Abstract
Hepatitis C virus (HCV)-related research has been hampered by the lack of appropriate small-animal models. It has been reported that tree shrews, or tupaias (Tupaia belangeri), can be infected with serum-derived HCV. However, these reports do not firmly establish the tupaia as a reliable model of HCV infection. Human CD81, scavenger receptor class B type I (SR-BI), claudin 1 (CLDN1), and occludin (OCLN) are considered essential receptors or coreceptors for HCV cell entry. In the present study, the roles of these tupaia orthologs in HCV infection were assessed. Both CD81 and SR-BI of tupaia were found to be able to bind with HCV envelope protein 2 (E2). In comparison with human CD81, tupaia CD81 exhibited stronger binding activity with E2 and increased HCV pseudoparticle (HCVpp) cell entry 2-fold. The 293T cells transfected with tupaia CLDN1 became susceptible to HCVpp infection. Moreover, simultaneous transfection of the four tupaia factors into mouse NIH 3T3 cells made the cells susceptible to HCVpp infection. HCVpp of diverse genotypes were able to infect primary tupaia hepatocytes (PTHs), and this infection could be blocked by either anti-CD81 or anti-SR-BI. PTHs could be infected by cell culture-produced HCV (HCVcc) and did produce infectious progeny virus in culture supernatant. These findings indicate that PTHs possess all of the essential factors required for HCV entry and support the complete HCV infection cycle. This highlights both the mechanisms of susceptibility of tupaia to HCV infection and the possibility of using tupaia as a promising small-animal model in HCV study.
Hepatitis C virus (HCV) is a major cause of liver disease. A total of 170 million individuals worldwide are estimated to be infected with HCV and are at risk of developing cirrhosis and hepatocellular carcinoma (32, 33). Unfortunately, there is presently no effective HCV vaccine available, and current treatments are far from satisfactory (22, 28). The development of antiviral therapies and effective vaccines has been hampered greatly by the lack of a convenient small-animal model. Chimpanzees (Pan troglodytes) are the only nonhuman primate host serving as an HCV infection model. However, experiments using chimpanzees are both expensive and ethically problematic. To date, three small-animal models of HCV infection have been reported: the immunotolerized rat model, Trimera mouse model, and uPA/SCID mouse model (16, 25, 39). However, these models are difficult to prepare, and the abnormal immune status of each greatly limits their application.
The tree shrew or tupaia (Tupaia belangeri) is a small, squirrel-like mammal that is closely related to primates (6). Since the 1980s, tupaias have been used as an animal model of various infectious agents and their associated diseases. Tupaia has been shown to be susceptible to a variety of human viruses, including herpes simplex virus and hepatitis B virus (HBV) (9, 38). Early in 1998 it was reported that inoculation with HCV RNA-positive serum could lead to short-term viremia and the appearance of anti-HCV IgG in tupaia (40). Furthermore, primary tupaia hepatocytes (PTHs) can be infected in vitro with serum derived from chronic hepatitis C patients (2, 18, 21, 36), although it is not clear whether the viral RNA measured is due to de novo production and/or from the virus inoculum. Recently, independent observations showed that inoculating tupaia with hepatitis C patient serum or viral particles reconstituted from full-length HCV cDNA caused mild hepatitis and intermittent viremia (1). However, these reports do not firmly establish the tupaia as a reliable model of HCV replication and pathogenesis. Importantly, patient sera often exhibit very weak infectivity (15, 36). Although these results show promise, additional work has to be conducted to evaluate the value of pursuing the tupaia system in HCV research.
The expression and distribution of virus-specific receptors are crucial for a virus’ host range and tissue tropism. HCV is a multiple-receptor virus, and various molecules are associated with the entry of HCV into host cells (5, 19, 36). The HCV envelope glycoprotein 1 (E1) and E2, forming a noncovalent heterodimer, are candidate ligands for cellular receptors (11, 27). Human CD81 (29, 43), scavenger receptor class B type I (SR-BI/ClaI) (31, 42), claudin 1 (CLDN1) (12), and occludin (OCLN) (30) are considered essential receptors or coreceptors for HCV cell entry. To investigate whether tupaia can serve as a small-animal model for HCV infection, tupaia CD81, SR-BI, CLDN1, and OCLN cDNAs were cloned for functional characterization in mediating HCV infection. HCV pseudoparticles (HCVpp) and cell culture-produced HCV (HCVcc) were used to infect PTHs and other cell lines stably transfected with one or all of these molecules. The elucidation of the role of these tupaia orthologs in mediating HCV infection will promote the development of the tupaia model as one which is relevant to human infection.
MATERIALS AND METHODS
Animals.
Adult tupaias captured in the wild in Yunnan Province, China, were quarantined for at least 2 months before experiments. All animals were detected to be negative for HBV and HCV by real-time PCR. The procedures used in the handling and care of the animals were approved by the Animal Ethical Committee of the Second Military Medical University, Shanghai, China.
Isolation of PTHs.
PTHs were isolated from adult tupaias by a two-step collagenase perfusion, as described previously (23, 38). Briefly, tupaias were anesthetized by the intramuscular injection of ketamine (0.5 mg/kg body mass) and xylazine (0.1 mg/kg). Livers were perfused in situ for 5 min via the portal vein with Hanks solution containing 5 mM EGTA and perfused with Hanks solution containing 5 mM CaCl2 and 0.5 mg/ml collagenase (Invitrogen) for 10 min. Freshly isolated hepatocytes were seeded at 5 × 105 cells/ml of medium (2 ml per well) on collagen-coated six-well plates and were maintained in Williams’ E medium (Invitrogen) supplemented with penicillin and streptomycin (100 U/ml) (Invitrogen), bovine insulin (5 mg/liter) (Sigma), 2% dimethyl sulfoxide (DMSO) (Sigma), 3.5 × 105 M hydrocortisone hemisuccinate (Sigma), and 10% fetal calf serum (Invitrogen). The cells were incubated at 37°C in a humidified 5% CO2 atmosphere; the medium was changed every 2 or 3 days.
Cloning of tupaia CD81, SR-BI, CLDN1, and OCLN and lentivirus plasmid construction.
Total RNA was extracted from PTHs using RNeasy reagent (Qiagen). Reverse transcription was performed using AMV RTase (Promega) with oligo15T primers. The products were used to amplify the cDNAs encoding CD81, SR-BI, CLDN1, and OCLN using the Expand High FidelityPLUS PCR system (Roche). Corresponding oligonucleotide primers were designed according to human-specific sequences. The products from two independent reactions were inserted into the pGEM-T vector (Promega) and sequenced. The appropriate cDNAs were then inserted into the lentivirus vector pLenti-6 (Invitrogen) for the packaging of recombinant lentivirus.
Four amino acids in the extracellular loop 2 (EL2) of human OCLN were mutated so as to be identical to the tupaia sequence (S203A, L207I, Y213N, and N217S) using a site-directed gene mutagenesis kit (Stratagene). The EL2-deleted tupaia OCLN DNA sequence (residues 199 to 243) was obtained by gene splicing using overlap extension PCR (SOE PCR). The resulting human-tupaia chimeric OCLN and EL2-deleted tupaia OCLN DNA fragments were inserted into the vector pLenti-6.
The cDNAs of these four receptors derived from Huh7.5 cells (provided by C. Rice, Rockefeller University, New York, NY) were also cloned and used for lentivirus plasmid construction. Murine CD81 cDNA was obtained from the livers of BALB/c mice by RT-PCR as described above.
Expression of human, tupaia, and mouse CD81 LEL.
DNA sequences encoding the human, tupaia, or mouse CD81 large extracellular loops (LEL) were amplified by PCR and then inserted separately into the BamHI/XhoI sites of pET32a (Novagen). Each was used to prepare a thioredoxin (TRX)-CD81 LEL fusion protein in Escherichia coli. Expression of the fusion proteins was induced by isopropyl-β-d-thiogalactopyranoside. The proteins were purified using a nickel-chelating Sepharose resin (Qiagen). Purified Trx-CD81 LEL fusion proteins were analyzed by SDS-PAGE and Western blotting using anti-human CD81 monoclonal antibodies (MAbs) 5A6 (Santa Cruz Biotechnology), JS81 (BD Pharmingen), and 1.3.3.22 (Santa Cruz Biotechnology) to probe the CD81 LEL.
Preparation of rabbit anti-human CD81 LEL IgG.
Male rabbits (2 to 2.5 kg each) were immunized by subcutaneous injection with 500 μg of the human CD81 LEL fused with TRX emulsified in complete Freund's adjuvant (Sigma) on day 0 and boosted with 100 μg of fusion protein on days 14 and 28. Serum samples were tested for reactivity with CHO cells expressing human or tupaia CD81 by fluorescence-activated cell sorting (FACS) at defined time points. The rabbits were bled on day 35, and total IgG was purified using a Protein A Sepharose 4 fast flow column (GE Healthcare).
DNA immunization of mice with human or tupaia SR-BI expression plasmid.
Antibodies against human and tupaia SR-BI were raised by DNA immunization of BALB/c mice as Zeisel et al. described (42). The animals received 3 applications of 100 μg pLenti-6 (Invitrogen) lentivirus expression plasmid containing the full-length human or tupaia SR-BI by using intramuscular injection at 2-week intervals. The mice were bled on day 14 after the last immunization, and the immune sera and the preimmune control sera were analyzed for specificity to SR-BI with CHO cells transduced with lentivirus containing the human or tupaia SR-BI gene by flow cytometry.
Expression of HCV E2 protein.
The DNA sequence encoding the carboxy terminus truncated E2 (residues 364 to 661 in the HCV polyprotein) of strain H77 (genotype 1a, GenBank accession no. AF009606) was synthesized by SOE PCR using optimized codons of highly expressed mammalian genes. The resulting DNA fragment was sequenced and inserted into the pCI-neo vector (Promega). E2 and mock plasmids were transfected into 293T cells using Lipofectamine 2000 (Invitrogen); after 60 h, the supernatant was decanted, and the cells were removed from the tissue culture dishes by a phosphate-buffered saline (PBS)-EDTA treatment, resuspended in PBS supplemented with proteinase inhibitor cocktail (Roche), and lysed by ultrasonication. Lysates were clarified by centrifugation and then concentrated 10-fold by Centricon ultrafiltration (Millipore). E2 expression was assessed by Western blot analysis using goat anti-E2 polyclonal antibodies (Biodesign International).
EIA for binding of E2 to recombinant human, tupaia, and mouse CD81.
An enzyme immunoassay (EIA) was performed to evaluate the binding of E2 to the recombinant human, tupaia, and mouse CD81 LEL as described previously (13, 14). Enzyme-linked immunosorbent assay (ELISA) plates (Nunc) were coated overnight with 1 μg of purified TRX fusion protein per well. The plates were then blocked with milk buffer (5% nonfat dry milk and 0.05% Tween 20 in PBS) for 1 h and incubated with a dilution series of HCV E2 at 4°C overnight. After extensive washing, bound E2 was detected using the conformation-dependent anti-E2 MAb H53 (provided by J. Dubussion, Institut Pasteur, Lille, France) diluted 1:200 in milk buffer, for 1 h at room temperature, followed by horseradish peroxidase (HRP)-conjugated anti-mouse IgG and tetramethylbenzidine substrate. Absorbance was measured at 450 nm using an EIA reader.
Generation of lentivirus encoding human or tupaia CD81, SR-BI, CLDN1, and OCLN.
Lentiviruses (vesicular stomatitis virus [VSV] pseudoparticles) were generated using a ViraPower lentiviral support kit (Invitrogen) according to the manufacturer's instructions. Briefly, HEK 293T cells were transfected with the VSV glycoprotein expression plasmid and HIV gag/pol (pLP1), HIV rev (pLP2), and transfer vector pLenti6 vectors encoding the corresponding human or tupaia genes using Lipofectamine 2000 (Invitrogen). Supernatants containing the pseudoparticles were harvested 48 h after transfection and filtered through 0.45-μm-pore-size membranes before use.
Lentivirus infection.
CHO cells were infected with lentiviruses encoding human or tupaia CD81 and SR-BI for the assessment of HCV E2 binding. HepG2 cells were infected with lentiviruses encoding human or tupaia CD81, and 293T cells were infected with lentivirus encoding human or tupaia CLDN1 to assay HCVpp infection. NIH 3T3 cells were infected with a number of different lentivirus combinations for the assessment of HCVpp infection. The cells were seeded at 2 × 105 cells/well in a 24-well plate, and then lentivirus supernatants were added (200 μl/well). At 60 h postinfection, the CD81 levels were assayed by flow cytometry by the use of a Cell Lab Quanta SC instrument (Beckman Coulter) using the anti-CD81 MAb 5A6 (Santa Cruz Biotechnology) and rabbit anti-human CD81 LEL polyclonal antibodies. SR-BI, CLDN1, and OCLN were detected by Western blot analysis using an anti-SR-BI MAb (BD Pharmingen), rabbit anti-human CLDN1 polyclonal antibodies (Cell Signaling Technology), and an anti-human OCLN MAb (Invitrogen).
Assay of E2 binding with cell surface-associated CD81 and SR-BI.
HCV E2 binding to cell surface-associated CD81 or SR-BI was measured using a FACS-based assay, as described previously (13, 14). Cells were detached using 2 mM EDTA in PBS and washed twice in PBS supplemented with 2% fetal calf serum and 0.05% NaN3 (wash buffer). The cells (3 × 105) were then incubated with crude cell extract containing E2 for 1 h at room temperature in wash buffer and washed twice with PBS. The cells were then incubated for 1 h at 4°C with diluted goat anti-E2 polyclonal antibodies (Biodesign International). After labeling with fluorescein isothiocyanate (FITC)-conjugated rabbit anti-goat IgG (Jackson ImmunoResearch), E2 binding (mean fluorescence intensity [MFI]) was quantified by flow cytometry.
HCVpp production and infection.
HCVpp were generated by the cotransfection of HEK 293T cells with an HCV envelope protein expression vector and a packaged plasmid based on the HIV-1 strain NL4-3 (Invitrogen) as described previously (3, 20). Briefly, 293T cells were cotransfected with expression plasmids encoding the HCV envelope glycoproteins, HIV gag/pol (pLP1), HIV rev (pLP2), and pLenti6 encoding luciferase. HCV envelope expression plasmids used here included genotype 1a strain H77 (provided by F. L. Cosset, INSERM U758, Lyon, France), genotype 1b strain Con-1 (provided by C. Rice, Rockefeller University, New York, NY), and genotypes 2a (clone UKN2A1.2), 2b (clone UKN2B2.8), 3a (clone UKN3A1.28C), and 4a (clone UKN4.21.16) (provided by J. K. Ball, The University of Nottingham, United Kingdom). Supernatants containing HCVpp were harvested at 48 h posttransfection, filtered through 0.45-μm membranes, and used for infection.
Huh7.5, HepG2, 293T, and NIH 3T3 cells engineered to express one or more human or tupaia molecules were seeded into 96-well plates at a density of 1 × 104 cells/well and incubated overnight at 37°C. The HCVpp supernatant (20 μl) was added to each well, and the plates were incubated for 5 h. The supernatants were then removed, and the cells were incubated in regular medium for 72 h at 37°C. Following incubation, the cells were washed once in PBS and then lysed in 50 μl of lysis buffer (Promega). Luciferase activity was quantified using the Bright Glow luciferase assay system (Promega).
Human CD81 MAbs JS81, 1.3.3.22, 5A6, and rabbit anti-human CD81 LEL IgG polyclonal antibodies were used to inhibit the HCVpp infection of HepG2 cells expressing human or tupaia CD81. Before infection, human CD81 monoclonal (5 μg/ml) or polyclonal antibodies (50 μg/ml) were added to the cell culture plates, which were incubated for 1 h at 37°C. The supernatants were then removed, and the plates were washed twice with PBS. HCVpp infection was then performed as described above. Mouse anti-human or -tupaia SR-BI serum was used to inhibit the HCVpp infection of Huh7.5 cells. Huh7.5 cells were preincubated in the presence of anti-SR-BI serum or preimmune control serum at a 1:50 dilution for 1 h at 37°C, the medium was removed, and the cells were infected with HCVpp.
HCVpp infection of PTHs.
Freshly isolated PTHs were seeded at 2 × 105 cells/ml of medium (0.5 ml per well) on collagen-coated 24-well plates. Aliquots of the supernatant (100 μl) containing HCVpp were added to each well, and the plates were incubated for 5 h. The medium was then removed and replaced with normal culture medium. On day 4 postinfection, the cells were lysed, and the luciferase activity was quantified. Anti-CD81 antibodies and anti- SR-BI sera were used to inhibit the HCVpp infection as described above.
Generation of HCVcc and infection of PTHs.
Plasmid pFLJ6/JFH1, containing the full-length chimeric genomic cDNA for HCV J6 and JFH-1 (kindly provided by C. Rice, Rockefeller University, New York, NY), was used to generate HCVcc as described previously (24, 37, 45). Briefly, the plasmid was linearized and used as the template for transcription using an in vitro MEGAscript kit (Promega). The in vitro-transcribed RNA was delivered to Huh7.5 cells by electroporation. Viral stocks were obtained by harvesting the culture supernatants on days 8 to 12 after transfection and titrating them with Huh7.5 cells. Three days later, HCVcc-infected cells were examined by immunofluorescence analysis. The cells were washed, fixed with cold methanol, probed with anti-HCV-positive sera, washed, and probed with FITC-conjugated anti-human IgG (Jackson ImmunoResearch). Stained foci were counted in quadruplicate wells, and the virus titer of focus-forming units (FFU)/ml was calculated.
Freshly isolated PTHs were infected with the culture supernatant of HCVcc, and Huh7.5 and HepG2 cells were used as positive and negative controls, respectively. The cells were separately seeded in a 24-well plate, and following an overnight culture, a 50-μl HCVcc supernatant with 5 × 105 FFU/ml was added to each well. The medium was removed after 5 h, the wells were washed with 1 ml culture medium for a total of five times, 500 μl medium was added per well, and the plate was placed in a 5% CO2-humidified incubator at 37°C. The medium was changed every 24 h, and the culture was maintained 8 days. The HCV RNA load in recovered supernatants was determined using a commercial real-time quantitative RT-PCR kit (PG Biotech, Shenzhen, China). To detect whether PTHs could produce infectious progeny, the supernatant was added to freshly seeded Huh7.5 cells in 96-well plates at 50 μl/well. At day 3 postinfection, immunofluorescence analysis of HCV protein expression was performed as described above.
Nucleotide sequence accession number.
The nucleic acid sequences of tupaia CD81, SR-BI, CLDN1, and OCLN produced in this study have been deposited in GenBank under accession numbers EF581831, EU379936, EF584564, and FJ809937, respectively.
RESULTS
Cloning of tupaia CD81, SR-BI, CLDN1, and OCLN.
The corresponding amino acid sequences between human and tupaia CD81, SR-BI, CLDN1, and OCLN shared homologies of 96, 87, 93, and 88%, respectively. The protein sequences of tupaia and human CD81 LEL differed in six residues (Fig. 1A). The four CD81 LEL residues critical for HCV envelope binding, Ile182, Phe186, Asn184, and Leu162 (10), were identical between humans and the tupaia. The protein sequences of tupaia and mice CLDN1 EL1 were identical. Three of 53 residues in EL1 (31, 46, and 74) differed between human and tupaia CLDN1s (Fig. 1B). Residue 102 (valine) in human OCLN was not present in tupaia OCLN, while the EL2 regions of human and tupaia OCLN differed by 4 residues (Fig. 1C).
FIG. 1.
Sequence alignments of human, tupaia, and mouse CD81 LEL, claudin 1 EL1, and occludin EL2.
HCV E2 protein can bind tupaia CD81 LEL.
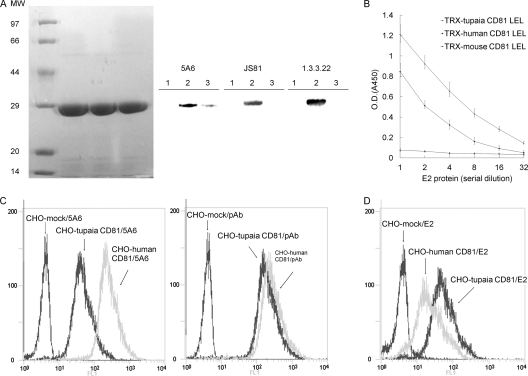
Human, tupaia, and mouse CD81 LEL were expressed as recombinant fusion proteins. All of the MAbs (JS81, 5A6, and 1.3.3.22) recognized the TRX-human CD81 LEL, whereas only 5A6 weakly detected the TRX-tupaia CD81 LEL, and none reacted with the TRX-mouse CD81 LEL at the expected size (29 kDa) (Fig. 2A). The HCV E2 protein bound readily to human and tupaia CD81 LEL, but not with the mouse CD81 LEL (Fig. 2B). The HCV E2 protein showed higher binding activity with the tupaia CD81 LEL than with the human CD81 LEL.
FIG. 2.
Tupaia CD81 binds HCV E2. (A) TRX-CD81 LEL fusion proteins were expressed and purified. Equal amounts of the fusion proteins were separated by SDS-PAGE and stained with Coomassie brilliant blue or immunoblotted with the anti-CD81 MAbs JS81, 5A6, and 1.3.3.22. Lane 1, TRX-mouse CD81 LEL; lane 2, TRX-human CD81 LEL; lane 3, TRX-tupaia CD81 LEL. MW, molecular weight (in thousands). (B) The interaction between HCV E2 and TRX-CD81 LEL fusion proteins was analyzed by EIA. Values are the means ± standard deviations of three independent experiments. O.D., optical density. (C) CHO cells were infected with lentiviruses containing human or tupaia CD81 sequences or mock lentivirus. Three days later, the cells were probed with the anti-CD81 MAb 5A6 (left) or polyclonal antibodies (pAb) (right) and analyzed for CD81 expression by flow cytometry. (D) HCV E2 binding to CD81-transduced CHO cells was detected with anti-E2 polyclonal antibodies by flow cytometry. In panels C and D, the values on the y axis indicate counts, and those on the x axis CD81 expression. The experiment was repeated at least three times, and one representative result is shown.
Binding of HCV E2 to cell surface-expressed human or tupaia CD81 was analyzed using a FACS-based assay. Rabbit anti-human CD81 polyclonal antibodies or MAb 5A6 bound CHO cells infected with lentivirus encoding human or tupaia CD81. No CD81 was detected on mock lentivirus-infected CHO cells (Fig. 2C). MAb 5A6 showed weaker binding to CHO cells expressing tupaia CD81, which was consistent with the Western blotting results. By an assay using human CD81 polyclonal antibodies, expression levels of human and tupaia CD81 were similar. HCV E2 could bind CHO cells expressing human or tupaia CD81 but not mock-infected CHO cells (Fig. 2D). Consistent with the EIA data, E2 protein could bind CHO cells expressing tupaia CD81 more strongly than those expressing human CD81 (Fig. 2D). The mean fluorescence intensity (MFI) of tupaia CD81-transduced CHO cells was about 2.5-fold higher than that of human CD81-transduced CHO cells.
HCV E2 can bind tupaia SR-BI.
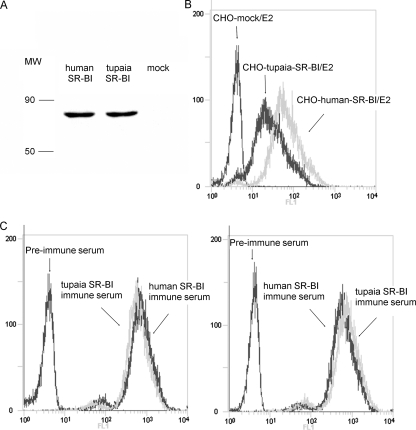
CHO cells were transduced with lentiviruses to express human or tupaia SR-BI. Expression of SR-BI was confirmed by Western blotting (Fig. 3A). An assay using flow cytometry showed that HCV E2 could bind with human SR-B1 and, to a lesser extent, with tupaia SR-BI (Fig. 3B). The MFI of tupaia SR-BI-transduced CHO cells was about 2-fold higher than that of human SR-BI-transduced CHO cells. Moreover, when the hypervariable region 1 (HVR1) was deleted from the E2 protein, no obvious binding could be detected between E2 and tupaia or human SR-BI (data not shown). Anti-human and -tupaia SR-BI sera were produced by DNA immunization of BALB/c mice. Human SR-BI immune serum reacted with human and tupaia SR-BI-expressing CHO cells with similar intensities but did not react with CHO cells that transduced with mock lentivirus, and similar results were obtained with tupaia SR-BI immune serum (Fig. 3C).
FIG. 3.
HCV E2 binds tupaia SR-BI. (A) CHO cells were infected with lentiviruses containing human or tupaia SR-BI sequences or mock lentivirus. Three days later, SR-BI expression was identified by immunoblotting with an anti-SR-BI MAb. MW, molecular weight (in thousands). (B) SR-BI- or mock-transduced CHO cells were incubated with the E2 extract, and E2 binding was assayed by flow cytometry using anti-E2 polyclonal antibodies. The experiment was repeated at least three times, and one representative result is shown. (C) CHO cells described in the legend to panel A were incubated with anti-SR-BI polyclonal serum collected from mouse that received DNA immunization with human or tupaia SR-BI expression plasmid or with preimmune control serum and analyzed for cell surface SR-BI expression by flow cytometry. In panels B and C, the values on the y axis indicate counts, and those on the x axis E2 or SR-BI expression, respectively.
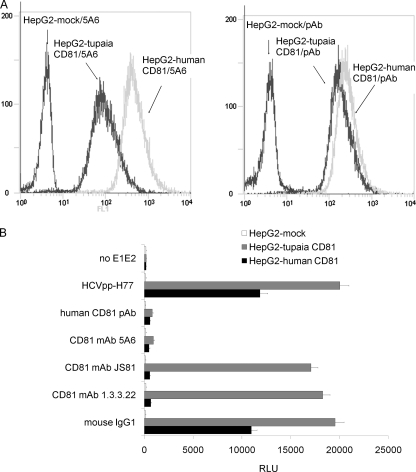
Tupaia CD81 supports HCVpp infection.
HepG2 cells were infected with recombinant lentiviruses to express membrane-associated human and tupaia CD81 (Fig. 4A). HepG2 cells expressing tupaia CD81 were more susceptible to HCVpp infection than those expressing human CD81, since the luciferase activity obtained from tupaia CD81- or human CD81-tranduced cells was 200- or 120-fold higher than that of mock transduced HepG2 cells (Fig. 4B). The preincubation of tupaia CD81-expressing HepG2 cells with the anti-CD81 MAb 5A6 or polyclonal antibodies decreased luciferase activity by 85 and 95%, respectively, while preincubation with the MAbs JS81 and 1.3.3.22, which were not able to bind tupaia CD81, did not block HCVpp infectivity. All three MAbs and polyclonal antibodies could block HCVpp infection of human CD81-expressing HepG2 cells (Fig. 4B).
FIG. 4.
Tupaia CD81 supports HCVpp infection. (A) HepG2 cells were infected with lentiviruses containing human or tupaia CD81 sequences or mock lentivirus. Three days later, the cells were probed with the anti-CD81 MAb 5A6 (left) or polyclonal antibodies (right), and CD81 expression was assessed by flow cytometry. The values on the y axis indicate counts, and those on the x axis CD81 expression. (B) Human or tupaia CD81- or mock-transduced HepG2 cells were incubated with anti-CD81 MAbs (5A6, JS81, and 1.3.3.22; 5 μg/ml), polyclonal antibodies (50 μg/ml), or mouse IgG1 (5 μg/ml) for 1 h prior to the addition of HCVpp or the control (no E1E2). At 72 h postinfection, the cells were harvested, and the level of luciferase activity (relative light units [RLU]) was determined. The values are the means ± standard deviations of results from three independent experiments.
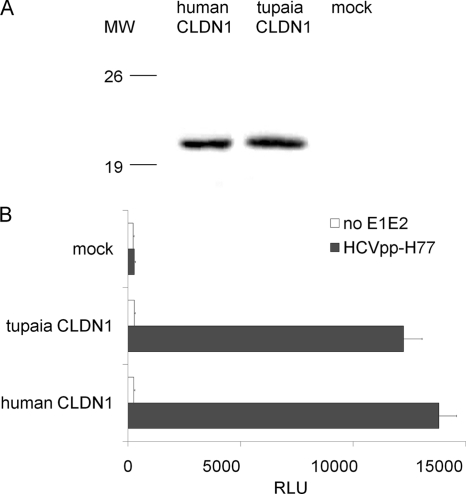
Tupaia CLDN1-expressing 293T cells became permissive for HCVpp infection.
To investigate whether tupaia CLDN1 could support HCV cell entry, 293T cells were infected with a lentivirus encoding either human or tupaia CLDN1. CLDN1 was detectable with anti- human CLDN1 polyclonal antibodies (Fig. 5A). Both tupaia and human CLDN1 specifically enhanced 293T susceptibility to HCVpp more than 80-fold (Fig. 5B).
FIG. 5.
Tupaia CLDN1 supports HCVpp infection. (A) 293T cells were infected with lentiviruses containing human or tupaia CLDN1 sequences or mock lentivirus, and the expression of CLDN1 was assayed by immunoblotting. MW, molecular weight (in thousands). (B) Transduced 293T cells were infected with HCVpp or control (no E1E2). Three days later, HCVpp infection was determined by measuring the level of luciferase activity. The values are the means ± standard deviations of three independent experiments.
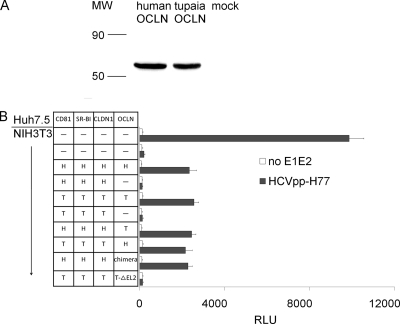
Tupaia OCLN rendered HCVpp infectious to mouse NIH 3T3 cells.
After the infection of NIH 3T3 cells with lentivirus containing human or tupaia OCLN cDNA, the corresponding proteins were detected with an anti-human OCLN MAb (Fig. 6A). NIH 3T3 cells were transduced with various combinations of lentiviruses encoding human or tupaia CD81, SR-BI, CLDN1, and OCLN, and the expression of each molecule was confirmed by Western blot analysis (data not shown). NIH 3T3 cells expressing all four human factors could be infected by HCVpp; the omission of OCLN abolished HCVpp cell entry. Similar results were observed with NIH 3T3 cells expressing four tupaia factors or lacking tupaia OCLN (Fig. 6B). The following combinations also could be infected by HCVpp: NIH 3T3 cells engineered to express human factors (CD81, SR-BI, and CLDN1) together with tupaia OCLN or with human-tupaia chimeric OCLN (in which EL2 from human OCLN was replaced by that from tupaia). NIH 3T3 cells expressing three tupaia factors together with human OCLN could also be infected by HCVpp. In contrast, NIH 3T3 cells expressing three tupaia factors (CD81, SR-BI, and CLDN1) together with tupaia OCLN lacking EL2 could not be infected with HCVpp. These results demonstrate that tupaia OCLN facilitates HCV cell entry in a pattern similar to that of human OCLN.
FIG. 6.
Tupaia OCLN facilitates the HCVpp infection of mouse NIH 3T3 cells. (A) NIH 3T3 cells were infected with lentiviruses containing human or tupaia OCLN sequences or mock lentivirus. Three days later, OCLN expression was assayed by immunoblotting with an anti-human OCLN MAb. MW, molecular weight (in thousands). (B) NIH 3T3 cells were infected with several combinations of lentiviruses and then with HCVpp. Three days later, the HCVpp infection levels were determined by measuring cellular luciferase activity. Naïve Huh7.5 and NIH 3T3 cells were used as positive and negative controls, respectively. H, human; T, tupaia; chimera, human-tupaia chimeric OCLN, in which the EL2 of humans was replaced by that of tupaia; T-ΔEL2, tupaia OCLN lacking EL2. The values are the means ± standard deviations of three independent experiments.
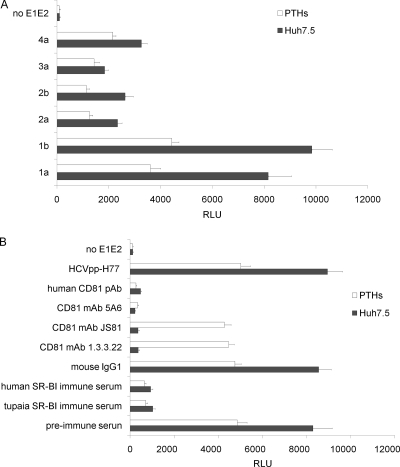
HCVpp can infect PTHs.
HCVpp of diverse genotypes were used to infect PTHs. PTHs were permissive to infection with all the tested HCVpp, and their susceptibility to different HCVpp was parallel with that of Huh7.5 cells (Fig. 7A). Pretreatment of PTHs with the anti-CD81 MAb 5A6 or polyclonal antibodies decreased HCVpp infectivity to less than 5% of mouse IgG1-pretreated PTHs (Fig. 7B). The anti-CD81 MAbs JS81 and 1.3.3.22 did not inhibit the HCVpp infection of PTHs, whereas it strongly abolished the HCVpp infection of Huh7.5 cells (Fig. 7B). Preincubation of Huh7.5 cells and PTHs with anti-human SR-BI mouse serum decreased luciferase activity by more than 85%, and so did the anti-tupaia SR-BI serum, while the preimmune sera did not influence the infectivity of HCVpp to Huh7.5 cells and PTHs (Fig. 7B).
FIG. 7.
PTHs can be infected by HCVpp. (A) HCVpp of diverse genotypes and control (no E1E2) were used to infect PTHs; Huh7.5 cells were used as a positive control. Three days after infection, the cells were lysed and assayed for luciferase activity. The values are the means ± standard deviations of three independent experiments. (B) Inhibition of HCV infection by anti-CD81 antibodies. PTHs and Huh7.5 cells were incubated with the human anti-CD81 MAbs 5A6, JS81, and 1.3.3.22, mouse IgG1 at 5 μg/ml, polyclonal antibodies at 50 μg/ml, human or tupaia SR-BI immune serum, or preimmune control serum at 1:50 dilution for 1 h prior to the addition of HCVpp. The values are the means ± standard deviations of three independent experiments.
HCVcc can infect PTHs and produce infectious virions.
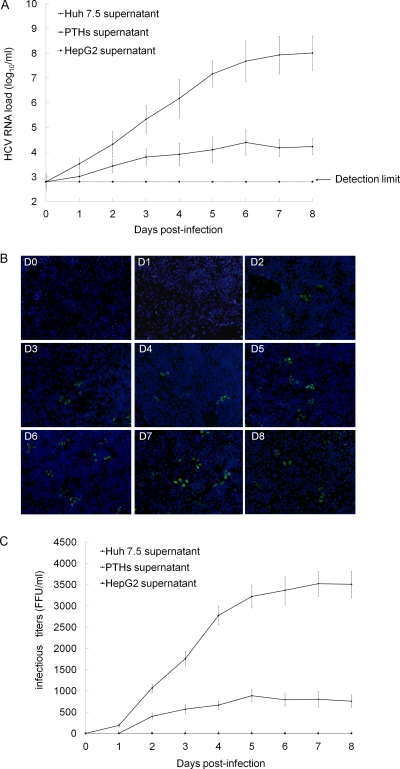
PTHs and Huh7.5 and HepG2 cells were incubated with J6/JFH-1 chimeric HCVcc for 5 h, and then the supernatants were collected every day. HCV RNA was detectable in the culture supernatants of PTHs by real-time quantitative RT-PCR analysis and maintained at a relatively stable level (103 to 104.5 copies/ml) during the observation period of 8 days. The detected RNA level peaked to 104.5 copies/ml on the 6th day of postinfection. In Huh7.5 cell culture, HCV RNA was robustly produced. As the negative control, HCV RNA in HepG2 supernatant was under the detection limit (8,000 copies/ml) all along (Fig. 8A).
FIG. 8.
PTHs can be infected by HCVcc. (A) The PTHs, Huh7.5 or HepG2 cells were incubated with J6/JFH-1 chimera HCVcc for 5 h, the media were removed, and the wells were washed five times with the culture medium. Fresh medium was added at 500 μl per well, and the plate was placed in a 5% CO2-humidified incubator at 37°C. The medium was changed every 24 h. The HCV RNA level in supernatants collected at days 0 to 8 was determined by real-time quantitative RT-PCR using a commercial kit (PG Biotech, Shenzhen, China). The supernatant at day 0 refers to the medium recovered from the cell culture at 3 h after washing the well and replacing the medium. (B) Culture medium from PTHs infected with HCVcc was collected every day for a total of 8 days and was used to infect freshly seeded Huh7.5 cells. Three days later, new HCV protein expression was detected by immunofluorescence analysis. The experiment was repeated at least three times, and one representative result is shown. (C) After detection by immunofluorescence analysis, the stained foci were counted in quadruplicate wells, and the mean infectious titers (FFU/ml) of culture medium collected from HCVcc-infected PTHs, Huh7.5, and HepG2 cells were calculated.
The PTH culture supernatant was further used to infect freshly plated Huh7.5 cells. Newly synthesized HCV proteins in the cells were detectable when the cells infected with the supernatants were recovered on days 2 to 8 (Fig. 8B). The infectious titers ranged from around 400 to 900 FFU/ml (Fig. 8C). The much higher infectivity titers were detected in supernatants of Huh7.5 cells. The supernatants from HCVcc-infected HepG2 cells were not found to be infectious all the time.
DISCUSSION
The present study demonstrates that tupaia CD81, SR-BI, CLDN1, and OCLN mediate HCV cell entry. PTHs are susceptible to infection by HCVpp and HCVcc, with the latter resulting in the full cycle of virus replication and appearance of infectious progeny virus. These results should promote the development of the tupaia system as an effective small-animal model for HCV infection and primary tupaia hepatocytes as an in vitro counterpart in the study of the biology of HCV.
The human CD81 LEL can bind HCV E2, and residues Ile182, Phe186, Asn184, and Leu162 of LEL have been reported to be the key sites for the binding of human CD81 with the HCV E2 protein (10); these four sites are conserved between the human and tupaia. Using three different human CD81 MAbs, only the 5A6 clone recognized the tupaia CD81 LEL, although the observed binding was weak. The EIA and flow cytometry assay for E2-CD81 binding also showed that HCV E2 bound to the tupaia CD81 LEL much more strongly than to the human CD81 LEL. These results are in contrast to a recent finding that the binding of the tupaia CD81 LEL glutathione S-transferase (GST) fusion protein to the HCV E2 protein was markedly reduced compared with that of human CD81 LEL GST (35). In this recent report, the recombinant human CD81 LEL and tupaia CD81 LEL were obtained from different research groups, while in the present study, both CD81 LEL were prepared under the same conditions. In addition, the divergence may be related to the genotype of the E2 proteins used for CD81 binding. The 1a genotype of the H77 isolate was used in the present study, while the genotype of the E2 protein was not presented in the recent report.
HCVpp, containing the E1 and E2 glycoproteins, is widely used to investigate HCV cell entry (3, 20). In the present study, tupaia CD81 could mediate HCVpp entry into HepG2 cells. Moreover, the level of the infection of tupaia CD81-expressing HepG2 cells with HCVpp was much higher than that of human CD81-expressing HepG2 cells; this difference may be due to differing binding activities for CD81 and E2. In addition, human CD81 MAb 5A6 and the rabbit anti-human CD81 LEL can inhibit HCVpp infection. Therefore, tupaia CD81 most likely possesses binding motifs similar to those of human CD81. Our results also suggest that, in addition to the identified motifs, other residues in the LEL may influence the binding activity of CD81 to HCV E2 and the ability to mediate HCV entry.
SR-BI cooperatively interacts with CD81 during HCV entry into host cells (42). HCV E2 can bind SR-BI, and the HVR1 region of E2 is responsible for this binding activity (31, 42). In addition, HCV is wrapped with lipoproteins which may mask HCV recognition by cell surface receptors except SR-B1, which recognizes very-low-density lipoprotein (VLDL); thus, SR-BI may enrich HCV on the hepatocyte membrane as a low-density lipoprotein receptor (LDL-R) (5, 19, 36). The present results show that HCV E2 is able to bind with tupaia SR-BI molecules, although with a reduced efficiency compared with the human ortholog, which is consistent with a recent report (7). Moreover, the E2-tupaia SR-BI binding also depended largely on the existence of HVR1 in the E2 protein. The amino acid sequence of tupaia SR-BI obtained in the present study is identical to that reported by Barth et al. (2); they reported that polyclonal antibodies against tupaia SR-BI inhibited the binding of PTHs with soluble E2 and HCV-like particles but could not block the infection of PTHs by serum-derived HCV. HCVpp-based models have demonstrated that the level of SR-BI needed for efficient infection varies between HCV genotypes and subtypes (17). Thus, it is possible that very few viruses infected PTHs in the presence of SR-BI antibodies, since evidence of replication was detectable only by RT-PCR. Under these conditions, early detection of HCV RNA by real-time quantitative RT-PCR may be useful in determining whether SR-BI antibodies interfere with the HCV infection of tupaia liver cells. In the present study, preincubating Huh7.5 cells or PTHs with anti-tupaia or anti-human SR-BI serum blocked HCVpp infection, which indicates that, just as with human SR-BI, tupaia SR-BI also has an important role in mediating HCV cell entry and that common or similar motifs exist in the two molecules that confer the ability to mediate HCV infection.
CLDN 1, a tight junction protein, is a putative receptor for HCV entry, and its functional domain is contained principally in the EL1 region (12). The amino acid sequences of tupaia and human CLDN1 share a high level of homology (93%). Only three residues (residues 31, 46, and 74) are different between human and tupaia CLDN1 EL1; I32 and E48, which have been recognized as key sites for HCV infection (12), are conserved in both the tupaia and human proteins. Tupaia and mouse cells share the same EL1 sequence, and it has been reported that the latter also supports HCV entry into host cells. The highly conserved residues W30-GLW51-C54-C64 in CLDN1 are required for HCV entry (8), and these are identical between the human and tupaia CLDN1. 293T cells transduced to express tupaia CLDN1 became susceptible to HCVpp, indicating that tupaia CLDN1 also supports HCV infection.
More recently, OCLN, another tight junction protein, has been proven critical for HCV entry into mouse cells (30). The tupaia and human OCLN amino acid sequences shared 88% homology. The key functional region for HCV entry is located in the OCLN EL2 (30). Tupaia and human EL2 are 91% homologous, with 4 of 45 residues in this region being different. Mouse and human EL2 are 86% homologous, with six residues being different. NIH 3T3 cells were cotransduced with human CD81, SR-BI, and CLDN1 and tupaia OCLN or with the four tupaia molecules and then subjected to HCVpp infection. Tupaia OCLN was found to mediate HCV entry. The EL2 domain is also a key domain for mediating HCV cell entry since EL2-deleted tupaia OCLN could not support HCVpp infection. When the EL2 domain of human OCLN was replaced by tupaia EL2, the chimera could also mediate the entry of HCVpp into NIH 3T3 cells. These results indicate that tupaia OCLN possesses functions related to HCV entry that are indistinguishable from those of human OCLN.
Furthermore, HCVpp of diverse genotypes could enter the PTHs, and this infectivity was similar to that in Huh7.5 cells, suggesting that PTHs possess all of the factors necessary for HCV entry into host cells. Like HepG2-tupaia CD81, PTHs preincubated with the anti-human CD81 MAb 5A6 or polyclonal antibodies did not become infected with HCVpp. As mentioned above, anti-human or -tupaia SR-BI serum blocked HCVpp infection of PTHs, just like the sera acted on the infectivity of HCVpp in Huh7.5 cells. These results suggest that the HCVpp infection of PTHs is also dependent on the expression of CD81 and SR-BI on the cell surface. Zhao and coworkers reported that monoclonal anti-human CD81 antibodies 5A6 and 1D6 could bind with tupaia CD81 but did not interfere with the infection by serum-derived HCV of PTHs (44). It is possible that very few viruses could infect PTHs in the presence of anti-CD81, especially as the affinity of the MAb 5A6 clone to tupaia CD81 is low, as we showed here. HCV RNA may accumulate in the culture period of 5 days, which meant that the inhibition by anti-CD81 could not be distinguished by RT-PCR assay. It has been well demonstrated that serum-derived HCV infection of primary human hepatocytes is CD81 dependent (26). Together with this recent demonstration, our present results suggest that it could be deduced that CD81 also plays an important role in HCV entry to PTHs.
The infectivity of serum-derived HCV is low, and it has been noted that less than 15% of HCV RNA-positive sera are truly infectious (15, 36). This may be due to the presence of anti-HCV antibodies in the sera of chronically infected patients and the fact that the major particles containing HCV RNA are noninfectious. The HCVcc model supports the production of authentic infectious viral particles in vitro and in vivo (24, 37, 45). It has been reported that HCVcc are more infectious for human hepatocytes than serum-derived HCV are (26). In this study, J6/JFH-1 chimeric HCVcc was used to infect PTHs, and afterward HCV RNA could be detected in the culture supernatants. Moreover, the supernatant collected from the PTH culture could further infect Huh7.5 cells, which indicates that infected PTHs generated infectious viral particles. Huh7.5 cells were taken as a positive control, and they supported the robust production of infectious HCV particles. In comparison, the HepG2 cells did not support production of HCV RNA and infectious virus particles. We observed that PTHs began to die after 4 or 5 days of incubation, so some viral RNA may have been released from the dead cells. Huh7.5 cells are highly permissive for HCV replicon replication (4), and it has been demonstrated that this cell line contains an inactivating mutation in RIG-I (34), an important component for interferon response via double-stranded RNA-sensing machinery (41). This may be associated with the fact that Huh7.5 cells supported both HCV replication and infectious virion production much more efficiently than PTHs did. Previous reports have indicated that PTHs could be infected by patient serum-derived HCV, while the assay was exclusively performed at the RNA level (2, 8, 21, 44). Using the HCVpp and HCVcc models, the present study clearly demonstrates that PTHs support the complete replication cycle of HCV.
In summary, tupaia CD81, SR-BI, CLDN1, and OCLN were cloned, and data were presented to show that these tupaia molecules are able to mediate HCV infection in both human and tupaia cells. Further, the data showed that HCVpp and HCVcc can infect PTHs and that HCVcc-infected PTHs could generate infectious progeny virus. These findings provide evidence that tupaia is important for HCV research.
Acknowledgments
We are grateful to C. M. Rice, J. Dubuisson, F.-L. Cosset, and J. K. Ball for the provision of reagents. We thank J. Zhong for expert technical assistance and D. H. Yu for help with manuscript preparation.
This work was funded by research grants from 973 Programme of China (2009CB522501, 2009CB522502, and 2009CB522503), Important National Science and Technology Special Projects for Prevention and Treatment of Major Infectious Diseases (2008ZX10002-013), Natural Science Foundation of China (30671921, 30921006, and 30770094), New Drugs Creation Special Projects of China (2009ZX09103-688), and the Shanghai LAD Project (B901).
For all authors, there is no conflict of interest to disclose.
Footnotes
Published ahead of print on 22 December 2010.
REFERENCES
- 1.Amako, Y., et al. 2010. Pathogenesis of hepatitis C virus infection in Tupaia belangeri. J. Virol. 84:303-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth, H., et al. 2005. Scavenger receptor class B type I and hepatitis C virus infection of primary tupaia hepatocytes. J. Virol. 79:5774-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burlone, M. E., and A. Budkowska. 2009. Hepatitis C virus cell entry: role of lipoproteins and cellular receptors. J. Gen. Virol. 90:1055-1070. [DOI] [PubMed] [Google Scholar]
- 6.Cao, J., E. B. Yang, J. J. Su, Y. Li, and P. Chow. 2003. The tree shrews: adjuncts and alternatives to primates as models for biomedical research. J. Med. Primatol. 32:123-130. [DOI] [PubMed] [Google Scholar]
- 7.Catanese, M. T., et al. 2010. Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants. J. Virol. 84:34-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cukierman, L., L. Meertens, C. Bertaux, F. Kajumo, and T. Dragic. 2009. Residues in a highly conserved claudin-1 motif are required for hepatitis C virus entry and mediate the formation of cell-cell contacts. J. Virol. 83:5477-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darai, G., A. Rosen, J. Scholz, and H. Gelderblom. 1983. Induction of generalized and lethal herpesvirus infection in the tree shrew by intrahepatic transfection of herpes simplex virus DNA. J. Virol. Methods 7:305-314. [DOI] [PubMed] [Google Scholar]
- 10.Drummer, H. E., K. A. Wilson, and P. Poumbourios. 2002. Identification of the hepatitis C virus E2 glycoprotein binding site on the large extracellular loop of CD81. J. Virol. 76:11143-11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubuisson, J. 2000. Folding, assembly and subcellular localization of hepatitis C virus glycoproteins. Curr. Top. Microbiol. Immunol. 242:135-148. [DOI] [PubMed] [Google Scholar]
- 12.Evans, M. J., et al. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801-805. [DOI] [PubMed] [Google Scholar]
- 13.Flint, M., et al. 2000. Functional characterization of intracellular and secreted forms of a truncated hepatitis C virus E2 glycoprotein. J. Virol. 74:702-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flint, M., et al. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournier, C., et al. 1998. In vitro infection of adult normal human hepatocytes in primary culture by hepatitis C virus. J. Gen. Virol. 79(10):2367-2374. [DOI] [PubMed] [Google Scholar]
- 16.Galun, E., et al. 1995. Hepatitis C virus viremia in SCID→BNX mouse chimera. J. Infect. Dis. 172:25-30. [DOI] [PubMed] [Google Scholar]
- 17.Grove, J., et al. 2007. Scavenger receptor BI and BII expression levels modulate hepatitis C virus infectivity. J. Virol. 81:3162-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guitart, A., et al. 2005. Hepatitis C virus infection of primary tupaia hepatocytes leads to selection of quasispecies variants, induction of interferon-stimulated genes and NF-kappaB nuclear translocation. J. Gen. Virol. 86:3065-3074. [DOI] [PubMed] [Google Scholar]
- 19.Helle, F., and J. Dubuisson. 2008. Hepatitis C virus entry into host cells. Cell. Mol. Life Sci. 65:100-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu, M., et al. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. U. S. A. 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia, Z. S., et al. 2008. Scavenger receptor class B type I mediates cell entry of hepatitis C virus. J. Int. Med. Res. 36:1319-1325. [DOI] [PubMed] [Google Scholar]
- 22.Keam, S. J., and R. S. Cvetkovic. 2008. Peginterferon-alpha-2a (40 kD) plus ribavirin: a review of its use in the management of chronic hepatitis C mono-infection. Drugs 68:1273-1317. [DOI] [PubMed] [Google Scholar]
- 23.Köck, J., et al. 2001. Efficient infection of primary tupaia hepatocytes with purified human and woolly monkey hepatitis B virus. J. Virol. 75:5084-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindenbach, B. D., et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 25.Mercer, D. F., et al. 2001. Hepatitis C virus replication in mice with chimeric human livers. Nat. Med. 7:927-933. [DOI] [PubMed] [Google Scholar]
- 26.Molina, S., et al. 2008. Serum-derived hepatitis C virus infection of primary human hepatocytes is tetraspanin CD81 dependent. J. Virol. 82:569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Op De Beeck, A., L. Cocquerel, and J. Dubuisson. 2001. Biogenesis of hepatitis C virus envelope glycoproteins. J. Gen. Virol. 82:2589-2595. [DOI] [PubMed] [Google Scholar]
- 28.Pawlotsky, J. M. 2006. Therapy of hepatitis C: from empiricism to eradication. Hepatology 43:S207-S220. [DOI] [PubMed] [Google Scholar]
- 29.Pileri, P., et al. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 30.Ploss, A., et al. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457:882-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scarselli, E., et al. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seeff, L. B. 2002. Natural history of chronic hepatitis C. Hepatology 36:S35-S46. [DOI] [PubMed] [Google Scholar]
- 33.Shepard, C. W., L. Finelli, and M. J. Alter. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558-567. [DOI] [PubMed] [Google Scholar]
- 34.Sumpter, R., Jr., et al. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian, Z. F., et al. 2009. Interaction of hepatitis C virus envelope glycoprotein E2 with the large extracellular loop of tupaia CD81. World J. Gastroenterol. 15:240-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Hahn, T., and C. M. Rice. 2008. Hepatitis C virus entry. J. Biol. Chem. 283:3689-3693. [DOI] [PubMed] [Google Scholar]
- 37.Wakita, T., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter, E., R. Keist, B. Niederost, I. Pult, and H. E. Blum. 1996. Hepatitis B virus infection of tupaia hepatocytes in vitro and in vivo. Hepatology 24:1-5. [DOI] [PubMed] [Google Scholar]
- 39.Wu, G. Y., et al. 2005. A novel immunocompetent rat model of HCV infection and hepatitis. Gastroenterology 128:1416-1423. [DOI] [PubMed] [Google Scholar]
- 40.Xie, Z. C., et al. 1998. Transmission of hepatitis C virus infection to tree shrews. Virology 244:513-520. [DOI] [PubMed] [Google Scholar]
- 41.Yoneyama, M., et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 42.Zeisel, M. B., et al. 2007. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology 46:1722-1731. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, J., et al. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao, X., et al. 2002. Primary hepatocytes of Tupaia belangeri as a potential model for hepatitis C virus infection. J. Clin. Invest. 109:221-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong, J., et al. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]