Abstract
There are large-scale variations of the GC-content along mammalian chromosomes that have been called isochore structures. Primates and rodents have different isochore structures, which suggests that these lineages exhibit different modes of GC-content evolution. It has been shown that, in the human lineage, GC-biased gene conversion (gBGC), a neutral process associated with meiotic recombination, acts on GC-content evolution by influencing A or T to G or C substitution rates. We computed genome-wide substitution patterns in the mouse lineage from multiple alignments and compared them with substitution patterns in the human lineage. We found that in the mouse lineage, gBGC is active but weaker than in the human lineage and that male-specific recombination better predicts GC-content evolution than female-specific recombination. Furthermore, we were able to show that G or C to A or T substitution rates are predicted by a combination of different factors in both lineages. A or T to G or C substitution rates are most strongly predicted by meiotic recombination in the human lineage but by CpG odds ratio (the observed CpG frequency normalized by the expected CpG frequency) in the mouse lineage, suggesting that substitution patterns are under different influences in primates and rodents.
Keywords: genome evolution, isochore, substitution patterns, meiotic recombination, biased gene conversion
Introduction
In mammals, the genomic GC-content (fraction of G and C bases) is not homogeneous: it exhibits large-scale variations that have been called isochore structures (Bernardi et al. 1985; Bernardi 2000; Eyre-Walker and Hurst 2001; Lander et al. 2001; Mouse Genome Sequencing Consortium et al. 2002). These structures are linked with several genomic and functional features such as intron length, gene, and transposable element density (Duret et al. 1995; Duret and Galtier 2009). Several models have been proposed to explain these variations: some based on natural selection (Bernardi et al. 1985; Bernardi 2000, 2007; Lercher et al. 2003) and others on neutral processes like variations of mutation patterns (Filipski 1988; Wolfe et al. 1989; Eyre-Walker and Hurst 2001) or GC-biased gene conversion (later designated as gBGC) (Galtier et al. 2001; Galtier and Duret 2007; Duret and Galtier 2009). In humans, it has been well established that gBGC plays an important role in GC-content evolution (Meunier and Duret 2004; Duret and Arndt 2008).
GC-biased gene conversion is a mechanism associated with meiotic recombination that affects the fixation of single nucleotide mutations (Marais 2003; Duret and Galtier 2009). Meiotic recombination is initiated by a double-strand break in one chromosome of a chromosomal pair. This double-strand break is repaired by the invasion of the homologous region of the sister chromosome, which then serves as template for DNA synthesis and repair by gene conversion (the copy and paste of one DNA fragment into another). During this process, strands from two sister chromosomes are paired together, which may result in mismatches occurring if the corresponding locus is heterozygote. It has been shown that the mismatch repair mechanism is biased toward G and C bases: it will repair, for example, a G:T mismatch more often into G:C than into A:T (Brown and Jiricny 1988; Bill et al. 1998). This will lead to an unequal segregation of alleles, G and C alleles segregating at higher frequencies than A and T alleles, which will result in a fixation bias (Nagylaki 1983) favoring G and C alleles associated with meiotic recombination (Marais 2003; Montoya-Burgos et al. 2003): high recombination will increase A or T (weak or W) → G or C (strong or S) substitution rates and decrease S→W substitution rates. This fixation bias can be mistaken for natural selection (Nagylaki 1983; Pollard et al. 2006; Galtier and Duret 2007).
Most studies on the precise impact of meiotic recombination and gBGC on substitution patterns focused on primates and humans (Meunier and Duret 2004; Arndt et al. 2005; Webster et al. 2005; Duret and Arndt 2008; Pozzoli et al. 2008; Tyekucheva et al. 2008; Galtier et al. 2009), some generating genome-wide substitution patterns for the human lineage from multiple alignments (Duret and Arndt 2008). Using the same strategy, it is now possible to estimate substitution patterns in the rodent lineage and evaluate the impact of recombination and gBGC on substitution patterns.
Several studies have shown that GC-rich isochores are vanishing in primates and rodents: the GC-content of GC-rich regions is decreasing (Duret et al. 2002; Smith and Eyre-Walker 2002; Belle et al. 2004). The main hypothesis to explain this decline is that at the time of mammalian radiation, chromosomal rearrangements, especially fusions, caused chromosomal arms to become longer (Nakatani et al. 2007). As there is a minimum of one crossover per chromosomal arm per meiosis (Petronczki et al. 2003), longer chromosomal arms have lower meiotic recombination rates (Kaback 1996; de Villena and Sapienza 2001; Coop and Przeworski 2007). Mouse and rat genomes (murid rodents), for example, have longer chromosomal arms and lower meiotic recombination rates than primates (Jensen-Seaman et al. 2004; Li and Freudenberg 2009). These fusions are thought to have caused gBGC to be less prominent in mammals and GC-rich isochores to decline. This decline appears to be specific to primates and murid rodents (Romiguier et al. 2010). However, murid rodents appear to have different GC-content evolution compared with primates: they have more homogeneous GC-content, which has been called the minor shift (Mouchiroud et al. 1988; Robinson et al. 1997; Mouse Genome Sequencing Consortium et al. 2002; Smith and Eyre-Walker 2002; Gibbs et al. 2004).
Comparing substitution patterns in primates and murid rodents can help us understand the precise influence of chromosomal organization on substitution patterns and also how other factors influence substitution patterns. For example, it has been shown in the human lineage that replication timing (Chen et al. 2010) and CpG dinucleotide density (Walser et al. 2008; Walser and Furano 2010) are associated with substitution patterns.
In this study, we computed genome-wide substitution patterns in the mouse lineage from multiple alignments and analyzed to what extent they are predicted by meiotic recombination and other genomic factors. We repeated this analysis in the human lineage. We were able to show that gBGC is active in the mouse lineage but weak compared with the human lineage. Using powerful statistical methods, we were able to show that, in both lineages, different factors predict substitution patterns. In the human lineage, W→S substitution rates are mostly predicted by meiotic recombination, whereas in the mouse lineage, they are mostly predicted by CpG odds ratio.
Materials and Methods
Multiple Alignments and Substitution Patterns
We computed substitution patterns in both human and mouse lineages using genome-wide triple alignments as follows. We divided all human and mouse autosomes into 1 Mbp nonoverlapping windows. We retrieved the Pecan 10 amniotes multiple alignments available at the Ensembl database (version 56) corresponding to each window and restricted these to the analysis of the following species: human, chimpanzee, and macaque for the analysis of the human lineage, mouse, rat, and human for the analysis of the mouse lineage. For both analyses, we masked all exons from our alignments using the Ensembl database annotation (version 56, mouse genome version mm9, human genome version hg19). We did not mask repeated elements from our alignments.
We inferred substitution rates for each window as follows. We used a maximum likelihood-based method (Arndt et al. 2003; Arndt and Hwa 2005; Duret and Arndt 2008), which does not assume that the substitution process is time reversible, nor that sequence composition has yet reached equilibrium. It also takes into account the fact that the methylated cytosine of a CpG dinucleotide is hypermutable: C→T and G→A mutations occur approximately ten times more frequently in CpGs than in non-CpGs (Bird 1978; Giannelli et al. 1999). The method we used adds an additional rate parameter to represent this CpG substitution process. We assumed complementary rates to be equal (A→G = T→C = AT→GC) and computed 7 substitution rates: 2 transition rates (AT→GC, GC→AT), 4 transversion rates (AT→CG, AT→TA, GC→TA, GC→CG), and one CpG rate (CpG→TpG/CpA). AT→GC and AT→CG substitution rates were grouped together as Weak (W) → Strong (S) substitution rates. GC→AT and GC→TA substitution rates were grouped together as S→W substitution rates. A substitution pattern consists of all substitution rates. We computed for each substitution pattern an equilibrium GC-content or future GC-content (later designated as GC*), which is the expected final GC-content if the sequence evolves with a constant substitution pattern through time. It can be viewed as the summary value of the substitution pattern.
We computed the following genomic features in each window: GC-content, the distance to the telomere, the CpG dinucleotide odds ratio (the observed CpG frequency divided by the expected CpG frequency, later designated as CpGodds), exon density (proportion of base pairs occupied by exons in a window, later designated as Exons) as well as SINE, LINE, and LTR transposable element densities (later designated as SINEs, LINEs, and LTRs). We extracted crossover rates from high-quality genetic maps available for the human genome (International HapMap Consortium et al. 2007) and the mouse genome (Shifman et al. 2006). Crossover (CO) rates were computed as the weighted average of CO rates of chromosomal regions that overlap the window. We were able to extract sex-averaged CO rates in the human genome, sex-averaged as well as male- and female-specific CO rates in the mouse genome. Because in the mouse lineage the CO rates and the distance to the telomere exhibit a nonnormal distribution (supplementary fig. 1, Supplementary Material online), we computed the logarithm of each of the CO rates (later designated as LCO) as well as each of the distance to the telomere (designated as LDT) and used them for the remainder of the study. We computed replication-timing values (RepTime) from high-resolution replication-timing profiles available for mouse embryonic stem cells (Hiratani et al. 2008) and human embryonic stem cells (Ryba et al. 2010), as the weighted median of replication-timing values of chromosomal regions that overlap the window. All genomic positions in the genetic maps and replication-timing profiles were converted to the versions of the human genome (hg19) and mouse genome (mm9) from which the alignments were computed using the liftOver tool available at UCSC (http://genome.ucsc.edu/cgi-bin/hgLiftOver).
We filtered windows as follows: we discarded windows with less than 100 kbp of sites where all three species have an aligned nucleotide, windows which overlapped centromeric regions, as well as windows without enough information to compute CO rates or other genomic features. Substitution rates and genomic factors were normalized to have a mean of 0 and a standard deviation of 1.
Principal Component Regression
We analyzed the link between substitution patterns and nine genomic factors (GC-content, CO rates, distance to the telomere, Exons, RepTime, SINEs, LINEs, LTRs, CpGodds) using principal component regression (principal component analysis followed by linear regression) as described below. We first carried out principal component analysis in both human and mouse lineages on the nine genomic factors. In this step, all factors were projected on nine orthogonal axes or principal components. Each principal component is characterized by an eigenvalue that determines how much of the factor’s total variance this component explains and by an eigenvector, with one entry per factor, each entry determining how important the factor is within the principal component. Entries of an eigenvector were normalized such that the sum of the square of the entries is equal to 1. All principal components are independent and are ranked based on the proportion of the variance of the factors they explain. We performed two independent projections for the mouse and human lineages. We then performed linear regressions, using the principal components previously computed as factors and substitution rates computed in each lineage as variables. We calculated for each linear regression the R2 of this regression, as well as the R2 for each individual principal component.
All statistics were performed using R (http://www.r-project.org/). We used the R package pls to perform principal component regression (Mevik and Wehrens 2007). We used the R code of Drummond et al. (2006) to generate figures and tables for principal component regression.
Results
GC-Content Is Decreasing in the Mouse Genome
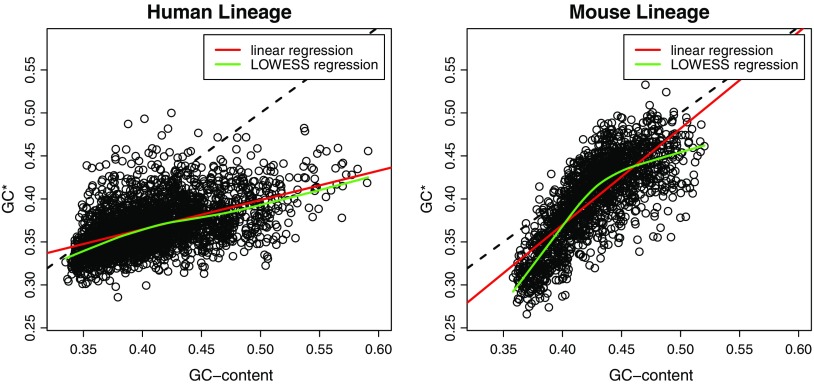
We computed substitution patterns and GC* (equilibrium GC-content) in both human and mouse lineages in 1 Mbp windows using triple alignments (for more details, see the Materials and Methods section). After filtering out windows without at least 100 kbp of sites where all three species of the triple alignments share a nucleotide and those overlapping centromeric regions, we obtained 1,594 windows containing more than 520 Mbp of analyzable sites in the mouse genome and 2,571 windows containing more than 1,800 Mbp of analyzable sites in the human genome. Results show that human and mouse GC-content are evolving very differently (fig. 1).
FIG. 1.—
Relationship between GC-content and GC* in the human (left panel) and mouse (right panel) lineages. The dashed line represents the GC-content = GC* relationship.
We found a linear relationship between GC-content and GC* in the human lineage. In GC-rich regions, GC-content is decreasing (GC* is lower than GC-content), whereas in GC-poor regions, GC-content is at equilibrium (GC* is equal to GC-content). In the mouse lineage, the relationship between GC-content and GC* is not linear, illustrated by the local LOWESS regression between the two variables (fig. 1). We see that the GC-content is decreasing in GC-rich regions but also in GC-poor regions. The GC-content in GC-intermediate regions (GC-content equal to 0.42) is at equilibrium.
Because substitution patterns appear to be different in both human and mouse lineages, we analyzed the influence of meiotic recombination and of other factors on substitution patterns in both lineages.
gBGC Is Active in the Mouse Lineage
We applied the same methodology as previous studies and analyzed the link between GC-content, GC*, and CO rates (Meunier and Duret 2004; Duret and Arndt 2008).
We observe a positive correlation between GC-content and CO rates in both lineages (table 1, supplementary tables 1–3, Supplementary Material online). To investigate the cause and effect relationship between GC-content and meiotic recombination in the mouse lineage, we calculated correlation coefficients between GC* and CO rates. We see that these correlations are stronger than between GC-content and CO rates (table 1, supplementary tables 1–3, Supplementary Material online). We repeated this analysis in the human lineage and found similar results. To compare our results with those of previous studies, correlation coefficients computed using CO in both human and mouse lineages are available in the supplementary material (Supplementary Material online).
Table 1.
Pearson Correlation Coefficients for Sex-Averaged CO Rates
| Human LCO |
Mouse LCO |
|||||
| R | R2 | P Value | R | R2 | P Value | |
| GC-content | 0.361 | 0.131 | <10−15 | 0.188 | 0.035 | <10−13 |
| GC* | 0.634 | 0.402 | <10−15 | 0.204 | 0.042 | <10−15 |
We draw two conclusions from these results. First, since GC* values are computed from substitution patterns and not from current GC-content, these results show that in the mouse lineage as well as the human lineage, meiotic recombination has an effect on GC-content evolution by acting on substitution patterns. This is consistent with the influence of gBGC on substitution patterns. We repeated this analysis in the mouse lineage for male- and female-specific CO rates, as well as using Spearman’s correlation coefficients and obtained similar results (supplementary tables 1–3, Supplementary Material online). We also obtained similar results when using LDT as it is known to be a proxy measure of meiotic recombination rates (Duret and Arndt 2008) (supplementary tables 4 and 5, Supplementary Material online). The correlation between LDT and recombination is negative, accordingly we observe negative correlations between LDT, GC-content, and GC* (supplementary tables 4 and 5, Supplementary Material online). Second, our results suggest that the influence of meiotic recombination on substitution patterns is weaker in the mouse lineage than in the human lineage because correlation coefficients are lower in the mouse lineage. Also, in the mouse lineage, the correlation coefficients between LCO and GC-content and between LCO and GC* are much closer than in the human lineage. One possible explanation is that the mouse genome has lower meiotic recombination rates than the human genome (human median CO rate = 1.34 cM/Mb, mouse median sex-averaged CO rate = 0.64 cM/Mb, supplementary fig. 1, Supplementary Material online).
Furthermore, in the mouse lineage, we can see that male-specific CO rates correlate more strongly with current GC-content or GC* than sex-averaged or female CO rates do (supplementary tables 1–3, Supplementary Material online). This indicates that male recombination has more influence on substitution patterns than female recombination in the mouse lineage, as previously observed in the human lineage (Webster et al. 2005; Duret and Arndt 2008). We therefore focused on male-specific CO rates in the mouse lineage for the remainder of the study.
Because meiotic recombination only predicts a small fraction of substitution rates in the mouse lineage, we investigated how other genomic factors, such as GC-content, replication timing, and transposable elements density, predict substitution rates in the mouse lineage and compared it with the human lineage.
Different Factors Predict Substitution Patterns in Both Human and Mouse Lineage
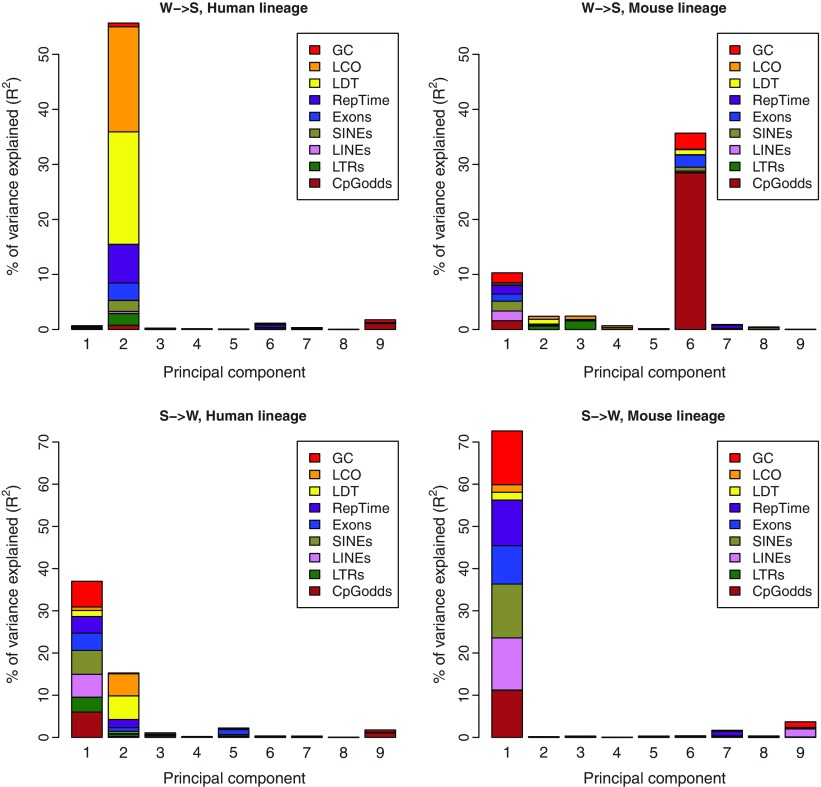
Because they have the most impact on GC* and GC-content evolution, we focused on W→S and S→W substitution rates in the human and mouse lineages and analyzed the link between them and nine genomic factors (GC-content, LCO, LDT, RepTime, Exons, SINEs, LINEs, LTRs, CpGodds). Because these genomic factors are intercorrelated, using multivariate linear regression will give unsatisfactory results. We therefore performed principal component regression. We first carried out one principal component analysis in each lineage on the 9 factors to transform them into 9 independent (or orthogonal) principal components 9 (designated as PC), each being a linear combination of the 9 genomic factors. Supplementary Figure 4 (Supplementary Material online) shows the eigenvectors of the first two principal components in the human and mouse lineages. We then used these components to build multivariate linear regressions for W→S and S→W substitution rates, where the substitution rate is the response variable and the components are the predictors and computed how much of the variable’s variance each principal component predicts (for details, see Materials and Methods).
We see that in both human and mouse lineages, substitution patterns are predicted by different factors. In the human lineage, W→S substitution rates are most strongly predicted by a component (PC2), which is mostly composed of LCO and LDT, two proxy measures of meiotic recombination (R2 = 0.55, fig. 2 and table 2). This result can be interpreted as reflecting the influence of gBGC on W→S substitution. In the mouse lineage, a component (PC6) which is dominated by CpG odds ratio rather than by measures of meiotic recombination, most strongly predicts W→S substitution rates (R2 = 0.35, fig. 2 and table 3). This result can be interpreted as reflecting gBGC only having a very limited impact on W→S substitution in the mouse lineage. Other principal components like the first component also explain a small proportion of the variance of the substitution rates in the mouse lineage (R2 = 0.10, fig. 2, table 3).
FIG. 2.—
Principal component regression for W→S substitution rates (top row) and S→W substitution rates (bottom row) in the human (left column) and the mouse (right column) lineages. The height of each bar represents how much of the variable’s variance the corresponding component explains. Each colored area is proportional to the relative importance of the corresponding factor inside a component.
Table 2.
Results of Principal Component Regression on W→S and S→W Substitution Rates in the Human Lineage
| Principal Components |
||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | All | |
| % of variance explained (R2) | ||||||||||
| W→S | 0.69*** | 55.68*** | 0.23* | 0.12* | 0.07* | 1.15*** | 0.34** | 0.03 | 1.77*** | 60.08*** |
| S→W | 36.98*** | 15.27*** | 1.09*** | 0.20* | 2.19*** | 0.34** | 0.33** | 0.03 | 1.77*** | 58.19*** |
| % contribution | ||||||||||
| GC | 16.5 | 1.2 | 0.2 | 1.6 | 0.1 | 10.8 | 1.9 | 40.3 | 27.4 | |
| LCO | 2.2 | 34.3 | 30.1 | 28.3 | 0.1 | 1.9 | 0.0 | 0.5 | 2.4 | |
| LDT | 3.9 | 36.7 | 9.2 | 16.5 | 2.0 | 16.1 | 0.0 | 9.1 | 6.5 | |
| RepTime | 10.6 | 12.6 | 8.3 | 1.4 | 13.3 | 39.0 | 11.6 | 1.6 | 1.5 | |
| Exons | 11.1 | 5.7 | 12.2 | 3.3 | 53.7 | 2.0 | 5.2 | 6.5 | 0.2 | |
| SINEs | 15.2 | 3.6 | 2.5 | 0.0 | 4.3 | 0.0 | 59.0 | 15.3 | 0.0 | |
| LINEs | 14.7 | 0.7 | 2.5 | 4.1 | 15.7 | 21.8 | 22.1 | 17.3 | 1.2 | |
| LTRs | 9.5 | 3.8 | 31.3 | 36.4 | 9.9 | 7.9 | 0.1 | 1.0 | 0.0 | |
| CpGodds | 16.2 | 1.4 | 3.6 | 8.4 | 0.8 | 0.5 | 0.0 | 8.4 | 60.4 | |
NOTE.—Factors that contribute for at least 20% of the component are indicated in bold. *P value < 0.05; **P value < 10−5; ***P value < 10−10.
Table 3.
Results of Principal Component Regression on W→S and S→W Substitution Rates in the Mouse Lineage
| Principal Components |
||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | All | |
| % of variance explained (R2) | ||||||||||
| W→S | 10.30*** | 2.40*** | 2.42*** | 0.68** | 0.14* | 35.67*** | 0.89** | 0.46* | 0.03 | 52.98*** |
| S→W | 72.61*** | 0.16* | 0.30** | 0.03 | 0.33** | 0.38** | 1.68*** | 0.34** | 3.69*** | 79.52*** |
| % contribution | ||||||||||
| GC | 17.5 | 0.1 | 0.0 | 0.6 | 8.6 | 8.2 | 13.2 | 15.4 | 36.4 | |
| LCO | 2.5 | 22.6 | 24.6 | 49.8 | 0.0 | 0.2 | 0.2 | 0.0 | 0.0 | |
| LDT | 2.6 | 36.1 | 8.1 | 42.7 | 5.4 | 2.6 | 2.4 | 0.1 | 0.0 | |
| RepTime | 14.9 | 5.7 | 0.0 | 0.1 | 0.7 | 0.1 | 62.7 | 14.7 | 1.2 | |
| Exons | 12.5 | 6.8 | 0.0 | 1.6 | 63.7 | 6.3 | 5.9 | 0.5 | 2.8 | |
| SINEs | 17.5 | 1.7 | 0.4 | 0.1 | 1.0 | 2.0 | 2.9 | 68.9 | 5.4 | |
| LINEs | 17.0 | 1.2 | 2.3 | 0.3 | 17.0 | 0.7 | 9.9 | 0.0 | 51.6 | |
| LTRs | 0.1 | 25.6 | 64.4 | 4.4 | 2.2 | 0.2 | 1.5 | 0.3 | 1.4 | |
| CpGodds | 15.3 | 0.3 | 0.0 | 0.4 | 1.4 | 79.8 | 1.4 | 0.1 | 1.3 | |
NOTE.—Factors that contribute for at least 20% of the component are indicated in bold. *P value < 0.05; **P value < 10−5; ***P value < 10−10.
In the human lineage, S→W substitution rates are most strongly predicted by the first two principal components (R2 = 0.37 and 0.15, respectively, fig. 2 and table 2). In contrast, in the mouse lineage, S→W substitution rates are most strongly predicted by the first principal component (R2 = 0.72, fig. 2 and table 3). In both lineages, the first component is composed by several factors (tables 2 and 3). Results for individual substitution rates and GC* can be found in supplementary tables 7 and 8 and supplementary figure 5 (Supplementary Material online).
These results are similar to what we observe when we analyze the data using a conventional linear regression-based method which computes the relative contribution to variability explained (RCVE) for each genomic factor (for more information, see supplementary materials, Supplementary Material online). In the human lineage, AT→GC and AT→CG rates are most strongly predicted by CO rates, whereas these rates are most strongly predicted by CpG odds ratio in the mouse lineage. Moreover, the regression slope between AT→GC or CG rates and CpG odds ratio is positive in the mouse lineage (supplementary figs. 2 and 3, Supplementary Material online).
Discussion
Choice of Outgroup
The method we use to infer substitution rates in one lineage uses triple alignments: it compares two sister species and uses an outgroup to infer the two sister’s ancestral state. To study the mouse lineage, we compared mouse and rat and used human as an outgroup. The mouse–rat–human divergence time is between 85 and 95 My. The mouse–rat divergence time is between 16 and 19 My (Poux et al. 2006; Huchon et al. 2007). Using human as an outgroup may cause to infer incorrect substitution rates in the long mouse lineage. However, human was chosen as an outgroup for the mouse lineage as the closest available high-coverage genome to mouse and rat. One of the closest related species to mouse and rat which complete genome has been published and aligned to other placentals is the guinea pig (Cavia porcellus). It is, however, a 6.79× low-coverage genome. Furthermore, the divergence time between mouse, rat, and guinea pig is around 60 My (Poux et al. 2006; Huchon et al. 2007), which is close to the mouse, rat, and human divergence time. Preliminary results obtained using guinea pig or kangaroo rat as outgroups were very similar to results obtained using human as an outgroup (data not shown). Moreover, mouse–rat–human triple alignments are much cleaner and contain more sites where the three species share a nucleotide than alignments with guinea pig or kangaroo rat. We therefore used mouse–rat–human triple alignments to infer substitution patterns in the mouse lineage. Moreover, the method we used to infer substitution rates (Arndt et al. 2003; Arndt and Hwa 2005; Duret and Arndt 2008) is based on maximum likelihood, which makes it robust to long lineage as it allows multiple substitutions at each site. It also imply time irreversibility and nonstationary state and infers one substitution pattern for each of the four branches of the rooted tree ([sister 1, sister 2], outgroup).
Potential Effects of Outgroup Choice, Different Timespans, and Density of Genetic Maps
Our results could be affected by the different timespans that substitution patterns reflect in both human and mouse lineages: human and chimpanzee diverged around 6 Ma, whereas mouse and rat diverged between 16 and 19 Ma (Poux et al. 2006; Huchon et al. 2007). CO rates computed in the mouse genome may not well reflect past recombination as mouse and rat genomes underwent frequent chromosomal rearrangements that affected their chromosomal recombination patterns. Moreover, the outgroup for the analysis of the mouse lineage is very distant, whereas the outgroup for the analysis of the human lineage is much closer: mouse and human diverged between 85 and 95 Ma, whereas human and macaque diverged between 27 and 33 Ma (Poux et al. 2006; Huchon et al. 2007). Another potential source of bias is the different densities of genetic maps available for human and mouse: the mouse maps contain between 10,000 and 11,000 markers on autosomes (approximately, one marker every 250 kbp), whereas the human map contains more than 3 million markers. To control for all these sources of bias, we performed the following analyses. We computed substitution patterns in the lineage between the human–macaque ancestor and human, using mouse as an outgroup (hereafter designated as the long human branch). At the same time, we computed new CO rates the following way: we generated a low-density human genetic map by sampling 11,000 random markers from the original map and recomputed CO rates as described in the Materials and Methods section. Results obtained for this long branch are very similar to results obtained with the original human branch. First, even though correlation coefficients between CO rates, GC-content and GC* are slightly lower for the long branch than for the original branch, the correlation between CO rates and GC* is stronger than the correlations between CO rates and GC-content (supplementary table 6, Supplementary Material online). Moreover, LDT correlates more strongly with GC-content and GC* in the long human branch than in the mouse lineage. Second, principal component regression results of the original branch and the long branch were very similar: in this branch, the second component is the main predictor of W→S substitution rates, whereas the first component is the main predictor of S→W substitution rates (supplementary fig. 7, Supplementary Material online). We therefore conclude that our results are not affected by the different timespans between human and mouse lineages nor by different density of genetic maps.
Furthermore, it has been shown that there is a cryptic variation of the mutation process in the human genome (Hodgkinson et al. 2009) that could cause a bias in our substitution pattern inference and affect our results. By conducting sequence evolution simulations, we were able to show that this cryptic variation is not likely to affect our results (for more information, see supplementary materials and figure 6, Supplementary Material online).
GC-Content Evolution and Chromosomal Organization
Our results show that both human and mouse lineages exhibit different modes of GC-content evolution. We were able to show that the erosion of GC-rich isochores is still ongoing in both lineages, confirming previous results (Duret et al. 2002; Smith and Eyre-Walker 2002; Belle et al. 2004). Moreover, it has been suggested that this murid shift was caused by recombination rates being less variable in the mouse genome (Eyre-Walker 1993). We do indeed observe that mouse CO rates are less variable than human CO rates (variance = 0.50 and 0.69 for mouse and human CO rates, respectively). These previous studies have shown, however, that the GC-content of GC-poor regions is increasing in murid rodents, whereas we show that the GC-content is decreasing in these regions. We can explain these differences by the small number of genes these studies relied on, analyzing the GC-content at synonymous positions (GC3).
It has been hypothesized that the decline of GC-rich isochores in primates and murid rodents has been caused by chromosomal fusions at the time of mammalian radiation, more than 80 Ma (Duret et al. 2002) However, since this decline is not shared across all mammals (Romiguier et al. 2010), it is likely that different factors influenced GC-content evolution in both human and mouse lineages. We therefore have to specifically compare primate and murid rodent GC-content evolution and substitution patterns.
Substitution Patterns Are under the Influence of Male-Specific Recombination
Our results show that, in the mouse lineage, male-specific CO rates is a better predictor of substitution patterns than female-specific CO rates, which can be interpreted as male-specific recombination having more impact on substitution patterns than female-specific recombination. This has been previously reported in the human lineage, which seems to indicate it is shared across mammals (Webster et al. 2005; Duret and Arndt 2008). We can put forward two hypotheses to explain these observations. First, the distribution of recombining regions along chromosomes is different for male- and female-specific recombination, both in the human genome and in the mouse genome (Myers et al. 2005; Paigen et al. 2008). Female recombining regions are more numerous and more homogeneously distributed along chromosomes than male recombining regions, however, male recombination hotspots are more active. This more heterogeneous distribution of recombination in males may lead to male-specific recombination rates better predicting substitution patterns than female-specific recombination rates. Second, meiotic recombination events cause the formation of Holliday Junctions that are solved either into crossovers or noncrossovers (Smith and Nicolas 1998; de Massy 2003; Baudat and de Massy 2007). Genetic maps available for the human and mouse genomes do not have enough resolution to show noncrossovers. It is possible that crossovers represent a greater proportion of recombination in males than in females. One alternative is to measure the frequency of double-strand breaks in genomic regions and use these as a proxy measure of meiotic recombination.
gBGC Is Weaker in the Mouse Lineage Compared with the Human Lineage
The effective population size of mice is around 30 times greater than that of humans: it is estimated to be around 20,000 in humans and around 600,000 in mouse (Keightley et al. 2005). gBGC should therefore be stronger in the mouse lineage compared with the human lineage because gBGC has a bigger impact in species with larger effective population sizes (Nagylaki 1983). However, the effect of gBGC appears to be weaker in mouse lineage compared with the human lineage. We cannot claim, however, that gBGC is generally absent in the mouse lineage as there are reported cases showing clear evidence of gBGC inside the mouse genome (Montoya-Burgos et al. 2003).
There are four possible explanations for this result. First, recombination rates are lower in the mouse genome compared with the human genome (Jensen-Seaman et al. 2004; Li and Freudenberg 2009), which will cause gBGC to be weaker in the mouse lineage compared with the human lineage. Second, it cannot be excluded that recombination events are repaired more often into crossovers than noncrossovers in the human genome compared with the mouse genome. This may cause CO rates to be a less accurate proxy of meiotic recombination in the mouse genome compared with the human genome. Third, it is possible that the heteroduplex length that forms during gene conversion is shorter in mouse than in human. This will cause gBGC to affect less bases in mouse compared with human. Finally, the mismatch repair mechanism could be less biased toward G and C bases in the mouse genome compared with the human genome. This will cause the fixation bias favoring G and C bases to be lower in mouse compared with humans.
We would like to point out that the fact that recombination rates evolve rapidly in mouse species could affect our results (Dumont et al. 2011). One way to solve this issue would be to study substitution patterns in the mouse lineage by comparing two closely related mouse species, using rat as an outgroup.
Substitution Patterns Are Predicted by a Combination of Factors in Both Human and Mouse Lineages
Principal component regression results show that S→W substitution rates in both lineages are mostly predicted by a component, which is a combination of different factors (GC-content, exon density, replication timing, transposable element densities). These results can be interpreted in different ways. First, it is possible that natural selections affect the fixation probabilities for the substitution rates we computed. Because we masked regions affected by natural selection in our windows (exons), we assume that it does not play a role on substitutions and that nucleotides are evolving neutrally in our windows. It is also possible that meiotic recombination influences the fixation probabilities of substitution rates through gBGC. However, because meiotic recombination is not the strongest predictor of these substitution rates and because it constitutes only a small fraction of this component, we assume that meiotic recombination has a low impact on fixation probabilities for S→W substitution rates. We then assume that these substitution rates are equal to mutation rates and therefore interpret these results as the influence of mutation on substitution patterns. We cannot tell, however, if the factors predicting S→W substitution rates have a direct impact on substitution patterns or if the associations we observe are not cause and effect associations.
CpG Odds Ratio Is the Main Predictor of W→S Substitution Rates in the Mouse Lineage
Our results show that in the mouse lineage, CpG odds ratio (the observed CpG frequency divided by the expected CpG frequency) is the main predictor of W→S substitution rates, which is not the case in the human lineage.
One might be tempted to interpret this results as due to CpG odds ratio being a proxy measure of meiotic recombination. A link between DNA methylation (which occurs on cytosines of CpG dinucleotides) and meiotic recombination has been described in the human genome (Sigurdsson et al. 2009). Moreover, in the mouse lineage, we observe an association between male CO rates and CpG odds ratio (partial correlation =0.14, P value < 10−7 when controlling for GC-content). However, our results show us that CpG odds ratio predicts W→S substitution rates independently of meiotic recombination.
First, it is possible that recombination decreases the CpG→TpG/CpA rate by protecting CpG dinucleotides from decaying into TpG or CpA dinucleotides. Because meiotic recombination is not the strongest predictor of the CpG →TpG/CpA substitution rate, meiotic recombination does not seem to protect CpG dinucleotides. Second, meiotic recombination could occur mostly in CpG-rich regions, for example, CpG islands. However, no link between recombination hotspots and CpG islands has been proposed in the mouse or the human genome (Myers et al. 2005; Paigen et al. 2008). Also, the DNA motif associated with hotspot activity is not CpG rich (Myers et al. 2008).
Furthermore, in principal component analysis results, meiotic recombination and CpG odds ratio contribute to two independent components, only the latter component predicts W→S substitution rates. This shows that in the mouse lineage, CpG odds ratio predicts substitution patterns independently of meiotic recombination.
We cannot tell, however, if CpG content has a direct influence on W→S substitution rates or if CpG content serves as a proxy measure for genomic factors, we did not include in our model or if there is no cause and effect relationship between CpG content and W→S substitution rates. Authors have proposed that, in the human lineage, CpG content and substitution rates are associated through different mechanisms such as chromatin opening linked to gene expression or error-prone repair of T:G mismatches by different DNA polymerases (Walser and Furano 2010). Moreover, they have found no evidence that this association is mediated through fixation probabilities of mutations. The relationship between CpG content, substitution rates, and other genomic factors needs to be further investigated in both human and mouse lineages.
We have found that in contrast with the human lineage, gBGC is weak in the mouse lineage and that CpG odds ratio, not meiotic recombination is the strongest predictor of W→S substitution rates. This reveals that isochore structures are evolving differently in both human and mouse lineages and seems to indicate that this is the result of substitution patterns being under different influences in those lineages.
Supplementary Material
Supplementary figures 1–7 and tables 1–8 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Brian Cusack, Laurent Duret, and Paz Polak for fruitful discussions and three anonymous referees for their useful comments. Y.C. is funded by a scholarship from the International Max Planck Research School for Computational Biology and Scientific Computing.
References
- Arndt PF, Burge CB, Hwa T. DNA sequence evolution with neighbor-dependent mutation. J Comput Biol. 2003;10(3–4):313–322. doi: 10.1089/10665270360688039. [DOI] [PubMed] [Google Scholar]
- Arndt PF, Hwa T. Identification and measurement of neighbor-dependent nucleotide substitution processes. Bioinformatics. 2005;21(10):2322–2328. doi: 10.1093/bioinformatics/bti376. [DOI] [PubMed] [Google Scholar]
- Arndt PF, Hwa T, Petrov DA. Substantial regional variation in substitution rates in the human genome: importance of GC content, gene density, and telomere-specific effects. J Mol Evol. 2005;60(6):748–763. doi: 10.1007/s00239-004-0222-5. [DOI] [PubMed] [Google Scholar]
- Baudat F, de Massy B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 2007;15(5):565–577. doi: 10.1007/s10577-007-1140-3. [DOI] [PubMed] [Google Scholar]
- Belle EMS, Duret L, Galtier N, Eyre-Walker A. The decline of isochores in mammals: an assessment of the GC content variation along the mammalian phylogeny. J Mol Evol. 2004;58(6):653–660. doi: 10.1007/s00239-004-2587-x. [DOI] [PubMed] [Google Scholar]
- Bernardi G. Isochores and the evolutionary genomics of vertebrates. Gene. 2000;241(1):3–17. doi: 10.1016/s0378-1119(99)00485-0. [DOI] [PubMed] [Google Scholar]
- Bernardi G. The neoselectionist theory of genome evolution. Proc Natl Acad Sci U S A. 2007;104(20):8385–8390. doi: 10.1073/pnas.0701652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi G, et al. The mosaic genome of warm-blooded vertebrates. Science. 1985;228(4702):953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Bill CA, Duran WA, Miselis NR, Nickoloff JA. Efficient repair of all types of single-base mismatches in recombination intermediates in Chinese hamster ovary cells. Competition between long-patch and G-T glycosylase-mediated repair of G-T mismatches. Genetics. 1998;149(4):1935–1943. doi: 10.1093/genetics/149.4.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol. 1978;118(1):49–60. doi: 10.1016/0022-2836(78)90243-7. [DOI] [PubMed] [Google Scholar]
- Brown TC, Jiricny J. Different base/base mispairs are corrected with different efficiencies and specificities in monkey kidney cells. Cell. 1988;54(5):705–711. doi: 10.1016/s0092-8674(88)80015-1. [DOI] [PubMed] [Google Scholar]
- Chen CL, et al. Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome Res. 2010;20(4):447–457. doi: 10.1101/gr.098947.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Przeworski M. An evolutionary view of human recombination. Nat Rev Genet. 2007;8(1):23–34. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- de Massy B. Distribution of meiotic recombination sites. Trends Genet. 2003;19(9):514–522. doi: 10.1016/S0168-9525(03)00201-4. [DOI] [PubMed] [Google Scholar]
- de Villena FPM, Sapienza C. Recombination is proportional to the number of chromosome arms in mammals. Mamm Genome. 2001;12(4):318–322. doi: 10.1007/s003350020005. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Raval A, Wilke CO. A single determinant dominates the rate of yeast protein evolution. Mol Biol Evol. 2006;23(2):327–337. doi: 10.1093/molbev/msj038. [DOI] [PubMed] [Google Scholar]
- Dumont BL, White MA, Steffy B, Wiltshire T, Payseur BA. Extensive recombination rate variation in the house mouse species complex inferred from genetic linkage maps. Genome Res. 2011;21(1):114–125. doi: 10.1101/gr.111252.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4(5):e1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D, Gautier C. Statistical analysis of vertebrate sequences reveals that long genes are scarce in GC-rich isochores. J Mol Evol. 1995;40(3):308–317. doi: 10.1007/BF00163235. [DOI] [PubMed] [Google Scholar]
- Duret L, Semon M, Piganeau G, Mouchiroud D, Galtier N. Vanishing GC-rich isochores in mammalian genomes. Genetics. 2002;162(4):1837–1847. doi: 10.1093/genetics/162.4.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A. Recombination and mammalian genome evolution. Proc Biol Sci. 1993;252(1335):237–243. doi: 10.1098/rspb.1993.0071. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Hurst LD. The evolution of isochores. Nat Rev Genet. 2001;2(7):549–555. doi: 10.1038/35080577. [DOI] [PubMed] [Google Scholar]
- Filipski J. Why the rate of silent codon substitutions is variable within a vertebrate’s genome. J Theor Biol. 1988;134(2):159–164. doi: 10.1016/s0022-5193(88)80199-1. [DOI] [PubMed] [Google Scholar]
- Galtier N, Duret L. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 2007;23(6):273–277. doi: 10.1016/j.tig.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Galtier N, Duret L, Glémin S, Ranwez V. GC-biased gene conversion promotes the fixation of deleterious amino acid changes in primates. Trends Genet. 2009;25(1):1–5. doi: 10.1016/j.tig.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Galtier N, Piganeau G, Mouchiroud D, Duret L. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics. 2001;159(2):907–911. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannelli F, Anagnostopoulos T, Green PM. Mutation rates in humans. II. Sporadic mutation-specific rates and rate of detrimental human mutations inferred from hemophilia B. Am J Hum Genet. 1999;65(6):1580–1587. doi: 10.1086/302652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428(6982):493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Hiratani I, et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6(10):e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson A, Ladoukakis E, Eyre-Walker A. Cryptic variation in the human mutation rate. PLoS Biol. 2009;7(2):e1000027. doi: 10.1371/journal.pbio.1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huchon D, et al. Multiple molecular evidences for a living mammalian fossil. Proc Natl Acad Sci U S A. 2007;104(18):7495–7499. doi: 10.1073/pnas.0701289104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen-Seaman MI, et al. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 2004;14(4):528–538. doi: 10.1101/gr.1970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback DB. Chromosome-size dependent control of meiotic recombination in humans. Nat Genet. 1996;13(1):20–21. doi: 10.1038/ng0596-20. [DOI] [PubMed] [Google Scholar]
- Keightley PD, Lercher MJ, Eyre-Walker A. Evidence for widespread degradation of gene control regions in hominid genomes. PLoS Biol. 2005;3(2):e42. doi: 10.1371/journal.pbio.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lercher MJ, Urrutia AO, Pavlícek A, Hurst LD. A unification of mosaic structures in the human genome. Hum Mol Genet. 2003;12(19):2411–2415. doi: 10.1093/hmg/ddg251. [DOI] [PubMed] [Google Scholar]
- Li W, Freudenberg J. Two-parameter characterization of chromosome-scale recombination rate. Genome Res. 2009;19(12):2300–2307. doi: 10.1101/gr.092676.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G. Biased gene conversion: implications for genome and sex evolution. Trends Genet. 2003;19(6):330–338. doi: 10.1016/S0168-9525(03)00116-1. [DOI] [PubMed] [Google Scholar]
- Meunier J, Duret L. Recombination drives the evolution of GC-content in the human genome. Mol Biol Evol. 2004;21(6):984–990. doi: 10.1093/molbev/msh070. [DOI] [PubMed] [Google Scholar]
- Mevik BH, Wehrens R. The pls package: principal component and partial least squares regression in R. J Stat Software. 2007;18(2):1–24. [Google Scholar]
- Montoya-Burgos JI, Boursot P, Galtier N. Recombination explains isochores in mammalian genomes. Trends Genet. 2003;19(3):128–130. doi: 10.1016/S0168-9525(03)00021-0. [DOI] [PubMed] [Google Scholar]
- Mouchiroud D, Gautier C, Bernardi G. The compositional distribution of coding sequences and DNA molecules in humans and murids. J Mol Evol. 1988;27(4):311–320. doi: 10.1007/BF02101193. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. Oct. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310(5746):321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40(9):1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- Nagylaki T. Evolution of a finite population under gene conversion. Proc Natl Acad Sci U S A. 1983;80(20):6278–6281. doi: 10.1073/pnas.80.20.6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Takeda H, Kohara Y, Morishita S. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 2007;17(9):1254–1265. doi: 10.1101/gr.6316407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, et al. The recombinational anatomy of a mouse chromosome. PLoS Genet. 2008;4(7):e1000119. doi: 10.1371/journal.pgen.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M, Siomos MF, Nasmyth K. Un ménage à quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112(4):423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- Pollard KS, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443(7108):167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Poux C, Chevret P, Huchon D, de Jong WW, Douzery EJP. Arrival and diversification of caviomorph rodents and platyrrhine primates in South America. Syst Biol. 2006;55(2):228–244. doi: 10.1080/10635150500481390. [DOI] [PubMed] [Google Scholar]
- Pozzoli U, et al. Both selective and neutral processes drive GC content evolution in the human genome. BMC Evol Biol. 2008;8:99. doi: 10.1186/1471-2148-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M, Gautier C, Mouchiroud D. Evolution of isochores in rodents. Mol Biol Evol. 1997;14(8):823–828. doi: 10.1093/oxfordjournals.molbev.a025823. [DOI] [PubMed] [Google Scholar]
- Romiguier J, Ranwez V, Douzery EJP, Galtier N. Contrasting GC-content dynamics across 33 mammalian genomes: relationship with life-history traits and chromosome sizes. Genome Res. 2010;20(8):1001–1009. doi: 10.1101/gr.104372.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, et al. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20(6):761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S, et al. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 2006;4(12):e395. doi: 10.1371/journal.pbio.0040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson MI, Smith AV, Bjornsson HT, Jonsson JJ. HapMap methylation-associated SNPs, markers of germline DNA methylation, positively correlate with regional levels of human meiotic recombination. Genome Res. 2009;19(4):581–589. doi: 10.1101/gr.086181.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KN, Nicolas A. Recombination at work for meiosis. Curr Opin Genet Dev. 1998;8(2):200–211. doi: 10.1016/s0959-437x(98)80142-1. [DOI] [PubMed] [Google Scholar]
- Smith NGC, Eyre-Walker A. The compositional evolution of the murid genome. J Mol Evol. 2002;55(2):197–201. doi: 10.1007/s00239-002-2316-2. [DOI] [PubMed] [Google Scholar]
- Tyekucheva S, et al. Human–macaque comparisons illuminate variation in neutral substitution rates. Genome Biol. 2008;9(4):R76. doi: 10.1186/gb-2008-9-4-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser JC, Furano AV. The mutational spectrum of non-CpG DNA varies with CpG content. Genome Res. 2010;20(7):875–882. doi: 10.1101/gr.103283.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser JC, Ponger L, Furano AV. CpG dinucleotides and the mutation rate of non-CpG DNA. Genome Res. 2008;18(9):1403–1414. doi: 10.1101/gr.076455.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MT, Smith NGC, Hultin-Rosenberg L, Arndt PF, Ellegren H. Male-driven biased gene conversion governs the evolution of base composition in human alu repeats. Mol Biol Evol. 2005;22(6):1468–1474. doi: 10.1093/molbev/msi136. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Sharp PM, Li WH. Mutation rates differ among regions of the mammalian genome. Nature. 1989;337(6204):283–285. doi: 10.1038/337283a0. [DOI] [PubMed] [Google Scholar]