Abstract
Chlamydia muridarum and Chlamydia trachomatis mouse models of genital infection have been used to study chlamydial immunity and vaccine development. To assess the protective role of CD4+ T cells in resolving C. trachomatis and C. muridarum genital tract infections, we used the female mouse model and evaluated infection in the presence and absence of CD4+ T cells. In contrast to C. muridarum infection, C. trachomatis infection was unaltered in the absence of CD4+ T cells. Mice infected with C. trachomatis developed protective immunity to re-challenge, but unlike C. muridarum infection, optimum resistance required multiple infectious challenges, despite the generation of adaptive serum and local chlamydial specific immune responses. Thus, understanding the chlamydial pathogenic and host immunologic factors that result in a diminished protective role for CD4+ T cells in C. trachomatis murine infection might lead to new insights important to human immunity and vaccine development.
The obligate intracellular bacterial pathogen Chlamydia trachomatis causes an estimated 92 million new cases of sexually transmitted infection each year worldwide [1]. Lasting and durable control of these infections, and prevention of the deleterious consequences of infection, will require the development of a vaccine. A limitation to the development of a vaccine relates to the incomplete understanding of the natural history of human chlamydial genital infection. Diagnosis of infection mandates treatment, and thus it is not fully understood which responses are necessary for protective immunity and when during the course of infection those responses mature and become effective. Therefore, Chlamydia infection of animals—primarily mice—has been used as a surrogate to study the natural history of genital infection and the development of adaptive immunity. Two mouse models of infection have been used extensively to study chlamydial genital infection and immunity: one model uses the mouse pathogen, Chlamydia muridarum; and the other uses the human pathogen, C. trachomatis. The need for adaptive immunity in resolving murine C. muridarum infection is unequivocal [2], but the role of adaptive immunity in resolving murine C. trachomatis infection is ambiguous [3, 4], even though primary infection elicits adaptive immune responses and a level of resistance to reinfection appears to develop [5–10].
In this study we sought to directly examine the contribution of adaptive immunity (CD4+ T cells) in resolving primary murine C. trachomatis genital infection. Because previous studies have clearly demonstrated that C. trachomatis genital infection of mice is highly dependent on mouse strain [5, 11], it was crucial to choose a mouse strain that supported a robust infection that persisted for sufficient time to allow for the development of adaptive immune responses. Accordingly, we chose to use the C3H/HeJ strain of mice. Mice of the C3H background have been shown to be susceptible to C. trachomatis genital infection, and the C3H/HeJ strain in particular has impaired innate immunity due to a nonfunctional Toll-like receptor (TLR) 4 [12]. Those combined attributes should favor the development of an infection of sufficient intensity and duration to delineate the role of adaptive immunity in resolving C. trachomatis genital infection. We show here that CD4+ T cell–mediated adaptive immunity is not required to control primary murine C. trachomatis genital infection.
MATERIALS AND METHODS
Mice
Female C3H/HeJ mice were purchased from the Jackson Laboratories and maintained in the animal facilities at the University of Arkansas for Medical Sciences (Little Rock). Mice aged 6–10 weeks were used for these studies. All experimental procedures were performed in accordance with institutional policies for animal health and well being and were approved by the Institutional Animal Care and Use Committee.
Bacteria
C. muridarum (Weiss strain; formerly known as C. trachomatis strain mouse pneumonitis) and C. trachomatis serovars A (strain 2497) [13], D (strain UW-3/CX), and L2 (strain 434) were grown in HeLa 229 cells and purified by density gradient centrifugation [14].
Genital Infection and Enumeration of Chlamydia
Mice were infected and chlamydiae were enumerated as previously reported [15]. In brief, at 10 and 3 days prior to infectious challenge, mice were injected subcutaneously with 2.5 mg of medroxyprogesterone acetate (SICOR Pharmaceuticals). Before infectious challenge, vaginal vaults of progesterone treated mice were gently swabbed with a calcium alginate tipped swab (Fisher Scientific) to remove excess mucous. Mice were challenged by placing 5 μL containing either 5 × 104 inclusion forming units (IFUs) of C. muridarum, or 1 × 105 IFUs of C. trachomatis serovar A, D, or L2 into the vaginal vault. Genital infection was assessed by swabbing the vaginal vault (rotating the swab 8 times clockwise and 8 times counter-clockwise), placing the swab into a tube containing 1.0 mL of SPG (0.25 M sucrose/10 mmol/L sodium phosphate/5 mmol/L l-glutamic acid; pH, 7.2) and two 4-mm glass beads, shaking for 5 min, and then inoculating 0.3 mL of appropriately diluted samples into duplicate wells of a 48-well plate containing HeLa 229 cell monolayers. Plates were centrifuged at 780 × g for 1 h at 37°C and rested for 30 min at 37°C, and the inoculum was removed and replaced with Dulbecco's Modified Eagles Medium containing 10% fetal bovine serum, 10 μg/mL gentamicin, and 1 μg/mL cycloheximide. After 30 h of incubation at 37°C, the supernatant was removed, cells were fixed with methanol, and the number of IFUs was visualized by indirect immunofluorescent staining using a chlamydial lipopolysaccharide mAb EVI-H1 [16].
Antibody Analysis
Serum and vaginal washes were collected and analyzed by enzyme-linked immunosorbent assay (ELISA) for Chlamydia-specific antibody, as described elsewhere [15]. All ELISAs were performed using elementary bodies (EBs) matched to the infecting Chlamydia strain (eg, serum and vaginal washes from serovar A–infected mice were analyzed using serovar A EBs) as ELISA antigen. Serum titers are expressed as the inverse of the highest serum dilution giving an absorbance (optical density at 405 nm [OD405]) of ≥0.25, which represents an OD405 value that is 3 standard deviations greater than the mean OD405 of preimmune serum. For vaginal washes, data are reported as the number of mice positive for Chlamydia antibody/total number tested. Positive responses are defined as an OD405 ≥ 0.1, which represents and OD405 value that is 3 standard deviations greater than the mean OD405 of preimmune vaginal washes.
CD4+ T Cell Depletion
Mice were depleted of CD4+ T cells as previously described [17, 18]. Four hundred micrograms of anti-CD4 mAb (clone GK1.5) was injected intraperitoneally according to the following schedule (day 0 represents day of infection): days -6, -5, -4, -1, 2, 5, 8, 11, 14, 17, 20, 23, 26, 29, 32, 35, 38, and 41. Following this treatment regimen, CD4+ T cells remain depleted throughout the test period, as noted by a >99% depletion of those cells by fluorescent-activated cell sorting analysis and greatly diminished antigen-specific antibody and delayed-type hypersensitivity responses [17, 18]. We have found that, if the aforementioned treatment regimen is followed, mice can be functionally depleted of CD4+ T cells (as assessed by greatly diminished antibody and cell-mediated immune (CMI) responses) for no longer than 50 days, after which time helper T cell functions return (antigen-specific antibody and CMI responses), even with continued anti-CD4 treatment. Therefore, all experiments using CD4-depleted mice are terminated 40–43 days after chlamydial infection.
Histological Analsysis
Mice were killed 7 and 14 days after primary infection. Genital tracts were removed and processed for histopathological analysis [15]. Thin sections were stained with hematoxylin and eosin and evaluated by a veterinary pathologist.
Statistical Analysis
Two-way analysis of variance with Bonferroni posttest was used to analyze the differences in IFU counts (infection) between groups. The Student t test was used to identify differences between serological responses.
RESULTS
Course of C. muridarum Genital Infection in C3H/HeJ (TLR4-Deficient) Mice
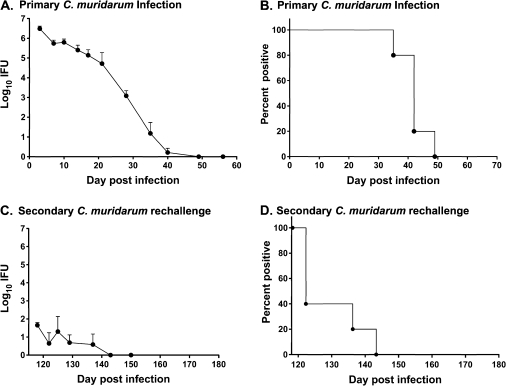
The primary objective of the study was to use the mouse model of C. trachomatis genital infection to assess the impact of CD4+ T cell responses in resolving infection. Because previous studies suggest that innate immunity alone can restrict the establishment of a robust C. trachomatis infection in some strains of mice [4], we selected the TLR4-deficient C3H/HeJ mouse strain to use in our studies. By using C3H/HeJ mice rather than the fully immunocompetent C3H/HeN strain of mice, we expected that the innate immune defect would facilitate infection, without negatively impacting the development of protective adaptive immunity. To show that the TLR4 defect did not negatively impact the development of protective adaptive immunity, we assessed infection and immunity to reinfection using the C. muridarum genital infection model. This infection model has clearly demonstrated the key role for adaptive immunity, and CD4+ T cells specifically, in infection resolution and immunity to reinfection [17, 18]. Genital challenge of C3H/HeJ mice with C. muridarum produced a robust infection, characterized by the shedding of >106 IFUs soon after challenge, with complete resolution by 5–7 weeks (Figure 1A and B). Mice that had resolved a primary genital infection were markedly protected from reinfection, as demonstrated by a significant reduction in bacterial shedding (>4 log10 IFU reduction; P < .001 at all time points) (Figure 1C) and shortened duration of infection (3–28 days) (Figure 1D). These results confirmed that the TLR4 defect did not negatively impact the course of infection or the development of adaptive immunity to reinfection.
Figure 1.
Course of primary (A and B) and secondary (C and D) Chlamydia muridarum infection in female C3H/HeJ mice. Medroxyprogesterone acetate–treated mice were challenged vaginally with C. muridarum, and the course of infection was monitored by enumerating chlamydiae recovered from cervicovaginal swabs collected at various times following challenge. Data are presented as mean inclusion forming units (IFUs) ± standard error of the mean for 6 mice (A and C) and as the percentage of culture-positive animals (B and D). “Day post infection” refers to the day after primary infectious challenge.
Course of Primary, Secondary and Tertiary C. trachomatis Genital Infection in C3H/HeJ (TLR4-Deficient) Mice
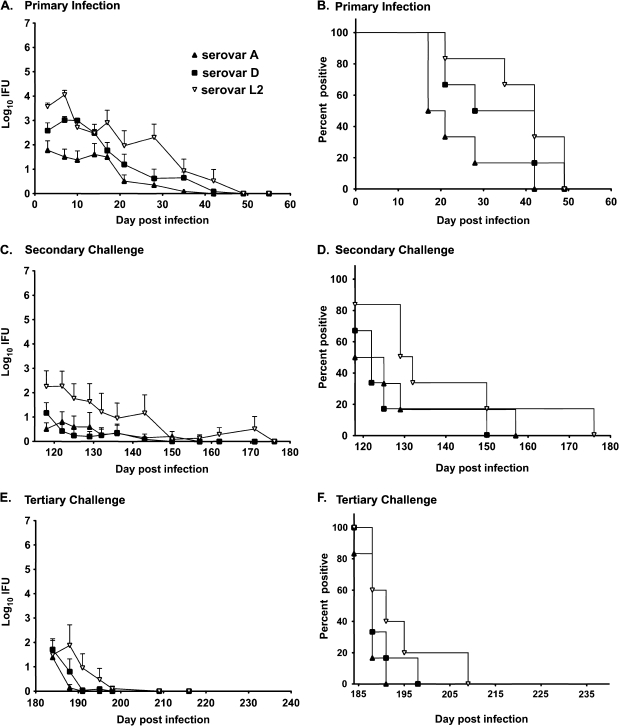
Because the TLR4-deficient C3H/HeJ mouse strain proved to be a useful model for assessing adaptive immunity to chlamydial genital infection, we next evaluated the ability of C3H/HeJ mice to resolve primary C. trachomatis infection and to resist reinfection. Mice were challenged with C. trachomatis serovar A, D, or L2, and the course of infection was monitored by enumerating IFUs. As a group, primary genital infection with C. trachomatis serovar A, D, or L2 resulted in shedding of fewer chlamydiae, compared with infection with C. muridarum (approximately 2–4 log10 IFU lower; P < .001) (Figure 1A and Figure 2A). Mice challenged with serovar A shed fewer chlamydiae than those challenged with either serovar D or L2 (Figure 2A; see legend for P values), and a large proportion of serovar A–infected mice resolved the infection more quickly than mice infected with serovar D or L2 (P < .05) (Figure 2B). Histological evaluation of genital tracts 7 and 14 days after primary infection revealed minimal inflammation and no evidence of lymphocyte clusters (data not shown), which was not unlike that which has been reported previously [19].
Figure 2.
Course of primary Chlamydia trachomatis genital infection (A and B), and secondary (C and D), and tertiary infectious challenge (E and F) in female C3H/HeJ mice. Medroxyprogesterone acetate–treated mice were challenged vaginally with either C. trachomatis serovar A (▴), D (▪), or L2 (▿), and infection was monitored by enumerating chlamydiae recovered from cervicovaginal swabs. Data are presented as mean inclusion forming units (IFUs) ± standard error of the mean for 6 mice per group (A, C, and E) and as the percentage of culture-positive animals (B, D, and F). “Day post infection” refers to the day following primary infectious challenge. For clarity, we have listed below the various statistical comparisons between the infection curves rather than on the figures. Primary infection curves: for A vs D, P < .01 for days 7 and 10; for A vs L2, P < .001 for days 3, 7, and 28, P < .01 for day 21, and P < .05 for days 10 and 17; for L2 vs D, P < .01 for day 28. Comparison of primary infection (1°) with secondary infection (2°) with the homologous serovar (eg, serovar A primary infections vs serovar A secondary infection): for 1° serovar A vs 2° serovar A, P < .05 for days 3 and 17; for 1° serovar D vs 2° serovar D, P < .001 for days 3, 7, 10, 14 and 17; for 1° serovar L2 vs 2° serovar L2, P was not significant at any time point. For comparison of primary infection vs tertiary infection with the homologous serovar for days 3, 7, 10, and 14 days post infection, P <0.01 for all serovars.
Significant protection from reinfection was observed when mice were rechallenged with the homologous C. trachomatis serovar (Figure 2). This was particularly evident when comparing primary infection to tertiary infection (eg, primary serovar A infection versus tertiary infection with serovar A) (Figure 2A, 2C, and 2E), although some protection (lower bacterial shedding) was observed on specific days post-secondary infection (see Figure 2 legend for P values). Thus, vaginal challenge of female C3H/HeJ mice with C. trachomatis (serovars A, D, or L2) results in an infection having a similar duration to that observed with C. muridarum infection, but far fewer bacteria are shed throughout the course of infection. Furthermore, the level of immunity elicited following primary C. trachomatis infection is far less than that elicited following C. muridarum infection. A single C. muridarum infection results in marked protective immunity, whereas 2 infections are necessary to stimulate significant immunity in the C. trachomatis infection model.
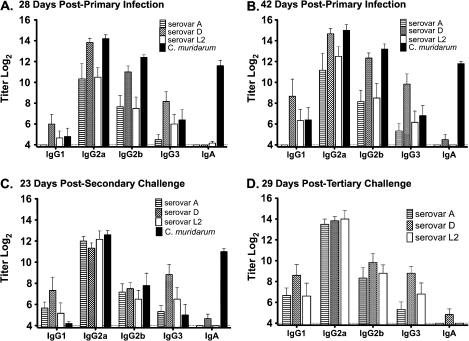
Characteristics of the Chlamydia-Specific Antibody Response
Serum and vaginal washes were collected and analyzed by ELISA using EBs matched to the infecting serovar as antigen. Although there were some differences in the magnitude of the responses among and between the serovars, compared with C. muridarum, in general the responses were remarkably similar. All infections resulted in high-titer immunoglobulin (Ig) G2a after primary infection, and the magnitude of those responses remained unchanged following secondary and tertiary infection (Figure 3B, 3C, and 3D). The IgG1, IgG2b, and IgG3 responses were somewhat more variable during primary infection, with lower titer responses in mice infected with either serovar A or L2. Overall, the antibody responses elicited by infection with C. trachomatis serovars and C. muridarum were consistent, with the exception of the serum IgA response. Notably, infection with C. trachomatis serovar A or L2 failed to elicit detectable serum anti-Chlamydia IgA even after 3 infectious challenges, and infection with serovar D produced a very unremarkable IgA response after primary infection, and this response did not increase in magnitude upon multiple reinfections. In contrast, C. muridarum infection elicited a substantial and long-lasting IgA response (Figure 3).
Figure 3.
Immunoglobulin class and subclass specificity of the anti-Chlamydia antibody response. Serum samples collected from individual mice (6 per group) infected with Chlamydia trachomatis serovar A ( ), D (
), D ( ), or L2 (□) or with Chlamydia muridarum (▪) were tested for anti-Chlamydia antibody using the homologous infecting Chlamydia strain as antigen in the enzyme-linked immunosorbent assay (eg, serum from serovar A–infected mice was tested against serovar A EBs). Data are presented as the mean anti-Chlamydia titer ± standard error of the mean for the indicated class and subclass immunoglobulins of serum collected 28 days after primary infection (A), 42 days after primary infection (B), 23 days after secondary challenge (C), and 29 days after tertiary challenge (D). Ig, immunoglobulin.
), or L2 (□) or with Chlamydia muridarum (▪) were tested for anti-Chlamydia antibody using the homologous infecting Chlamydia strain as antigen in the enzyme-linked immunosorbent assay (eg, serum from serovar A–infected mice was tested against serovar A EBs). Data are presented as the mean anti-Chlamydia titer ± standard error of the mean for the indicated class and subclass immunoglobulins of serum collected 28 days after primary infection (A), 42 days after primary infection (B), 23 days after secondary challenge (C), and 29 days after tertiary challenge (D). Ig, immunoglobulin.
Vaginal washes were also analyzed for Chlamydia-specific antibody (Table 1). As shown previously [15], vaginal washes from mice infected with C. muridarum were positive for anti-chlamydial IgA early after infection (14 days), with IgG2a and IgG2b becoming positive later (28 days). Conversely, only a fraction of the C. trachomatis–infected mice had detectable anti-chlamydial IgA at 28 days after primary infection, and none were positive for IgG2a or IgG2b. After tertiary infection, nearly all C. trachomatis–infected mice had anti-chlamydial IgG2a in vaginal washes, and most were also positive for IgA. Thus, although C. trachomatis infection does not stimulate heightened systemic anti-chlamydial IgA responses, infection does result in the local production of antigen-specific IgA, albeit delayed.
Table 1.
Anti-Chlamydia Antibody Responses in Vaginal Washes
| Antibody response (no. of animals positive/total no. tested) |
|||||||||
| Mouse infectiona | 14 days after primary challenge |
28 days after primary challenge |
49 days after tertiary challenge |
||||||
| IgG2a | IgG2b | IgA | IgG2a | IgG2b | IgA | IgG2a | IgG2b | IgA | |
| Chlamydia trachomatis A | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 6/6 | 0/6 | 4/6 |
| C. trachomatis D | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 2/4 | 6/6 | 3/6 | 5/6 |
| C. trachomatis L2 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 1/3 | 5/6 | 0/6 | 2/6 |
| Chlamydia muridarum | 0/3 | 0/3 | 3/3 | 3/3 | 1/3 | 3/3 | nt | nt | nt |
NOTE. Vaginal washes were analyzed for antibody responses to EBs of the infecting chlamydial strain. Immunoglobulin (Ig) G1 and IgG3 responses are not included as vaginal washes are routinely negative for those antibody subclasses.
Mice were infected with either C. trachomatis serovar A, D, or L2 or C. muridarum. To obtain an accurate depiction of vaginal wash antibodies, washes were obtained from independent groups of mice that had not been previously cultured, which is the basis for different group sizes for primary and tertiary infection time points.
The Role of CD4+ T Cells in Resolving C. trachomatis Genital Infection
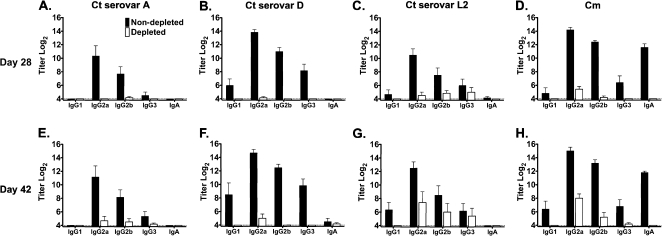
To directly test the role of CD4+ T cell–mediated adaptive immunity in resolving primary C. trachomatis genital infection, the course of infection was monitored in CD4+ T cell–depleted and –nondepleted C3H/HeJ mice. Depletion of CD4+ T cells has previously been shown to inhibit chlamydiae-specific cell-mediated immune responses (delayed type hypersensitivity) and T cell–dependent antibody responses [18, 20]. Using specific antibody responses as a measure of adaptive immunity, we show that CD4+ T cell depletion significantly inhibited the production of chlamydiae-specific antibody responses (Figure 4). Although Chlamydia-specific antibody responses began to develop by 42 days after infection (specifically in mice infected with C. muridarum and C. trachomatis serovar L2), those responses coincided with the diminished effectiveness of anti-CD4 depletion at this late time point.
Figure 4.
Serologic response of CD4+ T cell–depleted (□) and nondepleted (▪) (C3H/HeJ) mice after genital infection with Chlamydia trachomatis (Ct) A serovar (A and E), D serovar (B and F), or L2 serovar (C and G) or with C. muridarum (Cm; D and H) (5–6 mice per group). Antibody responses were measured at days 28 (A–D) and 42 (E–H) of infection. Serum samples were analyzed by enzyme-linked immunosorbent assasy for immunoglobulin (Ig) class/subclass antibody responses using the homologous infecting Chlamydia strain as antigen. Data are presented as the mean anti-Chlamydia titer ± standard error of the mean.
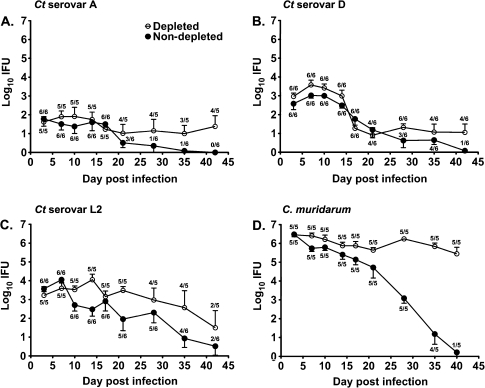
Depletion of CD4+ T cells did not significantly impact (P > .05) the course of C. trachomatis serovar A (Figure 5A), D (Figure 5B), or L2 (Figure 5C) genital infection. At no time point was bacterial shedding from CD4-depleted mice significantly different (P > .05) from untreated mice, regardless of the infecting serovar. Conversely, depletion of CD4+ T cells markedly impacted the ability of mice to resolve C. muridarum genital infection (Figure 5D) [17]. CD4-depleted C. muridarum–infected mice remain infected during the period of depletion and shed significantly greater numbers of bacteria during the typical period of infection resolution (days 21–40; P < .001).
Figure 5.
The course of Chlamydia trachomatis (Ct) serovar A (A), D (B), and L2 (C) or Chlamydia muridarum (D) genital infection in CD4+ T cell–depleted ( ) and nondepleted (•) C3H/HeJ mice. Depleted mice were treated with anti-CD4 prior to and throughout the course of infection. Data are presented as mean inclusion forming units (IFUs) ± standard error of the mean for 5–6 mice per group. The number of culture-positive mice/total mice is indicated for each time point.
) and nondepleted (•) C3H/HeJ mice. Depleted mice were treated with anti-CD4 prior to and throughout the course of infection. Data are presented as mean inclusion forming units (IFUs) ± standard error of the mean for 5–6 mice per group. The number of culture-positive mice/total mice is indicated for each time point.
DISCUSSION
A key observation emerging from numerous mouse model studies is that the characteristics of C. trachomatis genital infection are quite different from those of C. muridarum infection. For example, compared with C. muridarum genital infection C. trachomatis infection is characterized by the shedding of far fewer infectious bacteria and by less genital tract inflammation, and the magnitude and duration of infection are strongly influenced by mouse strain. To study adaptive immunity to C. trachomatis infection in the mouse we chose a strain of mice (eg, C3H/HeJ), which when challenged with C. trachomatis, produces an infection of sufficient magnitude and duration to elicit adaptive immune responses. Using the infection-susceptible C3H/HeJ strain of mice, we made 3 key observations important to understanding immunity to murine C. trachomatis infection: (1) unlike the unconditional requirement for CD4+ T cell–mediated responses in resolving C. muridarum genital infection, the resolution of murine C. trachomatis genital infection was far less dependent on CD4+ T cells; (2) although CD4+ T cells are not necessary to resolve primary infection, adaptive immune responses do develop, and a marked level of immunity develops following multiple genital infections; and (3) systemic and local anti-chlamydiae IgA responses are mostly absent or delayed.
Our results (Figure 3), as well as those of other investigators, demonstrate that genital infection of mice with C. trachomatis elicits significant and substantial cellular and humoral adaptive immune responses [3, 5–7, 9]. Yet unlike C. muridarum primary infection, which elicits long-lived protective immunity, the extent of the protective response elicited following primary C. trachomatis infection was not significantly different from that of naïve mice (Figure 2C and 2D). Immunity to C. trachomatis does, however, increase significantly following the resolution of a second genital infection (Figure 2E and 2F). The lack of correlation between the development of robust adaptive immune responses and inadequate protective immunity is rather curious. Why is it that C. trachomatis–infected mice do not exhibit greater immunity following a primary genital infection? The differing characteristics of genital infection may be key to understanding the difference between the striking protective immunity elicited by C. muridarum versus C. trachomatis infection.
C. muridarum produces a rapidly progressing highly inflammatory infection, whereas murine C. trachomatis infection produces nominal inflammation. In C. muridarum infection, the marked acute inflammation that is observed in genital tissues early during infection subsides and is replaced by mononuclear cells as infection resolves [15]. Mononuclear cells localize throughout the genital tissues, but a prominent feature is the clustering of CD4+ T cells [21]. C. trachomatis infection is less inflammatory, and it apparently does not produce the CD4+ T cell clusters, which perhaps explains why mice are not protected from reinfection after primary C. trachomatis infection.
The difference in the local inflammation elicited by C. muridarum and C. trachomatis is somewhat surprising given that they share a high degree of genomic synteny [22, 23]. However, one exception is a polymorphic region of the chromosome termed the plasticity zone. Among the genes found in the plasticity zone is the chlamydial cytotoxin, which shares homology with the large clostridial toxins. C. muridarum has 3 copies of the intact cytotoxin gene, whereas C. trachomatis serovars have multiple mutations in a single copy of the gene, resulting in a truncated cytotoxin [24]. Large clostridial toxins are known inducers of potent inflammatory responses [25–27]; therefore, the marked inflammatory response elicited by C. muridarum might result from the effects of the cytotoxin on the genital tissues. The cytotoxin, acting as a mucosal adjuvant, may then help to orchestrate the long-lived protective immunity that follows C. muridarum infection. Conversely, the inflammation elicited by the truncated C. trachomatis cytotoxin during primary infection may be insufficient to coordinate a potent protective response locally. Although the cytotoxin premise is intriguing, it is also notable that C. trachomatis induces a dominant regulatory T cell population that dampens T cell responses [28]. It is not known whether this response is specific for C. trachomatis or whether C. muridarum causes a similar response. However, an alternative explanation for the dampened protective immunity elicited after C. trachomatis infection is that C. trachomatis secretes or releases a specific factor(s) that targets these regulatory cells.
In addition to the possible role that the chlamydial cytotoxin may play in modulating the local inflammatory response, the cytotoxin has also been proposed to have a role in limiting the growth of C. trachomatis in mouse cells in vitro [29, 30]. In that model, interferon-γ–activated epithelial cells produce p47 GTPases that kill C. trachomatis; C. muridarum, however, survives in that environment, because the greater abundance of cytotoxin produced by C. muridarum inactivates and overcomes the detrimental effects of the p47 GTPases. Whether this protective phenomenon functions in vivo is not known. Furthermore, others have provided intriguing evidence to suggest that additional virulence factors contribute to the pathogenesis of C. trachomatis infection in the mouse [19].
One feature of the adaptive immune response that differs between genital C. muridarum infection and C. trachomatis infection is the delayed production of Chlamydia-specific IgA (Figure 3). C. muridarum infection elicits vigorous serum and local IgA responses within a few weeks after infection, whereas serum Chlamydia-specific IgA is undetectable (or very low in serovar D infection) and the local IgA response is delayed after C. trachomatis primary infection. The difference between the IgA response following C. trachomatis and C. muridarum infection is remarkable, but whether this difference contributes directly to the delay in the development of protective immunity is not known. Our previous studies using IgA-deficient mice show that IgA is not essential for protective immunity in the C. muridarum genital infection model [31], but that does not necessarily imply that it does not function in the C. trachomatis model of infection, and perhaps the local antibody response may be key in this model.
There are striking differences in immunity and pathogenesis between the C. muridarum and C. trachomatis genital infection models, but identifying which model best replicates chlamydial infection, pathogenesis, and immunity of women is not clear. Much like C. trachomatis infection of mice, women with genital C. trachomatis infection shed low to moderate numbers of infectious bacteria [32], signs of infection (ie, inflammation) are often minimal [33], infection can, but does not always, ascend to cause upper genital tract complication [34, 35], robust cellular and humoral immune responses develop [36], infection resolves in the absence of treatment [37], reinfection is common [38, 39], a level of immunity develops following multiple infections [40], and multiple serovars cause infection. However, there are also caveats that challenge the utility of the murine C. trachomatis model. For example, the absence of anti-chlamydial IgA in the C. trachomatis mouse infection model is quite different from what is observed in human infection [36, 41]. Furthermore, genital infection of mice with C. trachomatis is highly mouse strain dependent, and very substantial challenge doses of chlamydiae are required to produce productive infections [5]. Lastly, because C. trachomatis infection in the murine model resolves in the absence of adaptive immunity (Figure 4), it might be more difficult to define protective adaptive immune responses that develop during the natural course of infection. On the other hand, not only does the C. muridarum model replicate several aspects of human infection (ie, low infectious dose, ascending infection, resolution of infection without antibiotic treatment, and robust immune responses including IgA), but durable, long-lasting protective immunity develops after infection, facilitating the clear definition of protective mechanisms. Neither model of murine Chlamydia infection perfectly replicates human infection, but both have utility in the study of chlamydial pathogenesis and immunity.
Funding
The National Institutes of Health grant AI-038991 and the Arkansas Bioscience Institute.
Acknowledgments
We thank Dr Harlan Caldwell for critical review of the manuscript and thoughtful discussion of this work.
References
- 1.World Health Organization. Global prevalence and incidence of selected curable sexually transmitted diseases: overview and estimates. Geneva: World Health Organization; 1999. [Google Scholar]
- 2.Morrison RP, Caldwell HD. Immunity to murine chlamydial genital infection. Infect Immun. 2002;70:2741–51. doi: 10.1128/IAI.70.6.2741-2751.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyons JM, Ito JI, Morre SA. The influence of vaginally applied imiquimod on the course of Chlamydia trachomatis serovar D infection in a murine model. Infect Dis Obstet Gynecol. 2005;13:1–3. doi: 10.1080/10647440400025660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perry LL, Su H, Feilzer K, et al. Differential sensitivity of distinct Chlamydia trachomatis isolates to IFN-γ-mediated inhibition. J Immunol. 1999;162:3541–8. [PubMed] [Google Scholar]
- 5.Darville T, Andrews CW, Laffoon KK, Shymasani W, Kishen LR, Rank RG. Mouse strain-dependent variation in the course and outcome of chlamydial genital tract infection is associated with differences in host response. Infect Immun. 1997;65:3065–73. doi: 10.1128/iai.65.8.3065-3073.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyons JM, Morre SA, Airo-Brown LP, Pena AS, Ito JI. Comparison of multiple genital tract infections with Chlamydia trachomatis in different strains of female mice. J Microbiol Immunol Infect. 2005;38:383–93. [PubMed] [Google Scholar]
- 7.Lyons JM, Morre SA, Airo-Brown LP, Pena AS, Ito JI. Acquired homotypic and heterotypic immunity against oculogenital Chlamydia trachomatis serovars following female genital tract infection in mice. BMC Infect Dis. 2005;5:105. doi: 10.1186/1471-2334-5-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olivares-Zavaleta N, Whitmire W, Gardner D, Caldwell HD. Immunization with the attenuated plasmidless Chlamdyia trachomatis L2(25667R) strain provides partial protection in a murine model of female genitourinary tract infection. Vaccine. 2010;28:1454–62. doi: 10.1016/j.vaccine.2009.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramsey KH, Cotter TW, Salyer RD, et al. Prior genital tract infection with a murine or human biovar of Chlamydia trachomatis protects mice against heterotypic challenge infection. Infect Immun. 1999;67:3019–25. doi: 10.1128/iai.67.6.3019-3025.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramsey KH, DeWolfe JL, Salyer RD. Disease outcome subsequent to primary and secondary urogenital infection with murine or human biovars of Chlamydia trachomatis. Infect Immun. 2000;68:7186–9. doi: 10.1128/iai.68.12.7186-7189.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuffrey M, Falder P, Gale J, Taylor-Robinson D. Salpingitis in mice induced by human strains of Chlamydia trachomatis. Br J Exp Pathol. 1986;67:605–16. [PMC free article] [PubMed] [Google Scholar]
- 12.Poltorak A, He X, Smirnova I, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in the Tlr4 gene. Science. 1998;282:2085–88. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 13.Kari L, Whitmire WM, Carlson JH, et al. Pathogenic diversity among Chlamydia trachomatis ocular strains in nonhuman primates is affected by subtle genomic variations. J Infect Dis. 2008;197:449–56. doi: 10.1086/525285. [DOI] [PubMed] [Google Scholar]
- 14.Caldwell HD, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–76. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrison RP, Feilzer K, Tumas DB. Gene knockout mice establish a primary protective role for major histocompatibility complex class II-restricted responses in Chlamydia trachomatis genital tract infection. Infect Immun. 1995;63:4661–8. doi: 10.1128/iai.63.12.4661-4668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caldwell HD, Hitchcock PJ. Monoclonal antibody against a genus-specific antigen of Chlamydia species: location of the epitope on chlamydial lipopolysaccharide. Infect Immun. 1984;44:306–14. doi: 10.1128/iai.44.2.306-314.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrison SG, Morrison RP. A predominant role for antibody in acquired immunity to chlamydial genital tract reinfection. J Immunol. 2005;175:7536–42. doi: 10.4049/jimmunol.175.11.7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrison SG, Su H, Caldwell HD, Morrison RP. Immunity to murine Chlamydia trachomatis genital tract reinfection involves B cells and CD4+ T cells but not CD8+ T cells. Infect Immun. 2000;68:6979–87. doi: 10.1128/iai.68.12.6979-6987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sturdevant GL, Kari L, Gardner DJ, et al. Frameshift mutations in a single novel virulence factor alter the in vivo pathogenicity of Chlamydia trachomatis for the female murine genital tract. Infect Immun. 2010;78:3660–8. doi: 10.1128/IAI.00386-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrison SG, Morrison RP. Resolution of secondary Chlamydia trachomatis genital tract infection in immune mice with depletion of both CD4+ and CD8+ T cells. Infect Immun. 2001;69:2643–9. doi: 10.1128/IAI.69.4.2643-2649.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrison SG, Morrison RP. In situ analysis of the evolution of the primary immune response in murine Chlamydia trachomatis genital tract infection. Infect Immun. 2000;68:2870–9. doi: 10.1128/iai.68.5.2870-2879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Read TD, Brunham RC, Shen C, et al. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephens RS, Kalman S, Lammel C, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–9. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 24.Belland RJ, Scidmore MA, Crane DD, et al. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc Natl Acad Sci USA. 2001;98:13984–9. doi: 10.1073/pnas.241377698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savidge TC, Pan W-H, Newman P, O'Brien MO, Anton PM, Pothoulakis C. Clostridium difficile toxin B is an inflammartory enterotoxin in human intestine. Gastroenterology. 2003;125:413–20. doi: 10.1016/s0016-5085(03)00902-8. [DOI] [PubMed] [Google Scholar]
- 26.van Ginkel FW, Jackson RJ, Yoshino N, et al. Enterotoxin-based mucosal adjuvants alter antigen trafficking and induce inflammatory responses in the nasal tract. Infect Immun. 2005;73:6892–902. doi: 10.1128/IAI.73.10.6892-6902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev. 2005;18:247–63. doi: 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson M, Lycke N. A unique population of extrathymically derived alpha betaTCR+CD4-CD8- T cells with regulatory functions dominates the mouse female genital tract. J Immunol. 2003;170:1659–666. doi: 10.4049/jimmunol.170.4.1659. [DOI] [PubMed] [Google Scholar]
- 29.Nelson DE, Taylor LD, Shannon JG, et al. Phenotypic rescue of Chlamydia trachomatis growth in IFN-gamma treated mouse cells by irradiated Chlamydia muridarum. Cell Microbiol. 2007;9:2289–98. doi: 10.1111/j.1462-5822.2007.00959.x. [DOI] [PubMed] [Google Scholar]
- 30.Nelson DE, Virok DP, Wood H, et al. Chlamydial IFN-γ immune evasion is linked to host infection tropism. Proc Natl Acad Sci USA. 2005;102:10658–63. doi: 10.1073/pnas.0504198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrison SG, Morrison RP. The protective effect of anitbody in immunity to murine chlamydial genital tract reinfection is independent of immunoglobulin A. Infect Immun. 2005;73:6183–6. doi: 10.1128/IAI.73.9.6183-6186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geisler WM, Suchland RJ, Whittington WLH, Stamm WE. Quantitative culture of Chlamydia trachomatis: relationship of inclusion-forming units produced in culture to clinical manifestations and acute inflammation in urogenital disease. J Infect Dis. 2001;184:1350–4. doi: 10.1086/323998. [DOI] [PubMed] [Google Scholar]
- 33.Stamm WE. Chlamydia infections. In: Holmes KK, Sparling PF, Mardh P-A, et al., editors. Sexually transmitted diseases. 3rd ed. New York City: McGraw-Hill; 1999. pp. 407–422. [Google Scholar]
- 34.Cates JW, Brunham RC. Sexually transmitted diseases and infertility. In: Holmes KK, Sparling PF, Mardh P-A, et al., editors. Sexually transmitted diseases. 3rd ed. New York City: McGraw-Hill; 1999. pp. 1079–1088. [Google Scholar]
- 35.Haggerty CL, Gottlieb SL, Taylor BD, Low N, Xu F, Ness RB. Risk of sequelae after Chlamydia trachomatis genital infection in women. J Infect Dis. 2010;201:S134–55. doi: 10.1086/652395. [DOI] [PubMed] [Google Scholar]
- 36.Brunham RC. Human immunity to chlamydiae. In: Stephens RS, editor. Chlamydia: intracellular biology, pathogenesis, and immunity. Washington, DC: American Society for Microbiology Press; 1999. pp. 211–38. [Google Scholar]
- 37.Geisler WM. Duration of untreated, uncomplicated Chlamydia trachomatis genital infection and factors associated with chlamydia resolution: a review of human studies. J Infect Dis. 2010;201:S104–13. doi: 10.1086/652402. [DOI] [PubMed] [Google Scholar]
- 38.Dunne EF, Chapin JB, Rietmeijer CA, et al. Rate and predictors of repeat Chlamydia trachomatis infection among men. Sex Transm Dis. 2008;35:S40–S44. doi: 10.1097/OLQ.0b013e31817247b2. [DOI] [PubMed] [Google Scholar]
- 39.Hosenfeld CB, Workowski KA, Berman S, et al. Repeat infection with chlamydia and gonorrhea among females: a systematic review of the literature. Sex Transm Dis. 2009;36:478–89. doi: 10.1097/OLQ.0b013e3181a2a933. [DOI] [PubMed] [Google Scholar]
- 40.Batteiger BE, Xu F, Johnson RE, Rekart ML. Protective immunity to Chlamydia trachomatis genital infection: evidence from human studies. J Infect Dis. 2010;201:S178–89. doi: 10.1086/652400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brunham RC, Kuo C-C, Cles L, Holmes KK. Correlation of host immune response with quantitative recovery of Chlamydia trachomatis from the human endocervix. Infect Immun. 1983;39:1491–4. doi: 10.1128/iai.39.3.1491-1494.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]