Abstract
Production of type I interferons (IFNs; prominently, IFN-α/β) following virus infection is a pivotal antiviral innate immune response in higher vertebrates. The synthesis of IFN-β proceeds via the virus-induced assembly of the transcription factors IRF-3/7, ATF-2/c-Jun, and NF-κB on the ifnβ promoter. Surprisingly, recent data indicate that the NF-κB subunit RelA is not essential for virus-stimulated ifnβ expression. Here, we show that RelA instead sustains autocrine IFN-β signaling prior to infection. In the absence of RelA, virus infection results in significantly delayed ifnβ induction and consequently defective secondary antiviral gene expression. While RelA is not required for ifnβ expression after infection, it is nonetheless essential for fully one-fourth of double-stranded RNA (dsRNA)-activated genes, including several mediators of inflammation and immune cell recruitment. Further, RelA directly regulates a small subset of interferon-stimulated genes (ISGs). Finally, RelA also protects cells from dsRNA-triggered RIP1-dependent programmed necrosis. Taken together, our findings suggest distinct roles for RelA in antiviral innate immunity: RelA maintains autocrine IFN-β signaling in uninfected cells, facilitates inflammatory and adaptive immune responses following infection, and promotes infected-cell survival during this process.
Mammalian type I interferons (IFNs) are a family of powerful antiviral cytokines comprising primarily of a single IFN-β species and several IFN-α subtypes. All type I IFNs signal through the heterodimeric IFN-α receptor (IFNAR). Downstream of IFNAR, the Janus kinases Jak1 and Tyk2 are activated, resulting in phosphorylation of latent cytoplasmic signal transducers and activators of transcription 1 and 2 (STAT1 and STAT2) (46, 55). STAT1 and STAT2 translocate to the nucleus and, together with IRF-9, form a transcription factor called interferon-stimulated gene factor-3 (ISGF-3). ISGF-3 drives expression of genes containing interferon-stimulated response elements (ISREs) in their promoters, and over 400 genes are induced in this manner. Interferon-stimulated genes (ISGs) encode a diverse range of proteins that establish an antiviral state and modulate several aspects of the adaptive immune response (46).
Uninfected cells constitutively produce low levels of IFN-β, which acts in an autocrine manner to maintain basal expression of ISGs. Since several ISGs are themselves components of type I IFN production and signaling, it is thought that IFN-β autocrine signaling maintains a primed state of readiness that allows the cell to rapidly respond to an infection. Indeed, when IFN-β signaling components (such as IFNAR and STAT1) are ablated, basal expression levels of several ISGs collapse (50).
The production of type I IFNs (particularly IFN-β) occurs within minutes of virus entry and represents the host cell's primary innate response to many acute virus infections. In most cell types, the production of IFN-β is initiated by a pair of homologous RNA helicases, RIG-I and MDA-5, together called RIG-I-like receptors (RLRs) (66). RLRs are latent cytosolic proteins that detect virus genomes and/or replication intermediates to trigger signaling cascades that culminate in the rapid activation of at least three transcription factors: IRF-3/7, ATF-2/c-Jun, and NF-κB (1, 30). The promoter of the ifnβ gene contains four positive regulatory domains (PRDs I to IV) that serve as binding sites for these transcription factors as follows: IRF-3/7 binds to PRD I/III, ATF-2/c-Jun dimers interact with PRD IV, and NF-κB associates with PRD II (33).
In a long-standing model stemming from classic work by Thanos and Maniatis, virus recognition by RLRs is proposed to result in the coordinated assembly of IRFs, ATF-2/c-Jun, and NF-κB on the ifnβ promoter (52, 53). Together with other chromatin-remodeling proteins and coactivators, these transcription factors form an “enhanceosome” that drives robust expression of the ifnβ gene to start the interferon response (33, 52, 53). Subsequent studies using genetically deficient mice have since confirmed that IRF-3 and IRF-7 are critically required for IFN-β production after acute RNA virus infection (25, 43).
Surprisingly, more recent studies have found that NF-κB is almost completely dispensable for IFN-β production following RLR activation (38, 59). NF-κB refers to a family of homo- and heterodimeric transcription factors formed by combinations of five proteins: RelA, c-Rel, RelB, p50, and p52 (21). Virus infection results in the activation of primarily p50-RelA heterodimers, and studies using cells from p50−/−, rela−/−, and p50−/− rela−/− doubly deficient mice showed that IFN-β production proceeded relatively normally in the absence of these subunits (59). These findings raise two questions: (i) what is the role of NF-κB in regulating ifnβ gene expression and (ii) what is the function of virus-induced NF-κB, if not to activate ifnβ?
Our previous work showed that the NF-κB subunit RelA, while not essential for virus-stimulated ifnβ expression per se, is nonetheless required very early in the course of an infection for timely expression of key ISGs (57). rela−/− cells reproducibly show a significant early delay in the induction of ifnβ, but IFN-β production is restored to normal levels later in the infection (57). In the present study, we traced the defect in rela−/− cells to a stage prior to infection and showed that RelA is required for maintenance of autocrine IFN-β signaling. In the absence of RelA, constitutive IFN-β and ISG levels in uninfected cells are significantly reduced, and these cells therefore fail to mount an effective antiviral response early in infection. After infection, IRFs appear to take over IFN-β production, and RelA is instead required for the induction of proinflammatory gene expression and direct regulation of a subset of ISGs. Finally, we showed that RelA protects cells from double-stranded RNA (dsRNA)/IFN-induced necroptotic cell death. Together, these studies suggest a novel function for the NF-κB site in the ifnβ promoter and identify distinct roles for NF-κB RelA during the course of the innate antiviral response.
MATERIALS AND METHODS
Cells, viruses, and reagents.
Primary rela−/− and rela−/− ifnar−/− murine embryo fibroblasts (MEFs) were prepared from day 14.5 embryos generated from timed crosses of rela+/− and rela+/− ifnar−/− mice, respectively, as previously described (57, 58). ikkβ−/− MEFs were similarly derived from day 12.5 embryos of ikkβ+/− crosses. All experiments using these MEFs were performed on primary early passage (P < 6) cultures generated from at least four independent embryos per genotype, with littermate-derived wild-type MEFs as controls. myd88−/− trif−/− double-knockout MEFs were obtained from S. Akira (Osaka University, Japan). tradd−/− MEFs were provided by M. Pasparakis (University of Cologne, Germany) and T. Mak (University of Toronto, Canada). stat1−/− MEFs were obtained from L. Sigal (Fox Chase Cancer Center). All mice were on a C57BL/6 background. HeLa cells were purchased from the ATCC (Manassas, VA). Vesicular stomatitis virus (VSV) strains VSV-AV1 and VSV-GFP have been described before (13, 47). We used the following cytokines and chemicals: murine IFN-β (Pestka Biomedical Laboratories), necrostatin-1 (nec-1; Enzo Life Sciences), and z-VAD-fmk (Calbiochem). Poly(I:C) (Invivogen) was reconstituted in phosphate-buffered saline (PBS) at 2 mg/ml, denatured at 55°C for 30 min, and allowed to anneal to room temperature before use. Antibodies to RelA, P50, P52, c-Rel, and RelB were purchased from Santa Cruz Biotechnology. All other reagents were from Sigma-Aldrich, unless otherwise mentioned. All cells were cultured in high-glucose Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and antibiotics.
EMSA.
MEFs (1 × 106/condition) were washed twice with cold PBS and suspended in 400 μl hypotonic buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail [Roche]). After incubation for 15 min on ice, 25 μl 10% Nonidet P-40 was added for a further 10 min and nuclei were collected by centrifugation at 3,000 × g for 5 min at 4°C. Nuclear pellets were subsequently lysed in 30 μl buffer B (20 mM HEPES, pH 7.9, 400 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 5% glycerol, protease inhibitor cocktail) and incubated for 1 h at 4°C with brief intermittent mixing. Nuclear lysates were clarified by centrifugation at 10,000 × g for 5 min at 4°C. An oligonucleotide corresponding to two tandem copies of the PRD II element (5′-GGGAAATTCCGGGAAATTCC-3′, custom synthesized at the Fox Chase DNA Core Facility) was end labeled with [γ-32P]ATP by using T4 kinase (Promega). Ten micrograms of nuclear protein was incubated with radiolabeled PRD II oligonucleotide in electrophoretic mobility shift assay (EMSA) reaction buffer [2 μg poly(dI-dC), 12% glycerol, 20 mM HEPES, pH 7.0, 1 mM EDTA, 50 mM NaCl] for 30 min at room temperature and subjected to 5% nondenaturing PAGE in 0.5× Tris-borate-EDTA (TBE) buffer. For antibody supershift experiments, antibodies (200 ng) were added to nuclear extracts 15 min prior to the addition of radiolabeled oligonucleotide. Gels were vacuum dried and subjected to autoradiography.
DNA microarray expression analysis.
MEFs (5 ×106/condition) were stimulated in duplicate with poly(I:C) (6 μg/ml in Lipofectamine 2000) or murine IFN-β (200 U/ml). Total RNA was isolated in TRIzol reagent (Invitrogen, Carlsbad, CA) and purified using the RNeasy kit (Qiagen). Five hundred nanograms of total RNA was amplified and labeled using the Agilent low-RNA-input linear amplification kit. Labeled cRNA targets were hybridized onto Agilent 4 × 44k (4 arrays/chip and ∼44,000 transcripts/array) mouse whole-genome arrays. Microarray images were processed using Agilent Feature Extraction software (version 9.5). Identification of differentially expressed genes was performed with the LIMMA package (45) implemented in the R/Bioconductor platform (14). Biological pathways and networks were examined by Ingenuity Pathways Analysis (IPA) software.
Real-time PCR.
Total RNA extracted in TRIzol and purified using the Qiagen RNeasy kit was reverse transcribed into cDNA (Applied Biosystems). cDNAs were then subjected to real-time PCR analysis in an Applied Biosystems 7900HT sequence detection system using SYBR green I dye. All samples were run in triplicate. Results for target genes are presented after normalization to β-actin mRNA levels. PCR primers used have been described before (59).
RNA interference (RNAi).
HeLa cells (6 × 104/well) seeded into six-well dishes were transfected with pools of four distinct proprietary small interfering RNAs (siRNAs) (SMARTpool; Dharmacon) to each target mRNA at 20 nM using Oligofectamine (Invitrogen) as a transfection reagent. As controls, nontargeting siRNA duplexes (Dharmacon) were employed. Cells were used in experiments 72 h posttransfection.
Statistical analysis.
Student's t test was used for comparison between two groups, and P values of <0.05 were considered significant.
RESULTS
RelA controls constitutive expression of IFN-β and ISGs.
Since previous work from one of our laboratories showed that the NF-κB RelA subunit was not essential for virus-triggered production of IFN-β (59), we decided to take an unbiased DNA microarray-based approach to identify its role in antiviral innate immunity.
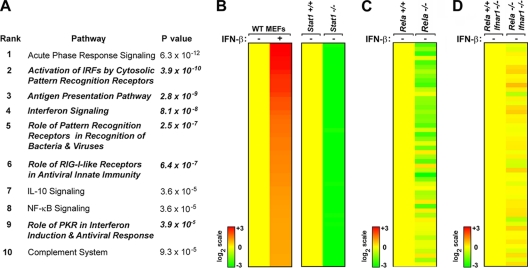
Genes whose basal expression level was downregulated ≥2-fold in rela−/− MEFs relative to that in rela+/+ MEFs were parsed by IPA software, and canonical biological pathways that were most significantly disrupted in the absence of RelA were identified. As expected, known NF-κB-regulated pathways (such as the acute-phase response) were found to be dependent on RelA (Fig. 1A). Remarkably, 6 of the top 10 most-disrupted pathways in rela−/− MEFs were linked to the type I IFN-dependent antiviral response (Fig. 1A, boldface/italic terms; see Table S1 in the supplemental material for list of genes in each canonical pathway). While it was possible that RelA controlled basal levels of each of these IFN-dependent pathway components independently of each other, we considered it more likely that RelA instead regulated constitutive expression of the ifnβ gene itself. Since IFN-β initiates autocrine signaling to support basal expression of ISGs, defective maintenance of ifnβ expression would be expected to collapse IFN-β-dependent autocrine signaling and explain the widespread defects seen in IFN-regulated pathways. Furthermore, this model also provides a function for the NF-κB site in the ifnβ gene and may explain why rela−/− MEFs have early defects in ifnβ induction following virus infection.
FIG. 1.
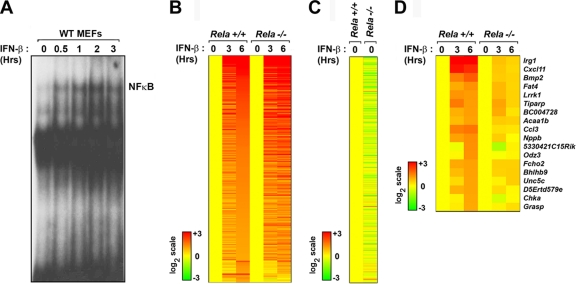
RelA controls basal expression of ISGs via regulation of autocrine IFN-β. (A) List of the top 10 most-disrupted (downregulated) canonical pathways in unstimulated rela−/− MEFs as generated by Ingenuity Pathways Analysis. Fisher's exact test was used to calculate a P value determining the probability that the downregulation of genes in the canonical pathway is explained by chance alone. (B) Wild-type (WT) MEFs were stimulated with IFN-β (200 U/ml for 6 h). All genes induced at a ≥2-fold level were subsequently evaluated for basal expression in stat1+/+ and stat1−/− MEFs. The top 50 genes that were most highly induced by IFN-β in WT MEFs (left) while simultaneously displaying the lowest basal levels in stat1−/− MEFs (right) were defined as the autocrine IFN-β signature. Expression in stat1+/+ MEFs was normalized to one (20; yellow). (C) Analysis of the autocrine IFN-β signature in unstimulated rela+/+ (normalized to yellow) and rela−/− MEFs. (D) Analysis of the autocrine IFN-β signature in unstimulated rela+/+ (normalized to yellow) and rela−/− MEFs on an ifnar1−/− background. Heat bars on the left represent relative expression levels on a log2 scale.
To determine whether RelA controls basal IFN-β production and consequent maintenance of ISGs, we first established that basal expression of ifnβ itself was reduced (by 1.95-fold; P < 5 × 10−8) in rela−/− MEFs compared to the amount in controls. Constitutive IFN-β is, however, produced in very small amounts and often cannot be reliably detected by direct means (17, 19, 50). We therefore undertook an indirect approach to evaluate basal IFN-β levels in a two-step process. First, we treated wild-type MEFs with recombinant IFN-β to identify all ISGs. A total of 461 genes were induced at a ≥2-fold level after 6 h of IFN-β treatment. Next, we examined the basal expression levels of these 461 genes in early-passage stat1+/+ and stat1−/− MEFs, since STAT1 is essential for IFN-β signaling (including autocrine maintenance of ISGs). Basal levels of ∼25% (119/461) of these genes were found to be reduced by ≥3-fold in stat1−/− MEFs compared to the levels for controls. Of these stringently selected genes, the expression patterns of 50 genes whose basal levels were most disrupted in the absence of STAT1 are shown in Fig. 1B (right panel). We defined these genes as the autocrine IFN-β signature (Table S2 in the supplemental material) and used their expression as a surrogate readout for the existence of autocrine IFN-β itself.
Next, we evaluated the behavior of the autocrine IFN-β signature in unstimulated rela+/+ and rela−/− MEFs. Of the 50 genes in this signature, 32 were expressed at significantly lower basal levels (<2-fold) in rela−/− MEFs than in rela+/+ MEFs (Fig. 1C). Importantly, when a similar analysis was performed on rela+/+ and rela−/− cells on an ifnar1−/− background, no such differences in the autocrine IFN-β signature were seen, and only two genes were basally expressed at <2-fold levels in rela−/− ifnar1−/− MEFs, compared to rela+/+ ifnar1−/− MEFs (Fig. 1D), demonstrating that the observed effects of RelA loss on ISG expression shown in Fig. 1C were directly attributable to type I IFN. Together, these results strongly suggest that RelA controls ifnβ expression to drive autocrine IFN-β signaling in unstimulated cells.
Key nodes of the IFN system show defective basal expression in rela−/− MEFs.
Autocrine IFN-β maintains a low level of ISG activity that is thought to allow a rapid antiviral response immediately after infection (50). This state of readiness hinges on the fact that several genes involved in the IFN response are themselves ISGs (50). Since our DNA microarray data now indicate that RelA may control constitutive autocrine IFN-β signaling, we reasoned that the delay in ifnβ induction seen after virus infection in these cells (57) may be due to reduced basal levels of key ISGs involved in the virus-induced production of IFN-β itself.
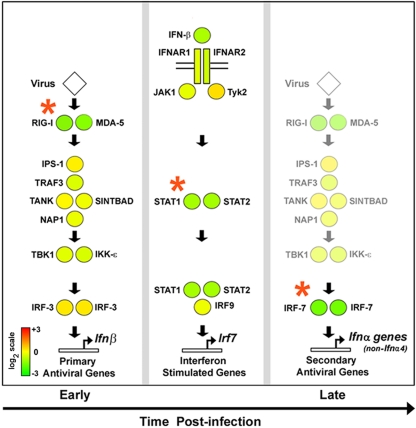
In a current model, the type I IFN system in most cells is activated in three overlapping steps. First, RLRs recognize incoming virus and induce IFN-β and other “primary” antiviral genes. Next, secreted IFN-β signals in an autocrine and paracrine manner to activate expression of ISGs, including that encoding IRF-7. Finally, IRF-7 in the infected cell contributes toward transactivating “secondary” antiviral genes (as well as increasing ifnβ expression, in a positive feedback loop) (1, 30, 49). Given the amplificatory nature of type I IFN signaling, we hypothesized that virus-induced production of IFNs would be particularly sensitive to basal levels of pivotal ISGs that directly dictate the magnitude and kinetics of the innate response. For the purposes of this analysis, we defined three such nodes that represent potential autocrine IFN-β-dependent bottlenecks in the type I IFN system. The molecules comprising these nodes were selected based on whether they are (i) known ISGs and (ii) nonredundant in antiviral response, as determined from studies in genetically deficient mice. The three nodes selected, in rough kinetic order of involvement, were (i) the RLRs RIG-I and MDA-5, (ii) STAT1 and STAT2, and (iii) IRF-7 (11, 25, 27, 29, 34, 37). As shown in Fig. 2, we found that all three nodes (represented by red asterisks) were significantly deficient (<2-fold) in basal expression in rela−/− MEFs. These data support the idea that rela−/− MEFs are delayed in virus-triggered induction of ifnβ as a result of reduced levels of key nodes involved in detection of virus and timely augmentation of IFN production.
FIG. 2.
Key nodes of the RLR antiviral response show defective basal expression in rela−/− MEFs. Circles representing key molecules involved in the RLR-triggered antiviral response were filled in with heat map color values representing basal expression levels in rela−/− MEFs, compared to rela+/+ MEFs. Basal expression in rela+/+ MEFs was normalized to one (20; yellow). The heat bar on the left represents relative expression levels on a log2 scale. Red asterisks indicate that nodes were significantly deficient (<2-fold) in their basal expression in rela−/− MEFs.
rela−/− MEFs show delayed IFN and ISG expression and increased susceptibility to virus infection.
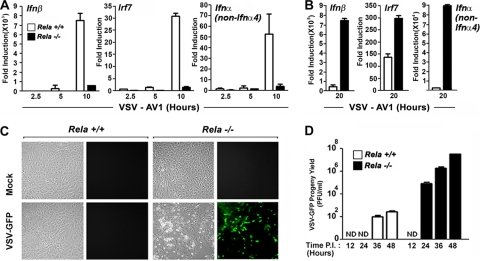
We next directly tested the functional consequences of basally defective ISG expression in rela−/− MEFs on their ability to mount an effective antiviral response. We infected rela+/+ and rela−/− MEFs with VSV and monitored induction of genes representative of the three stages of the antiviral response: primary, IFN induced, and secondary. For these experiments, we used a matrix protein mutant of VSV (VSV-AV1) that cannot prevent ifnβ mRNA export from the nucleus and therefore cannot inhibit the type I IFN response like wild-type VSV does (12, 47). As before, we found that induction of ifnβ was significantly delayed in rela−/− MEFs following VSV-AV1 infection at early time points (Fig. 3A) (57). Eventually, levels of ifnβ mRNA in these cells reached (and even greatly exceeded) those seen in rela+/+ cells, presumably because of increased RLR-mediated IRF-3/7 activation resulting from enhanced virus replication in rela−/− MEFs at later time points (Fig. 3B). Similarly, levels of irf7 mRNA and IRF-7-dependent secondary ifnα genes were also poorly induced in rela−/− MEFs early, but not later, in infection (Fig. 3A and B). Importantly, when green fluorescent protein (GFP) fluorescence and progeny virion output were evaluated in rela−/− MEFs infected with VSV-GFP at a multiplicity of infection (MOI) of 0.001, these cells were found to be very permissive to infection (Fig. 3C and D). Within 24 h postinfection, rela−/− MEFs produced ∼1 ×105 PFU/ml VSV-GFP, and they eventually generated between 1 × 107 and 1 × 108 PFU/ml VSV-GFP by 48 h postinfection. In contrast, rela+/+ MEFs remained mostly resistant to VSV-GFP and consistently restricted the virus yield to <1 × 103 PFU/ml after 48 h (Fig. 3D). These data demonstrate that impaired autocrine IFN-β signaling and basal ISG expression functionally translate into severe defects in the kinetics of the type I IFN response and consequent susceptibility to virus. Importantly, these findings also highlight the biological importance of autocrine IFN-β signaling during antiviral responses.
FIG. 3.
rela−/− MEFs show impaired early induction of ifnβ and IFN-β-dependent genes and are susceptible to virus. (A) rela+/+ and rela−/− MEFs were infected with VSV-AV1 (multiplicity of infection [MOI] = 10), and expression of ifnβ, irf7, and ifnα (non-ifnα4) genes was measured by real-time PCR at the indicated times postinfection. (B) rela+/+ and rela−/− MEFs infected with VSV-AV1 as described for panel A were evaluated for ifnβ, irf7, and ifnα (non-ifnα4) gene expression 20 h postinfection by real-time PCR. (C) rela+/+ and rela−/− MEFs were infected with VSV-GFP (MOI = 0.001) and photographed 24 h postinfection. (D) rela+/+ and rela−/− MEFs were infected with VSV-GFP (MOI = 0.001). Progeny virion yield in the supernatants was measured at the indicated times postinfection (P.I.) by standard plaque assay on BHK-21 cells. All experiments described for this figure were repeated at least three times with similar results. ND, not detected.
RelA cycles robustly in an I-κB kinase beta (IKK-β)-dependent manner in uninfected MEFs.
We next examined the mechanism by which RelA controls autocrine IFN-β signaling. In most primary cells, NF-κB is classically thought to remain latent in the cytoplasm through its interaction with I-κB proteins (21). Indeed, primary MEF cultures in general show very low basal NF-κB activity (when measured at discrete time points by techniques such as reporter assays or EMSA). To explain how RelA controls autocrine IFN-β production in unstimulated MEFs, however, we considered the possibility that the NF-κB activity in these cells may be more dynamic than previously believed. In one scenario, a small amount of NF-κB may respond to unknown autocrine stimuli in resting cells to shuttle to the nucleus and activate target genes. Since the I-κBs are themselves NF-κB targets (21), their rapid induction then would retain a quantum of NF-κB dimers in the cytoplasm, preventing nuclear accumulation of NF-κB. Indeed, ablation of I-κB protein expression results in constitutive NF-κB activation in unstimulated cells (5, 51). In such a scenario, the resultant dynamic equilibrium between nuclear and cytoplasmic NF-κB would ensure homeostatic control of basal NF-κB signaling but may often go undetected in single-time-point measurements of NF-κB activity.
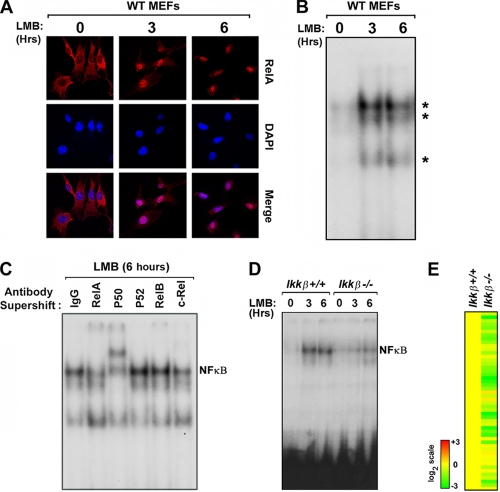
To determine whether NF-κB cycles between the cytoplasm and the nucleus in unstimulated cells, we therefore sought to examine NF-κB nuclear accumulation under conditions in which I-κB-dependent repression is alleviated. Reasoning that prevention of nuclear export would trap newly synthesized I-κB in the nucleus and allow accrual of active nuclear NF-κB, we incubated primary MEFs with the nuclear export inhibitor leptomycin B (LMB) and examined localization of RelA in these cells over a time course of 6 h. As depicted in Fig. 4A, RelA showed a predominantly cytoplasmic distribution in untreated cells but rapidly accumulated in the nucleus within 3 h of LMB treatment. By 6 h following the addition of LMB, virtually all RelA was nuclear (Fig. 4A). To test whether nuclear RelA trapped by LMB can interact with the PRD II element of the ifnβ promoter, we treated MEFs with LMB and incubated nuclear extracts prepared from these cells with a radiolabeled PRD II probe. EMSA performed on these extracts showed formation of at least three LMB-dependent complexes within 3 h of nuclear export inhibition (Fig. 4B). Subsequent antibody supershift experiments revealed that the primary complex is predominantly composed of p50-RelA dimers (Fig. 4C). Together, these results demonstrated that RelA cycles robustly through the nuclei of unstimulated cells, where it can interact with PRD II to drive basal transcription of ifnβ. In agreement, the IFN-β-dependent irf7 transcript accumulated upon LMB treatment in a RelA-dependent manner (see Fig. S2 in the supplemental material).
FIG. 4.
RelA shuttles through the nucleus of unstimulated cells to control autocrine IFN-β via IKK-β. (A) Wild-type MEFs were treated with the nuclear export inhibitor leptomycin B (LMB, 20nM) for 3 or 6 h and stained with antibodies to RelA. Cells were subsequently examined for RelA localization by confocal immunofluorescence microscopy. Nuclei were detected with DAPI (4′,6-diamidino-2-phenylindole). (B) Nuclear extracts from wild-type MEFs treated with LMB for 3 or 6 h were subjected to EMSA using a radiolabeled PRD II probe. (C) Nuclear extracts from wild-type MEFs treated with LMB for 6 h were preincubated with the indicated anti-NF-κB subunit antibodies. Supershift of NF-κB complexes was examined by EMSA. (D) Nuclear extracts from ikkβ+/+ and ikkβ−/− MEFs treated with LMB for three or 6 h were subjected to EMSA using a radiolabeled PRD II probe. Note that all LMB experiments were performed in serum-free medium to exclude nonspecific NF-κB activation by serum components. Experiments whose results are shown in panels A to D were repeated at least three times with similar results. (E) Basal expression of the autocrine IFN-β signature was evaluated in unstimulated ikkβ+/+ and and ikkβ−/− MEFs. Basal expression of the autocrine IFN-β signature in ikkβ+/+ MEFs was normalized to one (20; yellow). The heat bar on the left represents relative expression levels on a log2 scale.
Canonical (p50-RelA) NF-κB dimers are typically activated when IKKs, particularly IKK-β, target I-κB proteins for degradation by phosphorylation on key serines (21). We next asked whether IKK-β acts upstream of RelA to license constitutive NF-κB activity in unstimulated cells. We treated primary early-passage ikkβ+/+ and ikkβ−/− MEFs with LMB for up to 6 h and measured NF-κB binding to PRD II by EMSA. As shown in Fig. 4D, we observed strong NF-κB-PRD II complex formation in ikkβ+/+ MEFs within 3 h of LMB treatment. In ikkβ−/− MEFs, however, formation of this complex was not seen until 6 h posttreatment, and was severely reduced in magnitude (>80% at 3 h, as quantified by Image J software) (Fig. 4D). In agreement with a role for IKK-β in maintaining constitutive NF-κB activity, basal expression of the autocrine IFN-β ISG signature was also defective in ikkβ−/− MEFs (Fig. 4E). It is at present unclear how IKK-β is activated to support constitutive NF-κB activity (see Discussion). Collectively, these findings suggest that an IKK-β-NF-κB axis responds to unknown upstream signals to sustain autocrine IFN-β production.
RelA mediates inflammatory gene expression during RLR signaling.
RelA is among the first ifnβ enhanceosome components activated after virus infection (32). Later in the course of the RLR antiviral response, however, RelA is not required to drive ifnβ expression but nonetheless remains activated at least 12 h postinfection (59). These findings suggest that RelA may switch from controlling ifnβ early in an infection to regulating other RLR-responsive genes at later time points.
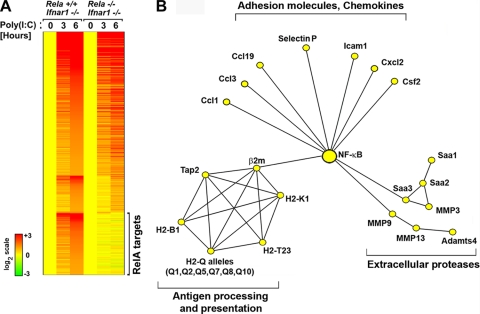
To identify these genes, we performed DNA microarray analyses on rela+/+ and rela−/− MEFs transfected with the dsRNA viral mimetic poly(I:C). Transfected poly(I:C) specifically activates RLRs in MEFs (28) and was used in these experiments instead of virus to negate gene induction biases caused by the increased permissiveness of rela−/− MEFs to virus. These experiments were done on an ifnar1−/− background to identify unique primary RelA targets in the absence of IFN-β-mediated secondary effects that may confound interpretation of results. For the purposes of these analyses, all genes induced ≥2-fold by transfected poly(I:C) in rela+/+ ifnar1−/− MEFs within 6 h were considered primary RLR-induced genes. A total of 596 genes were induced in this manner. Of these, 353 genes (including ifnβ; see Discussion) were induced to similar extents in the presence and absence of RelA and 243 were dependent on RelA to some extent for their expression. RelA-dependent genes were defined as those genes whose induction was <1.5-fold at either or both time points. By this criterion, 89 genes were dependent on RelA for full expression at 3 h but were induced normally at 6 h. A further three genes were activated normally at 3 h but showed diminished induction 6 h after RLR activation in the absence of RelA. Together, these genes were classified as partially RelA dependent and clustered accordingly (Fig. 5A). Remarkably, over one-fourth (151 out of 596) of RLR-induced genes were found to be obligate RelA targets at both time points and were categorized as fully RelA dependent (Fig. 5A).
FIG. 5.
Role of RelA in RLR-triggered primary antiviral gene expression. (A) rela+/+ and rela−/− MEFs (both on an ifnar1−/− background to identify only primary antiviral genes) were transfected with poly(I:C) (6 μg/ml) for 3 or 6 h. Total RNA from these cells was subjected to whole-genome microarrays, and RelA target genes were clustered based on whether they were partially or fully RelA dependent. Expression levels in untreated cells of both genotypes were normalized to one (20; yellow). The heat bar on the left represents relative expression levels on a log2 scale. (B) Ingenuity Pathway Analysis was used to identify highly connected networks based on functional and physical interactions from the Ingenuity Pathways Knowledge Base. The network depicted was obtained by merging three highest-scoring overlapping networks and removing genes or gene groups not directly regulated by poly(I:C) as per our microarray data.
Examination of the 151 RelA target genes revealed that a disproportionate number of them encoded molecules involved in aspects of immune cell recruitment and activation (see Fig. S1 in the supplemental material). Subsequent processing by Ingenuity Pathways Analysis of RLR-activated RelA targets identified three significant clusters: (i) chemokines and adhesion molecules, (ii) extracellular proteases, and (iii) modulators of antigen processing and presentation (Fig. 5B). In striking contrast to RelA's role before infection and during the early phase of RLR signaling, less than 5% of obligate RelA targets during the later RLR response (4/151; ifnar2, traf3, ikbke, and tank) were genes known to be involved in maintenance or amplification of the type I interferon response itself (see Fig. S1 in the supplemental material). Together, these findings demonstrate that RelA primarily controls the proinflammatory axis of RLR signaling to drive immune cell recruitment and activate adaptive immunity after infection.
RelA is required for direct induction of a small subset of ISGs.
In addition to regulating gene expression downstream of RLR activation, NF-κB has also been proposed to directly function in type I IFN signaling itself (10, 55). Previous studies have shown cell-specific activation of RelA-containing NF-κB complexes following stimulation with type I IFNs (62), but the transcriptional profile of RelA target ISGs remains unknown. To address this issue, we first confirmed that IFN-β activates NF-κB in MEFs (Fig. 6A). We then treated early-passage rela+/+ and rela−/− MEFs with recombinant murine IFN-β and subjected RNA from these cells to whole-genome microarray analyses. A total of 461 genes were induced ≥2-fold in rela+/+ MEFs following 6 h of treatment with IFN-β (Fig. 6B). Reassuringly (since our data thus far indicate that RelA controls autocrine IFN-β signaling), ∼75% of all ISGs were expressed at lower basal levels in rela−/− MEFs (Fig. 6C). After expression profiles were normalized to account for these basal differences, only 18 genes were found to be critically dependent on RelA for their induction by IFN-β. Immune response gene 1 (irg1), encoding a putative mitochondrial methylcitrate dehydratase, was identified as the primary RelA-dependent ISG. Also found to be RelA dependent were genes encoding the chemokines Cxcl11 and Ccl3 (Fig. 6D). Together, these data show that RelA is activated by IFN-β to induce a small subset of ISGs.
FIG. 6.
RelA directly controls expression of a small subset of ISGs. (A) Wild-type MEFs were treated with IFN-β (200 U/ml) for the indicated times, and nuclear extracts from these cells was examined by EMSA using a radiolabeled consensus NF-κB probe (Santa Cruz). This experiment was repeated at least three times with similar results. (B) rela+/+ and rela−/− MEFs were treated with IFN-β (200 U/ml) for 3 or 6 h. RNA extracted from these cells was subjected to whole-genome microarrays, and RelA target genes were clustered based on whether they were partially or fully RelA dependent. Expression levels in untreated cells of both genotypes were normalized to one (20; yellow). (C) Basal gene expression profiles of all ISGs in rela+/+ and rela−/− MEFs. Expression levels in untreated rela+/+ MEFs were normalized to one (20; yellow). (D) Expression profiles of fully RelA-dependent ISGs. Heat bars on the left represent relative expression levels on a log2 scale.
RelA protects cells from dsRNA-activated RIP1-dependent necroptosis.
During the course of the analyses described above, we noticed that rela−/− MEFs were more susceptible than rela+/+ MEFs to cell death following transfection of poly(I:C) (data not shown). Indeed, rela−/− MEFs also succumbed to poly(I:C) added directly to the medium, in the absence of any transfection reagent, while control rela+/+ MEFs were mostly resistant (Fig. 7A). Extracellular poly(I:C) can be taken up via class A scavenger receptors and endocytosis to stimulate RLRs and Toll-like receptor 3 (TLR3) in fibroblasts (9). At the highest dose of poly(I:C) used, ∼70% of rela−/− MEFs succumbed within 48 h of treatment, whereas rela+/+ cells were almost completely viable at this time (Fig. 7A). Interestingly, rela−/− MEFs on an ifnar1−/− background were partially resistant to poly(I:C)-triggered cell death when challenged at the same doses (Fig. 7A), but IFN-β by itself was largely nontoxic to rela−/− MEFs (see Fig. S3 in the supplemental material), suggesting that IFN-β functions to augment poly(I:C) toxicity. These results together indicate that RelA serves to protect cells during RLR and TLR responses.
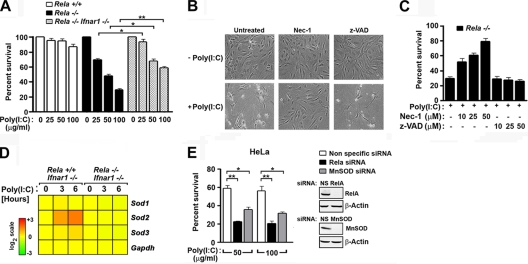
FIG. 7.
RelA protects cells from dsRNA-activated RIP1-dependent necroptosis. (A) rela+/+, rela−/−, and rela−/− ifnar1−/− MEFs were treated with poly (I:C) (0 to 100 μg/ml added directly to the medium) for 48 h, and viability was determined by trypan blue exclusion analysis. (B) rela−/− MEFs were treated with 100 μg/ml poly(I:C) in the presence of nec-1 (50 μM) or z-VAD (50 μM) and photographed 48 h posttreatment. (C) rela−/− MEFs were treated with 100 μg/ml poly(I:C) for 48 h in the presence of the indicated concentrations of nec-1 or z-VAD, and viability was determined by trypan blue exclusion analysis. Experiments whose results are shown in panels A to C were repeated at least three times with similar results. (D) Poly(I:C) induced expression of all murine sod genes in rela+/+ and rela−/− MEFs on an ifnar−/− background showing specific RelA-dependent induction of sod2. gapdh expression is shown as a control. (E) HeLa cells were transfected with nonspecific (NS), RelA-specific, or MnSOD-specific siRNA. After 72 h, cells were treated with the indicated doses of poly(I:C), and viability was determined 48 h posttreatment by trypan blue exclusion analysis. Knockdown of RelA and MnSOD was confirmed by immunoblotting (inset). *, P < 0.05; **, P < 0.005. This experiment was repeated at least three times with similar results.
Poly(I:C) can trigger apoptosis in susceptible cells by multiple mechanisms, including via the PKR-FADD-caspase 8 and RLR-IRF-3-Bax axes (3, 6, 15, 23). In rela−/− cells, however, poly(I:C) appears to activate a form of necrotic death that is independent of caspases but dependent on generation of reactive oxygen species (ROS) (31). During programmed necrosis (or “necroptosis”), the kinases RIP1 and RIP3 trigger generation of mitochondrial ROS to mediate cell death (7, 26, 56, 67). Further, the combination of IFN-γ and poly(I:C) activates necroptosis in susceptible L929 cells (24). We therefore asked whether poly(I:C) triggered RIP1-dependent necroptosis in the absence of RelA. We treated rela−/− MEFs with poly(I:C) in the presence or absence of the RIP1 kinase inhibitor necrostatin 1 (nec-1) (8) and evaluated cell death 48 h posttreatment. As shown in Fig. 7B and C, pretreatment with nec-1 almost completely inhibited poly(I:C)-triggered cytotoxicity, whereas pretreatment with the pancaspase inhibitor z-VAD had no discernible effect.
Among the 151 genes identified as dsRNA-induced obligate RelA targets was sod2, encoding the mitochondrial antioxidant enzyme manganese superoxide dismutase (MnSOD) (Fig. 7D; see also Fig. S1 in the supplemental material). Since accumulation of mitochondrial ROS appears to be essential for necroptosis (56), and since MnSOD scavenges ROS to protect cells from other stimuli such as tumor necrosis factor alpha (TNF-α) (48), we asked whether RelA protected cells from poly(I:C)-induced cell death by inducing sod2 to quench ROS. Accordingly, we ablated MnSOD expression in HeLa cells by RNAi and examined their ability to survive poly(I:C) treatment. HeLa cells were used in these studies because they are more amenable to RNAi-based experimentation than MEFs (4). We found that loss of RelA greatly sensitized HeLa cells to poly(I:C)-mediated cell death [60% survival with nonspecific siRNA compared to ∼20% survival with RelA siRNA after poly(I:C) treatment (Fig. 7E)]. Importantly, ablation of MnSOD also increased susceptibility to poly(I:C)-triggered cell death (∼2-fold) (Fig. 7E). It is noteworthy that silencing MnSOD expression did not have as profound an effect on cell viability as ablating RelA, suggesting that RelA regulates other cell survival genes in addition to sod2. Collectively, these results indicate that poly(I:C) activates RIP1-dependent necroptosis in rela−/− MEFs and identify sod2 as a potential prosurvival RelA target gene.
DISCUSSION
The production of IFN-β is widely considered to be the primary innate immune response to an acute RNA virus infection in most cells. In a current model, ifnβ is activated downstream of RLRs by the coordinated assembly of three transcription factors, IRF-3/7, NF-κB, and ATF-2/c-Jun (1, 30). Unexpectedly, previous work has shown that NF-κB is largely dispensable for RLR-triggered ifnβ induction (38, 59). In this study, we showed that RelA controls autocrine IFN-β and basal ISG expression prior to infection. In the absence of RelA, there is thus a significant delay in the induction of ifnβ and, consequently, severe defects in the activation of the type I IFN response. These defects are further exacerbated by reduced basal levels of genes representing key nodes of the type I IFN response that are themselves ISGs. Although other transcription factors (especially IRF-3/7) eventually compensate for RelA deficiency later in the course of the infection, the twin requirements for RelA in autocrine IFN-β signaling and early ifnβ induction are nonetheless critical for protection from acute RNA virus infection.
Low levels of constitutive IFN-β signaling have long been recognized as functionally capable of supporting basal ISG expression to maintain the uninfected cell in a primed state of antiviral readiness, but the source of this autocrine IFN-β has been mysterious (50). Taniguchi and colleagues have shown that IRF-3 and IRF-7, while critical for virus-induced IFN-β, are not required for constitutive IFN-β expression (19). Our data suggest that, instead, it is NF-κB which maintains constitutive IFN-β levels to promote autocrine signaling and maintain priming levels of ISGs. Interestingly, a recent study showed that c-Jun may also participate in controlling basal IFN-β levels in uninfected cells (16). Taken together, these findings support the idea that NF-κB and c-Jun sustain autocrine IFN-β, while IRF-3 and IRF-7 instead dominate IFN-β production following infection. To explain how NF-κB maintains autocrine IFN-β levels in the absence of any acute stimulus, we suggest that RelA is not static in unstimulated MEFs but can robustly shuttle through the nucleus in an IKK-β-dependent manner to regulate ifnβ. At present, upstream mechanisms that regulate IKK-β activity to promote RelA cycling are unclear. Our preliminary data from TRADD-deficient and MyD88/TRIF doubly deficient MEFs indicate that neither autocrine TNF-α nor TLR signaling is involved upstream of IKK-β (see Fig. S4 in the supplemental material). Thus, although exogenously supplied TNF-α can activate an autocrine IFN-β signal (65), basal TNF-α appears unable to do so.
Multiple control mechanisms likely ensure that constitutive NF-κB activity is tightly regulated. For example, the genes encoding I-κBs and the ubiquitin-editing enzyme A20 are themselves direct NF-κB targets that are rapidly induced to negatively regulate NF-κB activity (20, 21), and RelA itself has a functional nuclear export sequence that promotes expulsion to the cytoplasm for inhibition by I-κBs (18). The ifnβ promoter also contains a negative regulatory element (NRE) that partially overlaps with PRD II (36). The NRE is bound by an NF-κB repressor protein (NRF) that can directly interact with NF-κB subunits to inhibit their association with PRD II, and acute silencing of NRF triggers constitutive IFN-β production (35). The dynamic equilibrium established by such homeostatic mechanisms thus limits the amount of available nuclear NF-κB and secures low steady-state levels of IFN-β. Although these inhibitory mechanisms result in often undetectable levels of nuclear RelA or basal NF-κB activity, our in silico analyses demonstrate that constitutive NF-κB-driven transcription is nonetheless physiologically relevant in the maintenance of the type I IFN system. Notably, 6 of the top 10 biological pathways significantly perturbed in unstimulated rela−/− MEFs were related to IFN-β, indicating that a primary function of constitutive RelA in MEFs may be to support autocrine IFN-β signaling.
After infection, the requirement for RelA in promoting ifnβ induction appears to be surprisingly short-lived. Lomvardas and Thanos have shown that RelA is the first transcription factor that associates with the ifnβ promoter following Sendai virus infection of HeLa cells and precedes ATF-2 and IRF-3 recruitments by 2 and 4 h, respectively (32). Elegant work by Apostolou and Thanos has since shown that activated NF-κB, despite being found in rate-limiting amounts, rapidly accesses the ifnβ locus by a novel process of interchromosomal transfer from putative NF-κB “receptor centers” (2). In their model, certain specialized genomic loci containing readily accessible NF-κB binding sites serve as temporary receptors for incoming nuclear NF-κB, following which NF-κB is shuttled to a single ifnβ locus to kick-start monoallelic expression. Later in infection, feed-forward production of IRF-7 drives biallelic ifnβ expression to accelerate the type I IFN response (2). Consistent with this model, our work shown herein and elsewhere (57) indicates that RelA is required for virus-stimulated ifnβ expression very early after infection, before IRF-3 is activated. Previously, we posited that RelA may directly synergize with the coactivator CBP/p300 to drive early ifnβ transcription in the absence of IRF-3 (57). Our current data now suggest that RelA also contributes to early ifnβ induction by maintaining autocrine IFN-β-dependent ISG nodes before infection. When expression of these nodes are reduced, virus-triggered signaling leading to ifnβ induction is significantly delayed. In this regard, it is noteworthy that ifnβ induction levels by dsRNA for rela+/+ and rela−/− MEFs on an ifnar1−/− background are virtually indistinguishable (see Fig. S5 in the supplemental material). Since ifnar1−/− MEFs cannot support autocrine IFN-β signaling, these results suggest that RelA functions in dsRNA-driven ifnβ induction primarily by controlling autocrine IFN-β-dependent ISG nodes and only secondarily by direct activation of the ifnβ promoter after infection.
Later in the course of an infection, RelA switches to stimulating a predominantly proinflammatory set of genes. Approximately 25% of the RLR primary antiviral transcriptome was found to be RelA dependent. This subset was especially enriched in genes encoding (i) chemokines and adhesion molecules, (ii) matrix metalloproteinases and allied proteases involved in remodeling the extracellular matrix, activation of chemokine signaling, and recruitment of immune cells, and (iii) proteins involved in antigen processing and presentation, including a large number of classical and nonclassical major histocompatibility complex class I molecules. Considered together, these analyses strongly suggest that the RelA-dependent arm of the RLR response coordinates the recruitment of immune cells to the site of infection and promotes activation of adaptive immunity. Interestingly, rela−/− MEFs on an ifnar−/− background were still more permissive to virus than rela+/+ifnar−/− MEFs, indicating that among the RLR-induced primary RelA targets are genes with direct antiviral activity (see Fig. S6 in the supplemental material).
In contrast to these findings, a recent study by Schmolke and colleagues performed on influenza virus-infected cells concluded that almost all influenza virus-induced genes, including ifnβ, depend on NF-κB (44). Besides noting obvious differences in systems used (dsRNA transfection of MEFs in our case versus virus infection of human endothelial cells by Schmolke et al.), we wish to highlight an important distinction between the two studies. The experiments by Schmolke et al. were performed on cells with intact IFN-β signaling, whereas we carried out our microarray analyses on an ifnar1−/− background for the express purpose of identifying primary antiviral genes in the absence of autocrine IFN-β-dependent effects. It is therefore conceivable that the IKK-β mutant-expressing cells employed by Schmolke et al. (like ikkβ−/− and rela−/− MEFs) have compromised autocrine IFN-β signaling and are thus defective in their ability to establish feed-forward type I IFN signaling after infection. This distinction may at least partially explain why gross IFN-β-dependent defects were seen in their case but not ours. In support of this explanation, we have found that ikkβ−/− MEFs are significantly impaired in autocrine IFN-β signaling (Fig. 4E) and consequential virus-driven ifnβ and/or ISG expression (57).
Type I IFN has been shown to directly activate RelA in several cell types (62). We show here that RelA is required for the induction of a small subset of genes by IFN-β, one of which is cxcl11. Since cxcl11 was also identified as a STAT3-dependent ISG (64), this finding supports the hypothesis that STAT3 lies upstream of NF-κB during type I IFN signaling (40, 61, 63). Previous studies have shown that p50−/− rela−/− doubly deficient MEFs responded more robustly to type I IFN and consequently mounted and enhanced antiviral responses to viral challenge following IFN pretreatment (39, 60). Consistent with these findings, several antiviral ISGs were shown to be upregulated to a greater extent by IFN-β treatment in p50−/− rela−/− doubly deficient MEFs than in controls (39, 60). In support, p50 appears to be constitutively associated with the promoters of several ISGs that are negatively regulated by NF-κB during IFN-β signaling (60). Our own results now show that rela−/− singly deficient MEFs also exhibit these differences (see Fig. S7 in the supplemental material), suggesting that RelA itself also negatively regulates this subset of ISGs. In agreement, RelA has been shown to be constitutively associated with the promoters of some of these ISGs (60). The mechanisms by which NF-κB subunits negatively regulate ISG expression remain to be identified.
A previous report from one of our laboratories demonstrated that rela−/− MEFs were singularly susceptible to a novel form of ROS-mediated caspase-independent cell death induced by dsRNA (31). Here, we show that dsRNA triggers necroptotic cell death in rela−/− MEFs that is dependent on the kinase RIP1. Necroptosis has been best characterized in the context of TNF-α signaling, during which a kinase complex comprising RIP1 and RIP3 appears to function by altering mitochondrial metabolism and increasing ATP biogenesis (7, 22, 67). As a consequence, mitochondrial ROS accumulates to toxic levels that result in respiratory failure and eventual cell death. Importantly, NF-κB has been shown to upregulate antioxidant genes (e.g., sod2 and fhc) that quench ROS and allow continued cell survival following TNF-α stimulation (41, 42, 48). In the context of antiviral responses, it is possible that dsRNA similarly activates RIP1 to increase ATP synthesis and fuel antiviral enzyme systems (for example, 2′,5′-oligoadenylate synthase/RNase L). During this process, NF-κB may induce genes (such as sod2) that buffer mitochondria by scavenging ROS by-products to prolong survival of the infected cell and maximize secretion of IFN and other immune system modulators.
A recent whole-genome RNA interference screen for modulators of necroptosis identified several immune genes (including type I IFNs), suggesting that necroptosis may represent a host immune effector mechanism involved in pathogen recognition and clearance (24). In agreement with this hypothesis, it has been demonstrated that RIP3-deficient mice are severely defective in virus-induced programmed necrosis and are consequently more susceptible to vaccinia virus infections (7). Further, murine cytomegalovirus encodes a RIP3 inhibitor important for its pathogenesis (54). Thus, while NF-κB may initially protect cells from necroptosis to allow antiviral gene induction, the eventual induction of dsRNA-dependent necroptosis may facilitate uptake of viral antigens by dendritic cells and other professional antigen-presenting cells (APCs) in the context of pathogen- or damage-associated molecular patterns associated with necrotic death. Recent data from in vivo experiments would suggest that such dual stimulation of APCs is crucial for viral clearance (7, 54).
In summary, our results show that the NF-κB subunit RelA has distinct roles in the type I IFN antiviral innate immune response (shown schematically in Fig. 8). In uninfected cells, RelA cycles through the nucleus to maintain basal expression of ifnβ and sustain the IFN-β autocrine signaling. In the absence of RelA, cells are defective in basal expression of ISGs, including those encoding pivotal nodes required for feed-forward amplification of type I IFN signaling. Primarily for this reason, the RLR-triggered antiviral response is significantly delayed in rela−/− MEFs. Consequently, these cells are susceptible to RNA virus infections. Once IRF-3/7 is activated, RelA is not essential for ifnβ induction and instead regulates expression of proinflammatory genes that galvanize the adaptive immune response. Finally, RelA also regulates an antinecroptotic cell survival program that ensures maximal RLR- and TLR-driven antiviral gene expression in the infected cell. Together, these studies provide a comprehensive picture of the RelA-dependent transcriptional response during innate antiviral responses.
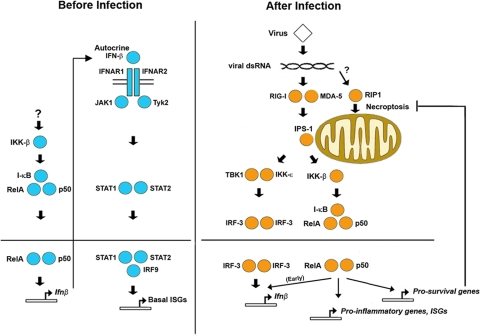
FIG. 8.
Distinct transcriptional roles for RelA in antiviral innate immune responses. Before infection (left, in blue), RelA is constitutively activated at low levels by an unknown IKK-β-dependent mechanism. Nuclear RelA-containing NF-κB maintains low basal ifnβ expression to sustain autocrine IFN-β signaling and support priming levels of basal ISGs. After infection (right, in orange), RelA is activated by RLRs to promote early ifnβ induction, activate proinflammatory and prosurvival (antinecroptotic) genes, and participate in IFN-β-induced induction of a subset of ISGs.
Supplementary Material
Acknowledgments
We thank Yutaro Kumagai, Taro Kawai, Shizuo Akira, Manolis Pasparakis, Tak Mak, Joan Durbin, Luis Sigal, Zhi-Wei Li, and Glen Barber for cells and viruses. We also thank Yuesheng Li for generating microarray data, Michael Slifker for in silico analyses, and Christoph Seeger for valuable comments.
This work was supported by an ACS Research Scholar Grant (RSG-09-195-01-MPC) to S.B. and a National Institutes of Health grant (R01 AI059715) to A.A.B. Additional funds were provided by the Fox Chase Cancer Center via institutional support of the Kidney Cancer Keystone Program.
S.H.B. and R.J.T. performed most of the experiments and participated in writing the manuscript. S.N. and J.W. performed real-time PCR analyses. A.A.B. oversaw experiments performed by J.W., generated rela−/−, ifnar1−/−, ikkβ−/−, and littermate control MEFs, and edited the paper. S.B. designed the experiments, interpreted data, and wrote the manuscript.
We declare that we have no conflict of interest.
Footnotes
Published ahead of print on 5 January 2011.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124:783-801. [DOI] [PubMed] [Google Scholar]
- 2.Apostolou, E., and D. Thanos. 2008. Virus infection induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell 134:85-96. [DOI] [PubMed] [Google Scholar]
- 3.Balachandran, S., et al. 1998. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J. 17:6888-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balachandran, S., E. Thomas, and G. N. Barber. 2004. A FADD-dependent innate immune mechanism in mammalian cells. Nature 432:401-405. [DOI] [PubMed] [Google Scholar]
- 5.Beg, A. A., W. C. Sha, R. T. Bronson, and D. Baltimore. 1995. Constitutive NF-kappa B activation, enhanced granulopoiesis, and neonatal lethality in I kappa B alpha-deficient mice. Genes Dev. 9:2736-2746. [DOI] [PubMed] [Google Scholar]
- 6.Chattopadhyay, S., et al. 2010. Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J. 29:1762-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho, Y. S., et al. 2009. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137:1112-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Degterev, A., et al. 2008. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 4:313-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeWitte-Orr, S. J., S. E. Collins, C. M. Bauer, D. M. Bowdish, and K. L. Mossman. 2010. An accessory to the ‘Trinity’: SR-As are essential pathogen sensors of extracellular dsRNA, mediating entry and leading to subsequent type I IFN responses. PLoS Pathog. 6:e1000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du, Z., et al. 2007. Non-conventional signal transduction by type 1 interferons: the NF-kappaB pathway. J. Cell Biochem. 102:1087-1094. [DOI] [PubMed] [Google Scholar]
- 11.Durbin, J. E., R. Hackenmiller, M. C. Simon, and D. E. Levy. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443-450. [DOI] [PubMed] [Google Scholar]
- 12.Faria, P. A., et al. 2005. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol. Cell 17:93-102. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez, M., M. Porosnicu, D. Markovic, and G. N. Barber. 2002. Genetically engineered vesicular stomatitis virus in gene therapy: application for treatment of malignant disease. J. Virol. 76:895-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentleman, R. C., et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gil, J., and M. Esteban. 2000. Induction of apoptosis by the dsRNA-dependent protein kinase (PKR): mechanism of action. Apoptosis 5:107-114. [DOI] [PubMed] [Google Scholar]
- 16.Gough, D. J., et al. 2010. Functional crosstalk between type I and II interferon through the regulated expression of STAT1. PLoS Biol. 8:e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gresser, I. 1990. Biologic effects of interferons. J. Investig. Dermatol. 95:66S-71S. [DOI] [PubMed] [Google Scholar]
- 18.Harhaj, E. W., and S. C. Sun. 1999. Regulation of RelA subcellular localization by a putative nuclear export signal and p50. Mol. Cell. Biol. 19:7088-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hata, N., et al. 2001. Constitutive IFN-alpha/beta signal for efficient IFN-alpha/beta gene induction by virus. Biochem. Biophys. Res. Commun. 285:518-525. [DOI] [PubMed] [Google Scholar]
- 20.Hayden, M. S., and S. Ghosh. 2008. Shared principles in NF-kappaB signaling. Cell 132:344-362. [DOI] [PubMed] [Google Scholar]
- 21.Hayden, M. S., and S. Ghosh. 2004. Signaling to NF-kappaB. Genes Dev. 18:2195-2224. [DOI] [PubMed] [Google Scholar]
- 22.He, S., et al. 2009. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 137:1100-1111. [DOI] [PubMed] [Google Scholar]
- 23.Heylbroeck, C., et al. 2000. The IRF-3 transcription factor mediates Sendai virus-induced apoptosis. J. Virol. 74:3781-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hitomi, J., et al. 2008. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 135:1311-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honda, K., et al. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772-777. [DOI] [PubMed] [Google Scholar]
- 26.Kasof, G. M., J. C. Prosser, D. Liu, M. V. Lorenzi, and B. C. Gomes. 2000. The RIP-like kinase, RIP3, induces apoptosis and NF-kappaB nuclear translocation and localizes to mitochondria. FEBS Lett. 473:285-291. [DOI] [PubMed] [Google Scholar]
- 27.Kato, H., et al. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19-28. [DOI] [PubMed] [Google Scholar]
- 28.Kato, H., et al. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato, H., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 30.Kawai, T., and S. Akira. 2008. Toll-like receptor and RIG-I-like receptor signaling. Ann. N. Y. Acad. Sci. 1143:1-20. [DOI] [PubMed] [Google Scholar]
- 31.Li, M., W. Shillinglaw, W. J. Henzel, and A. A. Beg. 2001. The Rela(p65) subunit of NF-kappaB is essential for inhibiting double-stranded RNA-induced cytotoxicity. J. Biol. Chem. 276:1185-1194. [DOI] [PubMed] [Google Scholar]
- 32.Lomvardas, S., and D. Thanos. 2002. Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110:261-271. [DOI] [PubMed] [Google Scholar]
- 33.Maniatis, T., et al. 1998. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb. Symp. Quant. Biol. 63:609-620. [DOI] [PubMed] [Google Scholar]
- 34.Meraz, M. A., et al. 1996. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84:431-442. [DOI] [PubMed] [Google Scholar]
- 35.Nourbakhsh, M., and H. Hauser. 1999. Constitutive silencing of IFN-beta promoter is mediated by NRF (NF-kappaB-repressing factor), a nuclear inhibitor of NF-kappaB. EMBO J. 18:6415-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nourbakhsh, M., K. Hoffmann, and H. Hauser. 1993. Interferon-beta promoters contain a DNA element that acts as a position-independent silencer on the NF-kappa B site. EMBO J. 12:451-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park, C., S. Li, E. Cha, and C. Schindler. 2000. Immune response in Stat2 knockout mice. Immunity 13:795-804. [DOI] [PubMed] [Google Scholar]
- 38.Peters, K. L., H. L. Smith, G. R. Stark, and G. C. Sen. 2002. IRF-3-dependent, NFkappa B- and JNK-independent activation of the 561 and IFN-beta genes in response to double-stranded RNA. Proc. Natl. Acad. Sci. U. S. A. 99:6322-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfeffer, L. M., et al. 2004. Role of nuclear factor-kappaB in the antiviral action of interferon and interferon-regulated gene expression. J. Biol. Chem. 279:31304-31311. [DOI] [PubMed] [Google Scholar]
- 40.Pfeffer, L. M., et al. 1997. STAT3 as an adapter to couple phosphatidylinositol 3-kinase to the IFNAR1 chain of the type I interferon receptor. Science 276:1418-1420. [DOI] [PubMed] [Google Scholar]
- 41.Pham, C. G., et al. 2004. Ferritin heavy chain upregulation by NF-kappaB inhibits TNFalpha-induced apoptosis by suppressing reactive oxygen species. Cell 119:529-542. [DOI] [PubMed] [Google Scholar]
- 42.Sakon, S., et al. 2003. NF-kappaB inhibits TNF-induced accumulation of ROS that mediate prolonged MAPK activation and necrotic cell death. EMBO J. 22:3898-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato, M., et al. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 44.Schmolke, M., D. Viemann, J. Roth, and S. Ludwig. 2009. Essential impact of NF-kappaB signaling on the H5N1 influenza A virus-induced transcriptome. J. Immunol. 183:5180-5189. [DOI] [PubMed] [Google Scholar]
- 45.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. [DOI] [PubMed] [Google Scholar]
- 46.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 47.Stojdl, D. F., et al. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263-275. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka, H., et al. 2002. E2F1 and c-Myc potentiate apoptosis through inhibition of NF-kappaB activity that facilitates MnSOD-mediated ROS elimination. Mol. Cell 9:1017-1029. [DOI] [PubMed] [Google Scholar]
- 49.Taniguchi, T., and A. Takaoka. 2002. The interferon-alpha/beta system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr. Opin. Immunol. 14:111-116. [DOI] [PubMed] [Google Scholar]
- 50.Taniguchi, T., and A. Takaoka. 2001. A weak signal for strong responses: interferon-alpha/beta revisited. Nat. Rev. Mol. Cell Biol. 2:378-386. [DOI] [PubMed] [Google Scholar]
- 51.Tergaonkar, V., R. G. Correa, M. Ikawa, and I. M. Verma. 2005. Distinct roles of IkappaB proteins in regulating constitutive NF-kappaB activity. Nat. Cell Biol. 7:921-923. [DOI] [PubMed] [Google Scholar]
- 52.Thanos, D. 1996. Mechanisms of transcriptional synergism of eukaryotic genes. The interferon-beta paradigm. Hypertension 27:1025-1029. [DOI] [PubMed] [Google Scholar]
- 53.Thanos, D., and T. Maniatis. 1995. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83:1091-1100. [DOI] [PubMed] [Google Scholar]
- 54.Upton, J. W., W. J. Kaiser, and E. S. Mocarski. 2010. Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe 7:302-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Boxel-Dezaire, A. H., M. R. Rani, and G. R. Stark. 2006. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity 25:361-372. [DOI] [PubMed] [Google Scholar]
- 56.Vandenabeele, P., W. Declercq, F. Van Herreweghe, and T. Vanden Berghe. 2010. The role of the kinases RIP1 and RIP3 in TNF-induced necrosis. Sci. Signal 3:re4. [DOI] [PubMed] [Google Scholar]
- 57.Wang, J., et al. 2010. NF-{kappa}B RelA subunit is crucial for early IFN-{beta} expression and resistance to RNA virus replication. J. Immunol. 185:1720-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, J., et al. 2007. Distinct roles of different NF-kappa B subunits in regulating inflammatory and T cell stimulatory gene expression in dendritic cells. J. Immunol. 178:6777-6788. [DOI] [PubMed] [Google Scholar]
- 59.Wang, X., et al. 2007. Lack of essential role of NF-kappa B p50, RelA, and cRel subunits in virus-induced type 1 IFN expression. J. Immunol. 178:6770-6776. [DOI] [PubMed] [Google Scholar]
- 60.Wei, L., et al. 2006. NFkappaB negatively regulates interferon-induced gene expression and anti-influenza activity. J. Biol. Chem. 281:11678-11684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, C. H., A. Murti, and L. M. Pfeffer. 1998. STAT3 complements defects in an interferon-resistant cell line: evidence for an essential role for STAT3 in interferon signaling and biological activities. Proc. Natl. Acad. Sci. U. S. A. 95:5568-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang, C. H., et al. 2000. IFNalpha/beta promotes cell survival by activating NF-kappa B. Proc. Natl. Acad. Sci. U. S. A. 97:13631-13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang, C. H., et al. 2001. Interferon alpha/beta promotes cell survival by activating nuclear factor kappa B through phosphatidylinositol 3-kinase and Akt. J. Biol. Chem. 276:13756-13761. [DOI] [PubMed] [Google Scholar]
- 64.Yang, C. H., et al. 2007. Identification of CXCL11 as a STAT3-dependent gene induced by IFN. J. Immunol. 178:986-992. [DOI] [PubMed] [Google Scholar]
- 65.Yarilina, A., K. H. Park-Min, T. Antoniv, X. Hu, and L. B. Ivashkiv. 2008. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat. Immunol. 9:378-387. [DOI] [PubMed] [Google Scholar]
- 66.Yoneyama, M., and T. Fujita. 2009. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 227:54-65. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, D. W., et al. 2009. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 325:332-336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.