication. Wild-type pBluescript OriLyt plasmids were cotransfected with BZLF1 into ZKO-293 cells with an RNase H1 expression plasmid or control vector. Plasmids were recovered from cells, digested with (A and B) or without (C and D) HindIII and DpnI enzymes, and analyzed by Southern blotting. Three identical, independent experiments were conducted in each case. One representative experiment is shown for each (A and C). Relative plasmid amounts were quantified using quantitative Southern blotting (B and D). In the cases where enzymes were used, results were calculated as the DpnI-resistant signal over the DpnI-digested signal and were normalized to the value obtained for vector controls (B). In cases where no enzymes were used, results were calculated as replicating (top band) plasmid over supercoiled (bottom band) signal and normalized to the value obtained for vector controls (D). Data averages from all three identical, independent experiments are shown in each case, with error bars representing the standard deviations. Statistical significance was calculated using a two-tailed, unpaired t test. (E) Western blot assays were used to monitor Zta and BALF2 protein expression levels in all transfected cells.
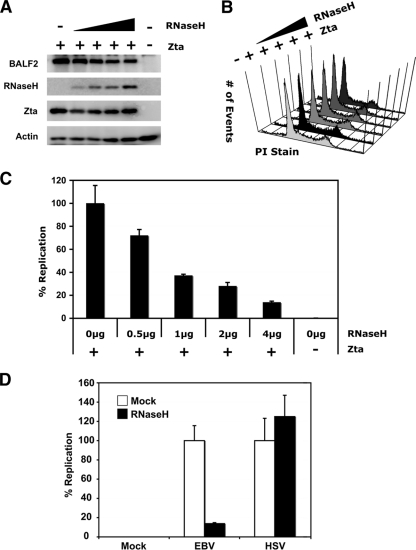
RNase H1 inhibits lytic replication of the EBV genome.
To determine the effect of overexpressed RNase H1 on replication of the EBV genome itself during lytic replication, ZKO-293 cells were cotransfected with or without BZLF1 and RNase H1-GFP expression plasmids or control vectors, and 48 h posttransfection viral DNA was isolated and quantified by real-time qPCR (Fig. 7). Expression of RNase H1-GFP inhibited lytic replication of the EBV genome in a dose-dependent manner (Fig. 7C). Western blot assays were used to monitor Zta, RNase H1-GFP, and BALF2 protein expression levels in all transfected cells (Fig. 7A). As before, overexpression of RNase H1-GFP did not have a significant effect on viral gene expression levels. Transfected cells were also subjected to cell cycle analysis by PI staining followed by flow cytometry (Fig. 7B). No significant change in the ZKO-293 cell cycle profile was detected in cells overexpressing RNase H1-GFP compared to the vector controls. These results suggest that the inhibition of EBV lytic DNA replication by RNase H1 is due to a specific effect on OriLyt rather than a global effect on cellular replication or transcriptional activity.
FIG. 7.
RNase H1 impairs EBV lytic replication. ZKO-293 cells were cotransfected with or without BZLF1 and RNase H1 expression plasmids or control vectors as shown. (A) Western blot assays were used to monitor Zta, RNase H1, and BALF2 protein expression levels in all transfected cells. (B) After 48 h, cells were analyzed for potential changes in viability and cell cycle. (C) At the same time, viral DNA was isolated and analyzed by real-time qPCR. Results were calculated as the signal obtained using EBV genome-specific (BNRF1) primers over the signal obtained using primers for cellular DNA (GAPDH) and normalized to the positive control expressing BZLF1 only. (D) 293 cells were transfected with RNase H1 (black bars) or vector control plasmids (white bars) and infected with HSV1 (as indicated) or mock infected 24 h posttransfection. At 10 h postinfection, cell were lysed and RNase H1 expression was confirmed by Western blotting (data not shown). DNA was isolated from cell lysates and analyzed by real-time qPCR. Results were calculated as the signal obtained using viral genome-specific (TKp) primers over the signal obtained using primers for cellular DNA (GAPDH) and normalized to the positive-control sample, which was infected with virus but not transfected with RNase H1.
RNase H1 overexpression does not impair HSV1 lytic replication.
To further demonstrate the specificity of RNase H1 for EBV replication, we tested the effect of the enzyme on replication of the alphaherpesvirus HSV1. 293 cells were transfected with RNase H1-GFP or a vector control and then infected 24 h later with the wild-type KOS strain of HSV1. Ten hours postinfection, viral and cellular DNAs were isolated and quantified by real-time qPCR (Fig. 7D). Whereas RNase H1 expression significantly reduced lytic replication of EBV, the same dose of RNase H1 had no inhibitory effect on lytic replication of HSV1 (Fig. 7D). This result supports the conclusion that RNase H1 expression inhibits EBV replication, without global inhibition of cellular replication or of the replication of related herpesviruses.
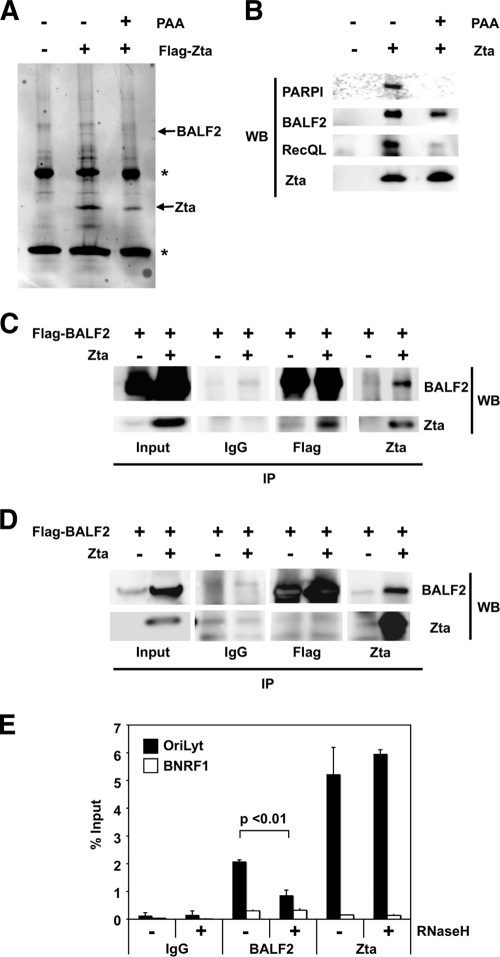
RNase H1 decreases recruitment of BALF2, but not Zta, to OriLyt.
The viral ssDNA binding protein BALF2 has been shown to be required for EBV lytic replication (19). We found, by mass spectrometry, that BALF2 was highly enriched in immunoaffinity-purified FLAG-Zta samples but not in the control FLAG immunopurifications of transfected ZKO-293 cells (Fig. 8A). This result was confirmed by Western blot analysis (Fig. 8B and C). BALF2 was also able to associate with Zta in the presence of phosphonoacetic acid (PAA), an inhibitor of the viral polymerase BALF5, suggesting that BALF2 is required at an early step during initiation of viral replication. In contrast, two other proteins also identified as FLAG-Zta binding partners by mass spectrometry, PARP1 and RecQL1, did not bind in the presence of PAA (Fig. 8A and B). The interaction between Zta and BALF2 was observed in ZKO-293 cells, but not in 293 cells, suggesting that additional viral proteins or lytic replication is required to facilitate this interaction (Fig. 8B to D). We next confirmed that BALF2 associated in vivo with OriLyt during viral reactivation by using a ChIP assay (Fig. 8E). BALF2 was enriched ∼20-fold relative to control IgG. BALF2 did not bind to a negative control BNRF1 region of the viral genome. Given that BALF2 is able to bind nonspecifically to ssDNA (73), we wondered whether RNase H1 overexpression, which should inhibit R-loop formation at OriLyt, would also affect the BALF2 association with OriLyt. To determine if RNase H1 interfered with BALF2 binding, we assayed BALF2 binding by ChIP assays in the presence of overexpressed RNase H1 (Fig. 8E). We found that RNase H1 expression reproducibly reduced BALF2 binding by more than 50% compared to vector-transfected treated cells. Importantly, RNase H1 overexpression had no effect on Zta recruitment to OriLyt. This result is consistent with the model that a stable RNA-DNA hybrid at OriLyt is required for strand unwinding and functional recruitment of the EBV single-stranded binding protein BALF2.
FIG. 8.
RNase H1 decreases recruitment of BALF2, but not Zta, to OriLyt. (A and B) ZKO-293 cells were transfected with FLAG-Zta or control FLAG expression vector and then subjected to FLAG IP. FLAG affinity-purified proteins were assayed by colloidal blue staining of the affinity-purified proteins (A) or by Western blotting (WB) for the viral BALF2 and Zta or the cellular, PARP1, and RecQL1 proteins (B). The immunoglobulin heavy and light chains are each marked by an asterisk. (C and D) ZKO-293 (C) or 293 (D) cells were transfected with FLAG-BALF2 and Zta or control vector and then subjected to FLAG, Zta, or control IgG IP. Affinity-purified proteins were probed for BALF2 or Zta by WB. (E) A ChIP assay was used to monitor BALF2 and Zta binding to OriLyt (black bars) or the BNRF1 negative-control region (white bars) in vivo in ZKO-293 cells transfected with FLAG-BALF2 and Zta in the presence of cotransfected RNase H1 (+) or vector control (−). Results from real-time qPCR were normalized to input DNA signal levels. Error bars represent the standard deviations of at least six qPCR replicate reactions run side by side. Three identical, independent experiments were conducted; data from one representative experiment are shown. Statistical significance was calculated using a two-tailed, unpaired t test.
DISCUSSION
In this work we reexamined the necessary cis requirements for function of the EBV OriLyt. By testing various truncations of the BamHI H fragment of the virus genome in plasmid replication assays, we found that one of the two divergent OriLyt transcript regions is necessary for DNA replication in cis (Fig. 1). Upon closer examination of the BHLF1 gene requirement, we found that its contribution to successful replication in cis is dependent on its ability to produce an RNA molecule (Fig. 2). The high G-rich content of the BHLF1 (and LF3) RNA is consistent with its ability to form a stable RNA-DNA hybrid, which could be detected with an antibody specific for RNA-DNA hybrids (Fig. 4). Replacement of the G-rich BHLF1 gene with the non-G-rich mCherry gene at OriLyt resulted in impairment of OriLyt function (Fig. 5). EBV OriLyt-driven replication, but not HSV1 replication, was also inhibited by RNase H1, an enzyme that specifically degrades the RNA component of an RNA-DNA hybrid (Fig. 6 and 7), and RNase H1 prevented BALF2 association with OriLyt (Fig. 8). Taken together, we propose that BHLF1 RNA forms a stable RNA-DNA hybrid essential for BALF2 loading and the initiation of lytic replication at OriLyt.
The BHLF1 and LF3 transcripts are among the most abundant RNAs expressed during the EBV lytic cycle. Despite their high prevalence, no clear functions for these transcripts have been described. These genes both have an open reading frame, which may potentially code for a highly repetitive protein (30). cDNAs from the BHLF1 gene have been mapped (47), but initial attempts to identify a BHLF1 (or LF3) protein product by hybrid selection, in vitro translation, and cloning were unsuccessful (27, 28, 30, 47). Antibody raised against a synthetic, Escherichia coli-produced peptide from the BHLF1 open reading frame recognized a TPA-inducible protein of ∼70 to 90 kDa (37, 45), and a similar result was observed for an antibody raised against a synthetic LF3 peptide (46). An LF3 gene study, however, showed that frameshifting events, at the beginning and end of the internal repeat 4 (IR4) region within the coding sequence, which would affect protein production, are frequent (78). It has also been shown that disruption of the BHLF1 open reading frame, by insertion of a hygromycin phosphotransferase-selectable marker into the unique RsrII site, does not effect lytic replication or virion production in W91 cells (39). Taken together, these studies suggest that BHLF1- and LF3-encoded proteins are not likely to be essential for lytic cycle replication.
Our study provides evidence that a major contribution of BHLF1, and perhaps LF3, is the production of a G-rich RNA required for OriLyt function in cis. Our data are consistent with the observation that the G-rich Kaposi's sarcoma-associated herpesvirus (KSHV) K5 RNA transcript is also necessary for KSHV OriLyt function (74). Our conclusion that disruption of BHLF1 transcription, and specifically the BHLF1 TATA box, impairs lytic replication is supported by a previous report showing the importance of BHFL1 transcription promoter elements (67). Our experiments also indicate that BHFL1 must be transcribed in cis, since the native BHLF1 gene products are expressed from the EBV bacmid genome present in the ZKO-293 cells used for our replication assays. Therefore, the simplest conclusion is that the 5′ end of the BHFL1 transcript is required in cis for OriLyt function.
A role for transcription in the selection and initiation of replication origins has been seen in a variety of contexts (29, 41). In the case of herepsviruses, both EBV and KSHV have transcription factors as their origin binding proteins, and it has also been shown that RNA polymerase II, TATA binding protein, and TATA binding protein-associated factors are recruited to HSV1 replication compartments (33, 50, 55). All known herpesvirus origins overlap bidirectional promoters and contain transcription factor binding sites important for replication (52). RNAs have also been identified at herpesvirus lytic origins; the best-characterized one exists at the human cytomegalovirus origin and forms a persistent RNA-DNA hybrid structure (49). An important RNA-DNA hybrid is also formed at the mitochondrial DNA (mtDNA) heavy strand replication origin (OH) during mtDNA replication, which, like herpesvirus replication, proceeds from a closed circular genome. For mtDNA replication, the RNA is transcribed within the origin but reanneals to its template DNA due to its high guanine content (31, 76, 77). The RNA in this structure, termed an R-loop, which is necessary for origin activation in cis, is thought to be processed by the mitochondrial RNase MRP into a primer functional for initiating mtDNA synthesis (31). Transcription also plays a role in the formation of RNA-DNA hybrids at the Ig class switch locus, where once again G-rich “R-loop initiation zones” within the RNA are required for R-loop formation (14, 16, 51, 58, 59, 80).
We considered whether BHLF1 RNA, one of the most-G-rich transcripts within the EBV genome (Fig. 4A), might generate an R-loop at OriLyt during lytic replication. Using specific antibodies, we found that a stable RNA-DNA hybrid molecule is formed at OriLyt during the early stages of lytic reactivation (Fig. 4B and C), suggesting an important interaction between BHLF1 RNA and its DNA template during the initiation of lytic replication. To further support this hypothesis, we tested the effect of human RNase H1, a nuclease with specificity against RNA-DNA hybrids, on OriLyt-dependent replication. We found that RNase H1 protein overexpression can inhibit OriLyt-dependent DNA replication (Fig. 6A to D and 7B).
We also have presented evidence that the EBV BALF2 protein is recruited to OriLyt and the site of the RNA-DNA hybrid molecule (Fig. 8E). BALF2 has been characterized as the EBV ssDNA binding protein required for herpesvirus replication (19, 72). BALF2 has been shown to interact with the other core EBV lytic replication proteins, including the BBLF4-BBLF2/3-BSLF1 helicase-primase complex, which in turn interacts with Zta (21). By using immunoprecipitation assays, we were able to confirm that BALF2 is found in complex with Zta in the context of lytic infection in ZKO-293 cells (Fig. 8A to C). This interaction is not dependent on function of the viral polymerase BALF5, since PAA treatment did not block the interaction. In contrast, the Zta association with the PARP-1 and the RecQL1 proteins was inhibited in the context of PAA treatment (Fig. 8B). We were unable to demonstrate the BALF2-Zta interaction in 293 cells in the absence of lytic replication (Fig. 8C). This result is consistent with the idea that the BBLF4-BBLF2/3-BSLF1 complex bridges the Zta-BALF2 interaction (21).
We found that RNase H1 overexpression also significantly affects the recruitment of BALF2, but not Zta, to OriLyt (Fig. 8E). This suggests that the ssDNA binding ability of BALF2 further recruits the protein to OriLyt and that RNase H1 expression inhibits lytic replication at least in part by decreasing the amount of OriLyt R-loop substrate available for BALF2 (and possibly other required replication proteins) to bind. We also showed that cellular DNA replication is not significantly affected by the same concentrations of RNase H1 (Fig. 7C), revealing a difference in sensitivities to RNase H1 between the virus and the host cell. Further studies will be required to determine whether this difference might be important for regulation of viral replication and whether this can be therapeutically exploited.
In addition to binding BALF2, we previously showed that Zta recruits the mitochondrial ssDNA binding protein (mtSSB) to promote viral replication but inhibit mtDNA replication (75). mtSSB plays a central role in mtDNA replication, recombination, and the stabilization of R-loops (61, 70). It will be important to determine whether mtSSB is involved in R-loop stabilization at OriLyt and to what extent EBV origin activation resembles initiation at the mitochondrial origin. Interestingly, HSV UL12.5 degrades mitochondrial DNA (12, 63), suggesting that other herpesviruses may also require mitochondrial factors to facilitate viral replication.
Transcription-dependent R-loops, like those examined at the Ig locus (22, 26, 60), are thought to facilitate recombination events. There are numerous links between herpesvirus lytic replication and DNA recombination (52). Relevant to this discussion is the fact that BALF2 has structural and functional similarities to the λ Red β recombination protein (54) and the HSV homologue ICP8, which has been shown to play a role in both strand invasion and R-loop stabilization in vitro (4, 42-44). Interestingly, HSV1 lytic replication was not affected by overexpression of human RNase H1 (Fig. 7D). This could be partially due to limited levels of ectopically expressed RNase H1. On the other hand, HSV1 origin activation is mediated by a DNA helicase, the UL9 protein, while the EBV origin initiator protein Zta is a bZIP transcription factor. Therefore, it may not be surprising that EBV origin activation may be more dependent on R-loop formation than is HSV. It will be important to determine whether R-loop formation plays any role in the HSV lytic cycle, either in facilitating initiation or in promoting subsequent recombination events that drive genome isomerization. An important future direction of investigation will be to determine whether R-loop formation is involved in DNA recombination during lytic replication and whether this is a common property of all herpesviruses.
Acknowledgments
We acknowledge the support of The Wistar Institute Cancer Center Core Facilities for Genomics and Flow Cytometry. We also thank Henri-Jacques Delecluse (Ludwig Maximilian University) for the ZKO-293 cells, Robert J. Crouch (NICHD) for the RNase H1-GFP expression plasmid, Perry J. Blackshear (NIEHS) for the RecQL1 rabbit polyclonal antibody, Ramin Shiekhattar (The Wistar Institute) for the RNA-DNA hybrid mouse monoclonal antibody, Nigel W. Fraser (University of Pennsylvania) for the KOS HSV1 virus, and Andreas Wiedmer, Christine M. Helfer, and Senem Sezgi for technical assistance.
This work was supported by an NIH grant (R01CA0856780) to P.M.L. A.J.R. was supported by a predoctoral fellowship on The Wistar Institute Cancer Biology Training Grant from the NIH (1T32 CA09171).
Footnotes
Published ahead of print on 29 December 2010.
REFERENCES
- 1.Baumann, M., R. Feederle, E. Kremmer, and W. Hammerschmidt. 1999. Cellular transcription factors recruit viral replication proteins to activate the Epstein-Barr virus origin of lytic DNA replication, oriLyt. EMBO J. 18:6095-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billaud, G., D. Thouvenot, and F. Morfin. 2009. Drug targets in herpes simplex and Epstein Barr virus infections. Infect. Disord. Drug Targets 9:117-125. [DOI] [PubMed] [Google Scholar]
- 3.Binne, U. K., W. Amon, and P. J. Farrell. 2002. Promoter sequences required for reactivation of Epstein-Barr virus from latency. J. Virol. 76:10282-10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boehmer, P. E. 2004. RNA binding and R-loop formation by the herpes simplex virus type-1 single-stranded DNA-binding protein (ICP8). Nucleic Acids Res. 32:4576-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boguslawski, S. J., et al. 1986. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods 89:123-130. [DOI] [PubMed] [Google Scholar]
- 6.Brousset, P., V. Butet, S. Chittal, J. Selves, and G. Delsol. 1992. Comparison of in situ hybridization using different nonisotopic probes for detection of Epstein-Barr virus in nasopharyngeal carcinoma and immunohistochemical correlation with anti-latent membrane protein antibody. Lab. Invest. 67:457-464. [PubMed] [Google Scholar]
- 7.Brousset, P., et al. 1993. Assessment of the methods for the detection of Epstein-Barr virus nucleic acids and related gene products in Hodgkin's disease. Lab. Invest. 69:483-490. [PubMed] [Google Scholar]
- 8.Cerritelli, S. M., and R. J. Crouch. 1998. Cloning, expression, and mapping of ribonucleases H of human and mouse related to bacterial RNase HI. Genomics 53:300-307. [DOI] [PubMed] [Google Scholar]
- 9.Cerritelli, S. M., et al. 2003. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell 11:807-815. [DOI] [PubMed] [Google Scholar]
- 10.Chevallier-Greco, A., et al. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho, M. S., and V. M. Tran. 1993. A concatenated form of Epstein-Barr viral DNA in lymphoblastoid cell lines induced by transfection with BZLF1. Virology 194:838-842. [DOI] [PubMed] [Google Scholar]
- 12.Corcoran, J. A., H. A. Saffran, B. A. Duguay, and J. R. Smiley. 2009. Herpes simplex virus UL12.5 targets mitochondria through a mitochondrial localization sequence proximal to the N terminus. J. Virol. 83:2601-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Countryman, J., and G. Miller. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. U. S. A. 82:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniels, G. A., and M. R. Lieber. 1995. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 23:5006-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng, Z., et al. 2002. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol. Cell 9:493-503. [DOI] [PubMed] [Google Scholar]
- 16.Dunnick, W., G. Z. Hertz, L. Scappino, and C. Gritzmacher. 1993. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 21:365-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feederle, R., et al. 2000. The Epstein-Barr virus lytic program is controlled by the operative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao, Y., P. R. Smith, L. Karran, Q. L. Lu, and B. E. Griffin. 1997. Induction of an exceptionally high-level, nontranslated, Epstein-Barr virus-encoded polyadenylated transcript in the Burkitt's lymphoma line Daudi. J. Virol. 71:84-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, Z., et al. 1998. The Epstein-Barr virus lytic transactivator Zta interacts with the helicase-primase replication proteins. J. Virol. 72:8559-8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez-Barrera, S., M. Garcia-Rubio, and A. Aguilera. 2002. Transcription and double-strand breaks induce similar mitotic recombination events in Saccharomyces cerevisiae. Genetics 162:603-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruffat, H., O. Renner, D. Pich, and W. Hammerschmidt. 1995. Cellular proteins bind to the downstream component of the lytic origin of DNA replication of Epstein-Barr virus. J. Virol. 69:1878-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammerschmidt, W., and B. Sugden. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55:427-433. [DOI] [PubMed] [Google Scholar]
- 25.He, X., and I. R. Lehman. 2000. Unwinding of a herpes simplex virus type 1 origin of replication (OriS) by a complex of the viral origin binding protein and the single-stranded DNA binding protein. J. Virol. 74:5726-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huertas, P., and A. Aguilera. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12:711-721. [DOI] [PubMed] [Google Scholar]
- 27.Hummel, M., and E. Kieff. 1982. Mapping of polypeptides encoded by the Epstein-Barr virus genome in productive infection. Proc. Natl. Acad. Sci. U. S. A. 79:5698-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeang, K. T., and S. D. Hayward. 1983. Organization of the Epstein-Barr virus DNA molecule. III. Location of the P3HR-1 deletion junction and characterization of the NotI repeat units that form part of the template for an abundant 12-O-tetradecanoylphorbol-13-acetate-induced mRNA transcript. J. Virol. 48:135-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohzaki, H., and Y. Murakami. 2005. Transcription factors and DNA replication origin selection. Bioessays 27:1107-1116. [DOI] [PubMed] [Google Scholar]
- 30.Laux, G., U. K. Freese, and G. W. Bornkamm. 1985. Structure and evolution of two related transcription units of Epstein-Barr virus carrying small tandem repeats. J. Virol. 56:987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, D. Y., and D. A. Clayton. 1996. Properties of a primer RNA-DNA hybrid at the mouse mitochondrial DNA leading-strand origin of replication. J. Biol. Chem. 271:24262-24269. [DOI] [PubMed] [Google Scholar]
- 32.Lee, S. S., and I. R. Lehman. 1997. Unwinding of the box I element of a herpes simplex virus type 1 origin by a complex of the viral origin binding protein, single-strand DNA binding protein, and single-stranded DNA. Proc. Natl. Acad. Sci. U. S. A. 94:2838-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leopardi, R., P. L. Ward, W. O. Ogle, and B. Roizman. 1997. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the U(L)13 proteinkinase. J. Virol. 71:1133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao, G., J. Huang, E. D. Fixman, and S. D. Hayward. 2005. The Epstein-Barr virus replication protein BBLF2/3 provides an origin-tethering function through interaction with the zinc finger DNA binding protein ZBRK1 and the KAP-1 corepressor. J. Virol. 79:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao, G., F. Y. Wu, and S. D. Hayward. 2001. Interaction with the Epstein-Barr virus helicase targets Zta to DNA replication compartments. J. Virol. 75:8792-8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieberman, P. M., and A. J. Berk. 1990. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J. Virol. 64:2560-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lieberman, P. M., J. M. Hardwick, and S. D. Hayward. 1989. Responsiveness of the Epstein-Barr virus NotI repeat promoter to the Z transactivator is mediated in a cell-type-specific manner by two independent signal regions. J. Virol. 63:3040-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lieberman, P. M., J. M. Hardwick, J. Sample, G. S. Hayward, and S. D. Hayward. 1990. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marchini, A., J. I. Cohen, F. Wang, and E. Kieff. 1992. A selectable marker allows investigation of a nontransforming Epstein-Barr virus mutant. J. Virol. 66:3214-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metzenberg, S. 1989. Relative rates of RNA synthesis across the genome of Epstein-Barr virus are highest near oriP and oriLyt. J. Virol. 63:4938-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami, Y., and Y. Ito. 1999. Transcription factors in DNA replication. Front. Biosci. 4:D824-D833. [DOI] [PubMed] [Google Scholar]
- 42.Nimonkar, A. V., and P. E. Boehmer. 2003. On the mechanism of strand assimilation by the herpes simplex virus type-1 single-strand DNA-binding protein (ICP8). Nucleic Acids Res. 31:5275-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nimonkar, A. V., and P. E. Boehmer. 2003. Reconstitution of recombination-dependent DNA synthesis in herpes simplex virus 1. Proc. Natl. Acad. Sci. U. S. A. 100:10201-10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nimonkar, A. V., and P. E. Boehmer. 2003. The herpes simplex virus type-1 single-strand DNA-binding protein (ICP8) promotes strand invasion. J. Biol. Chem. 278:9678-9682. [DOI] [PubMed] [Google Scholar]
- 45.Nuebling, C. M., and N. Mueller-Lantzsch. 1989. Identification and characterization of an Epstein-Barr virus early antigen that is encoded by the NotI repeats. J. Virol. 63:4609-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nuebling, C. M., and N. Mueller-Lantzsch. 1991. Identification of the gene product encoded by the PstI repeats (IR4) of the Epstein-Barr virus genome. Virology 185:519-523. [DOI] [PubMed] [Google Scholar]
- 47.Pfitzner, A. J., E. C. Tsai, J. L. Strominger, and S. H. Speck. 1987. Isolation and characterization of cDNA clones corresponding to transcripts from the BamHI H and F regions of the Epstein-Barr virus genome. J. Virol. 61:2902-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Portes-Sentis, S., A. Sergeant, and H. Gruffat. 1997. A particular DNA structure is required for the function of a cis-acting component of the Epstein-Barr virus OriLyt origin of replication. Nucleic Acids Res. 25:1347-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prichard, M. N., et al. 1998. Identification of persistent RNA-DNA hybrid structures within the origin of replication of human cytomegalovirus. J. Virol. 72:6997-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quadt, I., A. K. Gunther, D. Voss, M. Schelhaas, and D. Knebel-Morsdorf. 2006. TATA-binding protein and TBP-associated factors during herpes simplex virus type 1 infection: localization at viral DNA replication sites. Virus Res. 115:207-213. [DOI] [PubMed] [Google Scholar]
- 51.Reaban, M. E., J. Lebowitz, and J. A. Griffin. 1994. Transcription induces the formation of a stable RNA.DNA hybrid in the immunoglobulin alpha switch region. J. Biol. Chem. 269:21850-21857. [PubMed] [Google Scholar]
- 52.Rennekamp, A. J., and P. M. Lieberman. 2010. Initiation of lytic DNA replication in Epstein-Barr virus: search for a common family mechanism. Future Virol. 5:65-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rennekamp, A. J., P. Wang, and P. M. Lieberman. 2010. Evidence for DNA hairpin recognition by Zta at the Epstein-Barr virus origin of lytic replication. J. Virol. 84:7073-7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reuven, N. B., A. E. Staire, R. S. Myers, and S. K. Weller. 2003. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J. Virol. 77:7425-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rice, S. A., M. C. Long, V. Lam, and C. A. Spencer. 1994. RNA polymerase II is aberrantly phosphorylated and localized to viral replication compartments following herpes simplex virus infection. J. Virol. 68:988-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robertson, E. S. 2005. Epstein-Barr virus. Caister Academic Press, Wymondham, Norfolk, England.
- 57.Rooney, C. M., D. T. Rowe, T. Ragot, and P. J. Farrell. 1989. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J. Virol. 63:3109-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roy, D., and M. R. Lieber. 2009. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol. Cell. Biol. 29:3124-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roy, D., K. Yu, and M. R. Lieber. 2008. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol. Cell. Biol. 28:50-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roy, D., Z. Zhang, Z. Lu, C. L. Hsieh, and M. R. Lieber. 2010. Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol. Cell. Biol. 30:146-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruhanen, H., et al 2010. Mitochondrial single-stranded DNA binding protein is required for maintenance of mitochondrial DNA and 7S DNA but is not required for mitochondrial nucleoid organisation. Biochim. Biophys. Acta 1803:931-939. [DOI] [PubMed] [Google Scholar]
- 62.Ryon, J. J., et al. 1993. In situ detection of lytic Epstein-Barr virus infection: expression of the NotI early gene and viral interleukin-10 late gene in clinical specimens. J. Infect. Dis. 168:345-351. [DOI] [PubMed] [Google Scholar]
- 63.Saffran, H. A., J. M. Pare, J. A. Corcoran, S. K. Weller, and J. R. Smiley. 2007. Herpes simplex virus eliminates host mitochondrial DNA. EMBO Rep. 8:188-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sato, H., T. Takimoto, S. Tanaka, J. Tanaka, and N. Raab-Traub. 1990. Concatameric replication of Epstein-Barr virus: structure of the termini in virus-producer and newly transformed cell lines. J. Virol. 64:5295-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schepers, A., D. Pich, and W. Hammerschmidt. 1993. A transcription factor with homology to the AP-1 family links RNA transcription and DNA replication in the lytic cycle of Epstein-Barr virus. EMBO J. 12:3921-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schepers, A., D. Pich, and W. Hammerschmidt. 1996. Activation of oriLyt, the lytic origin of DNA replication of Epstein-Barr virus, by BZLF1. Virology 220:367-376. [DOI] [PubMed] [Google Scholar]
- 67.Schepers, A., D. Pich, J. Mankertz, and W. Hammerschmidt. 1993. cis-acting elements in the lytic origin of DNA replication of Epstein-Barr virus. J. Virol. 67:4237-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 69.Takada, K., N. Shimizu, S. Sakuma, and Y. Ono. 1986. trans activation of the latent Epstein-Barr virus (EBV) genome after transfection of the EBV DNA fragment. J. Virol. 57:1016-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takamatsu, C., et al. 2002. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 3:451-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takebe, Y., et al. 1988. SR alpha promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol. Cell. Biol. 8:466-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsurumi, T., et al. 1998. Overexpression, purification and helix-destabilizing properties of Epstein-Barr virus ssDNA-binding protein. J. Gen. Virol. 79:1257-1264. [DOI] [PubMed] [Google Scholar]
- 73.Tsurumi, T., et al. 1996. Epstein-Barr virus single-stranded DNA-binding protein: purification, characterization, and action on DNA synthesis by the viral DNA polymerase. Virology 222:352-364. [DOI] [PubMed] [Google Scholar]
- 74.Wang, Y., Q. Tang, G. G. Maul, and Y. Yuan. 2006. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: dual role of replication and transcription activator. J. Virol. 80:12171-12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wiedmer, A., et al. 2008. Epstein-Barr virus immediate-early protein Zta co-opts mitochondrial single-stranded DNA binding protein to promote viral and inhibit mitochondrial DNA replication. J. Virol. 82:4647-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu, B., and D. A. Clayton. 1995. A persistent RNA-DNA hybrid is formed during transcription at a phylogenetically conserved mitochondrial DNA sequence. Mol. Cell. Biol. 15:580-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu, B., and D. A. Clayton. 1996. RNA-DNA hybrid formation at the human mitochondrial heavy-strand origin ceases at replication start sites: an implication for RNA-DNA hybrids serving as primers. EMBO J. 15:3135-3143. [PMC free article] [PubMed] [Google Scholar]
- 78.Xue, S. A., M. D. Jones, Q. L. Lu, J. M. Middeldorp, and B. E. Griffin. 2003. Genetic diversity: frameshift mechanisms alter coding of a gene (Epstein-Barr virus LF3 gene) that contains multiple 102-base-pair direct sequence repeats. Mol. Cell. Biol. 23:2192-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Young, L. S., and A. B. Rickinson. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757-768. [DOI] [PubMed] [Google Scholar]
- 80.Yu, K., F. Chedin, C. L. Hsieh, T. E. Wilson, and M. R. Lieber. 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4:442-451. [DOI] [PubMed] [Google Scholar]
- 81.Zhang, Q., E. Holley-Guthrie, J. Q. Ge, D. Dorsky, and S. Kenney. 1997. The Epstein-Barr virus (EBV) DNA polymerase accessory protein, BMRF1, activates the essential downstream component of the EBV oriLyt. Virology 230:22-34. [DOI] [PubMed] [Google Scholar]
- 82.Zhang, Q., et al. 1996. Functional and physical interactions between the Epstein-Barr virus (EBV) proteins BZLF1 and BMRF1: effects on EBV transcription and lytic replication. J. Virol. 70:5131-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]