Abstract
Background. Human immunodeficiency virus type 1 (HIV-1) vaccines directed to the cell-mediated immune system could have a role in lowering the plasma HIV-1 RNA set point, which may reduce infectivity and delay disease progression.
Methods. Randomized, placebo-controlled trial involving HIV-1-infected participants who received a recombinant adenovirus serotype 5 (rAd5) HIV-1 gag vaccine or placebo. Sequence-based HLA typing was performed for all 110 participants who initiated analytic treatment interruption (ATI) to assess the role of HLA types previously associated with HIV prognosis. Plasma HIV-1 gag and pol RNA sequences were obtained during the ATI. Virologic endpoints and HLA groups were compared between treatment arms using the 2-sample rank sum test. A linear regression model was fitted to derive independent correlates of ATI week 16 plasma viral load (w16 PVL).
Results. Vaccinated participants with neutral HLA alleles had lower median w16 PVLs than did vaccinated participants with protective HLA alleles (P = .01) or placebo participants with neutral HLA alleles (P = .02). Factors independently associated with lower w16 PVL included lower pre-antiretroviral therapy PVL, greater Gag sequence divergence from the vaccine sequence, decreased proportion of HLA-associated polymorphisms in Gag, and randomization to the vaccine arm.
Conclusions. Therapeutic vaccination with a rAd5-HIV gag vaccine was associated with lower ATI week 16 PVL even after controlling for viral and host genetic factors.
Clinical Trials Registration. NCT00080106.
A robust cell-mediated immune response is vital for the resolution of acute infection with human immunodeficiency virus type 1 (HIV-1) and long-term control of disease progression [1–5]. HIV-1 vaccines directed to the cell-mediated immune system could have a role in lowering the plasma HIV-1 RNA set point [6], which may reduce infectivity and delay disease progression.
AIDS Clinical Trials Group (ACTG) protocol A5197 was a randomized, placebo-controlled trial to test the effect of a recombinant adenovirus serotype 5 (rAd5) HIV-1 gag therapeutic vaccine on plasma viral load (PVL) in participants undergoing an analytic treatment interruption (ATI) [7]. A total of 110 participants underwent a 16-week ATI after randomization in a 2:1 ratio to receive 3 doses of either vaccine or placebo. Although there was a trend toward a vaccine benefit, this difference failed to reach statistical significance for the prespecified coprimary ATI endpoints (PVL set point: mean of ATI week 12 and 16 PVL and time-averaged area under the curve). However, a secondary analysis based on PVL at ATI week 16 (w16 PVL) found that HIV-1 RNA levels were 0.5 log10 lower in the vaccine arm [7, 8].
The influence of HLA class I alleles on viral evolution, disease progression, and PVL is well known [9–14]. A number of “protective” alleles (HLA B*13, B*27, B*51, B*57, and B*5801) are associated with lower levels of viremia, delayed disease progression, and/or improved outcomes [10, 15–20]. “Unfavorable” HLA alleles associated with accelerated disease progression include the HLA-B*35-Px variants (B*3502, 3503, 3504, or 5301) [21, 22]. In this analysis, we describe the distribution of HLA alleles in ACTG A5197 and explore their impact on the w16 PVL response to a rAd5 HIV-1 gag vaccine.
This vaccine induced significant CD4+ and CD8+ HIV-specific T cell responses [7]. In addition to HLA and T cell activation, other factors that may play a role in vaccine effectiveness and viral rebound include pre-antiretroviral therapy (ART) PVLs [23–25], CD4+ T cell counts [23, 24], HLA-associated viral polymorphisms [14], preexisting Ad5 antibody titers [26], and sequence similarity of patient virus to the vaccine [27]. Using a hypothesis-driven approach, we created a multiple linear regression model to identify factors independently correlated with virologic rebound. Because the more robust vaccine effect was not noted until ATI week 16, we focused our analysis on the w16 PVL outcome.
METHODS
Patients and Study Design
Study design and patient inclusion criteria for ACTG A5197 have been described elsewhere [7]. Eligible participants were receiving ART and had CD4+ cell counts ≥500/mm3, plasma HIV-1 RNA levels ≤50 copies/mL with a history of PVL ≤500 copies/mL for 24 months prior to enrollment, and screening serum Ad5 antibody titers ≤200 units/mL. Participants were stratified at randomization by their highest pre-ART PVL (<30,000 copies/mL, ≥ 30,000 copies/mL, or unknown). Participants received a replication-defective rAd5 vaccine containing an HIV-1 gag insert or placebo at weeks 0, 4, and 26 (step I). Starting at week 39, 110 participants (n = 73 vaccine, n = 37 placebo) underwent a 16-week ATI (step II). Participants were encouraged to restart ART with any signs of immunodeficiency, for HIV-1 RNA >300,000 copies/mL at 3 consecutive study visits, or if the CD4+ cell count fell below 300 cells/mm3 at 2 consecutive visits or to <50% of baseline. All participants who entered the ATI were included in this analysis. “Dropouts” were defined as those who entered step II but did not complete the 16-week ATI. Participants dropped out of step II mainly as a result of reinitiation of ART for one of the reasons stated above, adverse events, or patient preference.
HLA Typing
HLA class I typing was performed in accordance with the sequence-specific oligonucleotide probing and sequence-based typing protocols recommended by the 13th International Histocompatibility Workshop (http://www.ihwg.org). Protective HLA alleles were defined a priori as HLA B*13, B*27, B*51, B*57, and B*5801. Unfavorable HLA alleles were defined as the HLA-B*35-Px variants (B*3502, 3503, 3504), or B*5301. The protective HLA group includes all individuals with at least 1 protective HLA allele. Those without a protective HLA allele but with at least 1 unfavorable HLA allele were categorized in the unfavorable HLA group. Participants with neither protective nor unfavorable HLA alleles were categorized in the neutral HLA group.
Intracellular Cytokine Staining Assay for Interferon-γ
Blood drawn at protocol-defined intervals was sent to Merck Research Laboratories by overnight courier. Vaccine-induced cell-mediated immune responses were evaluated by an intracellular cytokine staining (ICS) assay as described elsewhere [7, 28, 29]. Lymphocytes were exposed to Gag, Nef, Pol or mock peptide pools for 18 hours. Interferon-γ–producing CD4+ and CD8+ T cells were detected with use of multiparameter flow cytometry. Participants were categorized as responders to a peptide pool when >400 interferon-γ–producing cells per million lymphocytes were observed and this response was at least 3 times greater than the response to the mock peptide pool (except for CD8 response to pol2, which required >500 per 106 interferon-γ–producing cells and >4 times the response to the mock peptide pool) [7, 29].
Viral Sequencing from Plasma and Sequence Analysis
For most participants, plasma viral sequences were analyzed at 2 time points: at the time of initial detectable viremia (in general, ATI weeks 2–7) and at ATI week 16. Of the 110 participants who entered ATI, 105 had plasma available from at least 1 ATI time point. The Gag and reverse-transcriptase (RT) coding sequences were amplified from plasma HIV-1 RNA by nested RT polymerase chain reaction using gene-specific primers. Population sequencing was performed on an ABI 3730 automated DNA sequencer (Applied Biosystems). Chromatograms were analyzed using Sequencher (Genecodes). We calculated the number of amino acid mismatches between the vaccine or consensus HIV-1 subtype B sequence and the patient-derived sequence. HLA-associated polymorphisms in patient HIV-1 sequences were determined from a published list [14] derived from an analysis of >1200 chronically infected, treatment-naive individuals from several cohorts (HOMER, ACTG 5142/5128, and Western Australia [WAHCS]) using phylogenetically corrected methods [30] and a q-value correction for multiple tests (q < .05). The proportion of HLA-associated Gag polymorphisms in patient sequences was calculated by dividing the number of HLA-associated Gag polymorphisms observed in that sequence by the total number of amino acids in Gag that displayed an HLA-associated footprint at the population level for all HLA alleles expressed by the participant. The RT-coding region of pol was analyzed for the presence of the M184V mutation, which confers 3TC resistance. Laboratory assessments were performed by individuals who were blinded to treatment assignments. The sequences reported here have been deposited in GenBank (accession nos. GU560900–GU561307).
Statistical Analysis
The distribution of HLA alleles between treatment arms and the distribution of step II dropouts among the HLA groups were analyzed using the Fisher exact test. A comparison of PVL set point (log10 mean week 12 and 16 PVLs) between HLA groups was performed with the stratified (by trial arm) 2-sample rank sum test. Exact 2-sided P values and shift parameter estimates were provided for all comparisons. A comparison of the w16 PVL between vaccine and HLA groups was performed with the stratified (on protocol-defined strata) 2-sample rank sum test. As specified in the original protocol [7], participants without an HIV-1 RNA evaluation at either ATI week 12 or 16 of step II were assigned the worst ranks. Rank was assigned in reverse order of the length of time off ART. No adjustments were performed for multiple comparisons. Note that the comparisons presented here were conducted post hoc after viewing the data.
The distribution of the number of gag-specific CD4+ interferon-γ–producing cells and responders among HLA groups and vaccine arms was analyzed with the 2-sample Wilcoxon rank sum test and Fisher exact test, respectively. The Spearman correlation was used to determine the relationship between w16 PVL and patient-to-vaccine (or consensus subtype B) amino acid sequence mismatches. The distribution of the number and proportion of HLA-associated polymorphisms in Gag at ATI week 16 in different HLA groups was assessed with the Jonckheere-Terpstra trend test. Univariate linear regression was used to determine significant factors associated with the w16 PVL (observed data only). Predictor variables with P < .1 were added to the multiple linear regression model. Study arm and HLA group were added to the models irrespective of their univariate significance.
RESULTS
Distribution of HLA Alleles between Vaccine and Placebo Arms
Post hoc HLA typing of study participants revealed a significant imbalance in the distribution of favorable and neutral HLA alleles between arms. Of the 37 participants randomized to the placebo arm who entered the ATI, 6 (16%) were found to have a favorable HLA allele and 27 (73%) had neutral HLA alleles. In contrast, 32 (44%) of 73 participants in the vaccine arm who entered the ATI possessed at least 1 favorable HLA allele and 36 (49%) were found to have neutral HLA alleles.
Association between HLA Alleles and PVL Set Point
Compared with those with protective or neutral HLA alleles, the 9 participants with unfavorable HLA alleles had elevated PVL rebound during ATI and a higher dropout rate during step II. By week 16, 3 (33%) of 9 individuals with an unfavorable HLA allele had dropped out of that phase of the study and 2 others had missed the week 16 PVL measurement, compared with 7 (7%) of 101 participants with protective or neutral HLA alleles (P = .01). Given the high dropout rate by ATI week 16, we performed an analysis of the protocol-defined PVL set point and confirmed that, compared with individuals with unfavorable HLA alleles, participants with neutral HLA alleles had a 1.32 log10 lower PVL (P = .005) and those with protective HLA alleles had a 0.71 log10 lower PVL (P = .05) after adjusting for vaccine effect.
Viral Rebound in Participants with Neutral HLA Alleles
When participants were categorized as having protective, neutral, or unfavorable HLA alleles, the observed vaccine benefit was found predominantly among participants with neutral alleles (Table 1). The w16 PVLs were significantly lower in vaccinated participants with neutral HLA alleles than in those with protective HLA alleles (P = .01) or those with neutral HLA alleles who received placebo (P = .02). The high dropout rate during step II among individuals with unfavorable HLA alleles precluded a more detailed analysis in that group.
Table 1.
Plasma Human Immunodeficiency Virus Type 1 RNA Levels at Week 16 of the Analytic Treatment Interruption
| HLA group | Pa | Treatment arm, median | Pb | |
| Vaccine | Placebo | |||
| Protective | .01 | 4.6 (n = 32) | 4.4 (n = 6) | |
| Neutral | 4.0 (n = 36) | 4.5 (n = 27) | .02 | |
| Unfavorable | 5.7 (n = 5) | N/A (n = 4) | ||
NOTE. Data are median log10 plasma HIV-1 RNA copies/mL, with the number of participants contributing to the analysis shown in parentheses. The median is calculated on the basis of the assumption that the missing values are greater than the greatest observed value. When the number of missing values is more than half the sample size, the median is reported as “N/A”. Three of the 4 participants in the placebo group with unfavorable HLA alleles did not complete the week 16 evaluation, including 2 participants who met protocol-defined criteria for reinitiation of ART prior to ATI week 16. NA, not applicable.
Comparison of the vaccine/protective and vaccine/neutral subgroups.
Comparison of the vaccine/neutral and placebo/neutral subgroups.
Association between Gag-specific CD4+ IFN-γ Response and Viral Load
Participants with CD4+ IFN-γ responses to HIV-1 Gag at study week 38 had a 0.73 log10 lower PVL at week 16 of the subsequent ATI after adjusting for vaccine effect (P = .02). Vaccinated participants with neutral HLA alleles had a higher median number of Gag-specific CD4+ IFN-γ–producing cells at week 38 than did immunized participants with protective alleles or placebo recipients with neutral alleles (Table 2).
Table 2.
Number of log10 Gag-Specific CD41 IFN-g–Producing Cells per Million Lymphocytes at Study Week 38 Categorized by HLA Allele Grouping and Treatment Arm
| Treatment arm, median [IQR]a |
|||||
| HLA group | Pb | Vaccine | Placebo | Pc | |
| Protective | 10 | 2.21 [± 0.70] (n=31) | 2.25 [± 0.43] (n=5) | ||
| Neutral | 2.40 [± 0.40] (n=36) | 2.11 [± 0.51] (n=27) | .003 | ||
| Unfavorable | 2.1 [± 0.51] (n=5) | 2.17 [± 1.40] (n=4) | |||
NOTE
IQR, interquartile range.
Comparison of the vaccine/protective and vaccine/neutral subgroups.
Comparison of the vaccine/neutral and placebo/neutral subgroups.
Association of Patient Virus Divergence with Viral Rebound
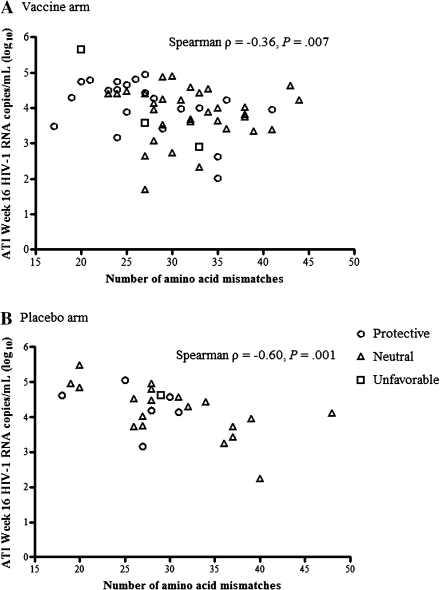
The number of predicted amino acid mismatches was calculated between the vaccine Gag sequence and the earliest available patient-derived HIV-1 sequence after ATI (median, 4 weeks). These early sequences represent viral populations least likely to have been shaped by vaccine-driven immune responses. Among vaccine recipients, we found a significant inverse correlation between the number of amino acid mismatches and the w16 PVL (Spearman ρ = –0.36; P = .007) (Figure 1a). Unexpectedly, we found a significant inverse correlation in the placebo arm as well (Spearman ρ = –0.60; P = .001) (Figure 1b). The number of amino acid mismatches correlated with the number of HLA-associated Gag polymorphisms (defined as in [14]) only in those with protective HLA alleles (Spearman ρ = 0.37; P = .03). No association was detected between the number of amino acid mismatches and the number of Gag-specific CD4+ or CD8+ IFN-γ–producing cells at study week 38. These correlations remained largely unchanged if the patient sequences were compared with the consensus subtype B Gag sequence instead of the vaccine (the vaccine shared 98% amino acid sequence homology with the consensus subtype B Gag sequence). To determine whether such a correlation existed outside of Gag, amino acid mismatches between consensus subtype B and patient-derived viral sequences were also calculated for RT. No correlation was found between the number of amino acid mismatches between patient virus and consensus RT sequence and the w16 PVL (Spearman ρ = –0.04; P = .71).
Figure 1.
Association between ATI week 16 virus load endpoint (log10 HIV-1 RNA copies/mL) and the number of amino acid mismatches between vaccine and subject Gag sequences in the vaccine (A) and placebo (B) arms.
HLA-associated Gag Polymorphisms in Plasma Virus
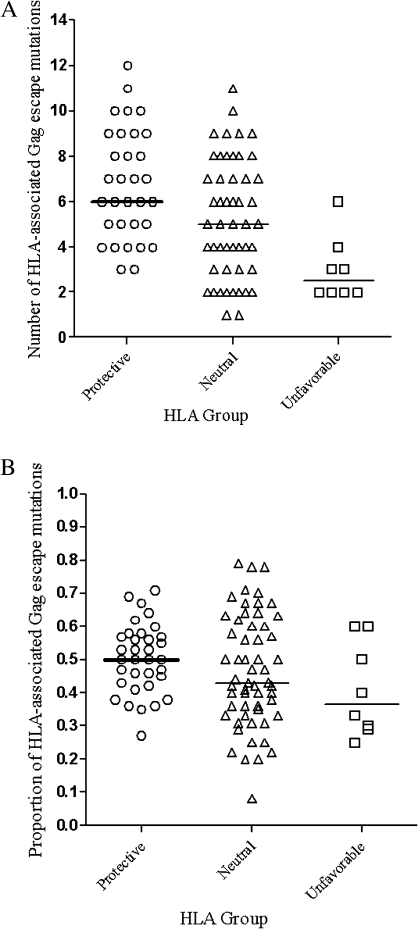
The number of HLA-associated polymorphisms in Gag was determined by sequencing plasma HIV RNA from ATI week 16. There was a significant difference in the number of polymorphisms by HLA group. Individuals with protective or neutral HLA alleles had more HLA-associated polymorphisms in Gag than did those with unfavorable HLA alleles (6 vs 2.5, P < .001; and 5 vs 2.5, P = .009, respectively) (Figure 2a). The distribution of polymorphisms was shifted from high to low in the direction of protective, neutral, and unfavorable HLA groups (Jonckheere-Terpstra test 1-sided P < .001). A similar trend was found when the proportion of HLA-associated Gag polymorphisms among HLA groups was compared (Figure 2b; Jonckheere-Terpstra test 1-sided P = .03).
Figure 2.
Number of HLA-associated Gag escape mutations (A) and proportion of HLA-associated Gag polymorphic sites showing escape mutations (B) per person by HLA group. The proportion of escape mutations was calculated as the number of HLA-associated polymorphisms in a viral sequence divided by the total number of possible HLA-associated polymorphic sites for each subject.
Factors Independently Associated with ATI Viral Load
Factors independently associated with a lower w16 PVL (n = 50) (Table 3) included a lower pre-ART PVL (P < .001), a lower proportion of HLA-associated polymorphisms in Gag (P < .001), a higher number of amino acid mismatches between vaccine and patient virus in Gag (P = .002), and randomization to the vaccine arm (P = .02). The sample available for the multiple linear regression model was limited by missing data for some participants, principally as a result of absence of pre-ART viral load data or dropping out prior to ATI week 16. There were no significant differences in the distribution of any of the factors included in the regression models between participants included or excluded from these analyses (data not shown). Presence of the M184V mutation was not significantly associated with w16 PVL (P = .60). Of the variables found to be significant in the univariate analysis, week 38 Gag-specific CD4+ interferon-γ response, Ad5 titer, and CD4 nadir did not reach statistical significance in the multiple linear regression model.
Table 3.
Factors Independently Associated with log10 Human Immunodeficiency Virus Type 1 RNA Levels at Analytic Treatment Interruption Week 16 Using Observed Data Only
| Variable | P | Coefficient (95% CI) |
| Pre-ART viral load, log10 copies/mL | <.001 | 0.53 (0.36–0.70) |
| Proportion of HLA-associated escape mutations | <.001 | 1.83 (0.99–2.66) |
| Patient-vaccine amino acid mismatches | .002 | –0.03 (–0.05 to –0.01) |
| Vaccine arm assignment | .02 | –0.31 (–0.56 to –0.06) |
NOTE. ART, antiretroviral therapy; CI, confidence interval.
DISCUSSION
In ACTG A5197, HIV-1–infected individuals who received a rAd5 HIV-1 gag therapeutic vaccine had a 0.5 log10 lower PVL after a 16-week ATI, compared with those who received placebo [7, 8]. In this post hoc exploratory analysis, we analyzed factors associated with vaccine response and identified variables associated with viral rebound at ATI week 16. We found that the vaccine effect on PVL was observed primarily in participants with neutral HLA alleles; those with unfavorable HLA alleles fared poorly regardless of study arm.
The beneficial effect of protective HLA alleles on PVL set point has been described elsewhere [17, 31, 32]. In the STEP trial, which used a rAd5 HIV-1 Gag-Pol-Nef vaccine, vaccine recipients with protective HLA alleles had significantly lower PVL set points after infection [33]. In the present study, vaccine-associated reduction in w16 PVL was not observed in participants with protective HLA alleles. A potential explanation may be that in this chronically infected population, patients with favorable HLA alleles had already developed maximal anti-Gag immune responses, or HIV-1 had already adapted to evade those responses so that little additional benefit was obtained from vaccination [34]. For example, in studies of HLA-matched transmission pairs, protective alleles were not associated with lower set point PVL in the newly infected persons when HLA-associated viral escape mutations were present [32, 35]. The finding that the vaccine effect on w16 PVL was greatest in individuals with neutral HLA alleles was surprising. It may be that such individuals elaborate suboptimal HIV-1–specific cell-mediated immune responses to the infecting virus and thus benefit from a vaccine-induced boost in immunity. The relatively small number of participants with an unfavorable HLA allele precludes broad generalizations, but the higher PVL set point in this group is consistent with the higher risk of clinical progression reported in previous studies [21, 22].
The chance imbalance of protective HLA alleles between vaccine and placebo arms in this study raises a cautionary note. On the basis of large studies of HLA allelic frequencies in US populations [36, 37] and the ethnic makeup of the A5197 study population, we determined the expected distribution of participants within each HLA group by treatment arm (Table 4). Because the vaccine effect was seen predominantly in those with neutral HLA alleles, the under representation of participants with neutral HLA alleles in the vaccine arm likely resulted in an underestimate of vaccine efficacy. The imbalance in HLA alleles in the placebo arm did not seem to have a substantial effect on the viral load outcome, because the participants with protective and neutral HLA alleles had similar w16 PVL results. These results suggest that HLA class I typing should be an integral part of the analysis of vaccine trials designed to stimulate cell-mediated immune responses. In certain circumstances, stratifying for protective and/or unfavorable HLA alleles at randomization may be appropriate. This concern is especially pertinent for small, early-phase trials of preventive or therapeutic vaccines.
Table 4.
Expected and actual HLA phenotypic frequencies by treatment arm and HLA group.
| HLA group | Vaccine arm |
Placebo arm |
||
| Expected | Actual | Expected | Actual | |
| Protective | 30% (22/73) | 44% (32/73) | 31% (11/37) | 16% (6/37) |
| Neutral | 62% (45/73) | 49% (36/73) | 61% (23/37) | 73% (27/37) |
| Unfavorable | 8% (6/73) | 7% (5/73) | 8% (3/37) | 11% (4/37) |
NOTE. Expected phenotypic frequencies based on A5197 ethnic distribution and calculated from HLA alleleic frequencies of U.S.-based cohorts of European Americans and African Americans (M. Carrington personal communication), and Hispanic and Asian Americans [1, 2].
A strong CD4-mediated Gag-specific IFN-γ response has been associated with natural control of HIV-1 replication during primary infection [38]. The strength of such responses correlates inversely with plasma HIV-1 RNA levels in untreated individuals [39]. Vaccination of HIV-1–infected participants with the rAd5 HIV-1 gag resulted in significantly increased HIV-specific CD4+ and CD8+ IFN-γ activity [7]. In this study, vaccinated participants with neutral HLA alleles tended to have a higher number of Gag-specific CD4+ IFN-γ–producing cells and a higher proportion of CD4+ IFN-γ responders, which in turn were associated with significantly lower w16 PVL. One potential explanation of these results could lie in the finding that CD8+ T cells lose the capacity to proliferate during chronic infection but this function is restored by vaccine-augmented CD4+ T helper cells [40]. However, in the current study, CD4+ IFN-γ responses did not predict w16 PVL in the multiple linear regression model. Additional analysis of other vaccine-induced cytokine responses may clarify their role in determining PVL during postvaccination ATI.
Autologous HIV-1 peptides induce stronger HIV-1–specific T cell responses than do consensus peptides [27]. We therefore hypothesized that greater similarity between vaccine and patient-derived Gag sequences would be associated with stronger T cell responses, resulting in lower viral rebound during ATI. No association was found, however, between the number of amino acid mismatches and the number of Gag-specific CD4+ or CD8+ IFN-γ–producing cells immediately before treatment interruption. Surprisingly, the number of Gag mismatches was negatively correlated with w16 PVL in both vaccine and placebo arms. No such correlation was found for RT. HIV-1 adapts to host immune responses at the population level, and consensus sequences reflect that adaptation [13]. Perhaps viruses that are closest in sequence to the clade consensus have a fitness advantage; greater divergence from the consensus subtype B Gag sequence could reflect greater immune pressure on the virus. Although the number of Gag mismatches was correlated with the number of HLA-associated Gag polymorphisms in participants with protective HLA alleles, the number of Gag mismatches was independently associated with lower w16 PVL on multivariate analysis. In vitro fitness studies in acute and chronically infected patients are needed to explore the meaning of this finding.
Mutations in epitopes targeted by HLA-restricted CD8 T lymphocytes may result in loss of viral fitness, but they also represent viral escape from immunological control, and their presence has been associated with higher PVL [32, 41–43]. In the current study, we found that the proportion of HLA-associated Gag polymorphisms was associated with higher w16 PVLs. This result is consistent with a previous study that reported a modest positive correlation between the proportion of escaped Gag sites and PVL [43].
As a post hoc analysis, this study has a number of limitations. The decision was made to use w16 PVL as the main outcome measure because of the more robust vaccine effect when compared with the protocol-defined endpoints. The small number of participants included in the multivariate linear regression model and in the study as a whole limits the generalizability of the results. The main factor for excluding patients from the model was a lack of information on pre-ART viral load, which was known for only two-thirds of participants but was one of the strongest predictors of viral rebound. This finding validates the decision to stratify according to the highest pre-ART viral load at randomization in the original clinical trial. No significant differences were found between those individuals included and excluded from the linear regression model. Despite these limitations, in-depth analyses of HIV-1 vaccine trials are crucial in identifying potential correlates of vaccine efficacy.
The ACTG A5197 study represents another step forward in furthering our understanding of the immunological and virological factors important for control of HIV-1 infection. In the exploratory analysis reported here, we identified a number of factors associated with viral rebound during the ATI following administration of a rAd5 HIV-1 gag vaccine. Importantly, therapeutic vaccination was associated with lower ATI week 16 PVL even after controlling for these host and viral factors. Additional analyses of ACTG A5197 and other therapeutic vaccine trials may provide a deeper understanding of the interplay between the HLA-restricted, cell-mediated immune response and the process by which HIV escape and virologic rebound occur. Such insights will be crucial for the successful development of therapeutic and preventative HIV vaccines.
Funding
This work was supported in part by the National Institutes of Health (NIH; grants nos. T32 AI07387 to J.L., U01 AI068636 to the AIDS Clinical Trials Group, U01 AI068634 to the ACTG Statistical and Data Management Center, P30 AI60354 to the Harvard University Center for AIDS Research, and K24 RR016482 to D.R.K.; and subcontracts from U01 AI068636 to the Harvard Virology Support Laboratory and the Case Immunology Support Laboratory). Z.L.B. was supported by a postdoctoral fellowship and a New Investigator Award from the Canadian Institutes of Health Research (CIHR). This project has been funded in part with federal funds from the National Cancer Institute, NIH, under contract no. HHSN261200800001E, and by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Acknowledgments
We thank the patients, ACTG A5197 team members, and participating ACTG clinical research sites for their efforts on behalf of this study, and Daniel Casimiro for critically reviewing the manuscript. We also thank Jennifer Sela, Pamela Rosato, and Yuko Yuki for their assistance with HLA typing. We are grateful for the help of Amanda Zadzilka, Holly Meyers, and the Frontier Science Sequence Support Group.
References
- 1.Schmitz JE, Kuroda MJ, Santra S, et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–60. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 2.Jin X, Bauer DE, Tuttleton SE, et al. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–8. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koup RA, Safrit JT, Cao Y, et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–5. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–10. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musey L, Hughes J, Schacker T, Shea T, Corey L, McElrath MJ. Cytotoxic-T-cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. N Engl J Med. 1997;337:1267–74. doi: 10.1056/NEJM199710303371803. [DOI] [PubMed] [Google Scholar]
- 6.Barouch DH. Challenges in the development of an HIV-1 vaccine. Nature. 2008;455:613–9. doi: 10.1038/nature07352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schooley RT, Spritzler J, Wang H, et al. AIDS clinical trials group 5197: a placebo-controlled trial of immunization of HIV-1-infected persons with a replication-deficient adenovirus type 5 vaccine expressing the HIV-1 core protein. J Infect Dis. 2010;202:705–16. doi: 10.1086/655468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schooley RWH, Spritzler J, Lederman M, et al. 15th Conference on Retroviruses and Opportunistic Infections. Boston, MA: the AIDS Clinical Trials Group. Therapeutic vaccination with a replication defective adenovirus type 5 HIV-1 gag vaccine in a prospective double-blinded, placebo-controlled trial (ACTG 5197) 3–6 February 2008. [Google Scholar]
- 9.Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–30. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiepiela P, Leslie AJ, Honeyborne I, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–75. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 11.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–43. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 12.Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–51. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 13.Kawashima Y, Pfafferott K, Frater J, et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458:641–5. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brumme ZL, John M, Carlson JM, et al. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS ONE. 2009;4:e6687. doi: 10.1371/journal.pone.0006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honeyborne I, Prendergast A, Pereyra F, et al. Control of human immunodeficiency virus type 1 is associated with HLA-B*13 and targeting of multiple gag-specific CD8+ T-cell epitopes. J Virol. 2007;81:3667–72. doi: 10.1128/JVI.02689-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leslie A, Kavanagh D, Honeyborne I, et al. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J Exp Med. 2005;201:891–902. doi: 10.1084/jem.20041455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altfeld M, Addo MM, Rosenberg ES, et al. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS. 2003;17:2581–91. doi: 10.1097/00002030-200312050-00005. [DOI] [PubMed] [Google Scholar]
- 18.Altfeld M, Kalife ET, Qi Y, et al. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8+ T cell response against HIV-1. PLoS Med. 2006;3:e403. doi: 10.1371/journal.pmed.0030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frater AJ, Brown H, Oxenius A, et al. Effective T-cell responses select human immunodeficiency virus mutants and slow disease progression. J Virol. 2007;81:6742–51. doi: 10.1128/JVI.00022-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthews PC, Prendergast A, Leslie A, et al. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J Virol. 2008;82:8548–59. doi: 10.1128/JVI.00580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao X, Nelson GW, Karacki P, et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N Engl J Med. 2001;344:1668–75. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- 22.Jin X, Gao X, Ramanathan M, Jr, et al. Human immunodeficiency virus type 1 (HIV-1)-specific CD8+-T-cell responses for groups of HIV-1-infected individuals with different HLA-B*35 genotypes. J Virol. 2002;76:12603–10. doi: 10.1128/JVI.76.24.12603-12610.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Moing V, Chene G, Carrieri MP, et al. Predictors of virological rebound in HIV-1-infected patients initiating a protease inhibitor-containing regimen. AIDS. 2002;16:21–9. doi: 10.1097/00002030-200201040-00004. [DOI] [PubMed] [Google Scholar]
- 24.Ioannidis JP, Havlir DV, Tebas P, Hirsch MS, Collier AC, Richman DD. Dynamics of HIV-1 viral load rebound among patients with previous suppression of viral replication. AIDS. 2000;14:1481–8. doi: 10.1097/00002030-200007280-00003. [DOI] [PubMed] [Google Scholar]
- 25.Fagard C, Oxenius A, Gunthard H, et al. A prospective trial of structured treatment interruptions in human immunodeficiency virus infection. Arch Intern Med. 2003;163:1220–6. doi: 10.1001/archinte.163.10.1220. [DOI] [PubMed] [Google Scholar]
- 26.Buchbinder SP, Mehrotra DV, Duerr A, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–93. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altfeld M, Addo MM, Shankarappa R, et al. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J Virol. 2003;77:7330–40. doi: 10.1128/JVI.77.13.7330-7340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tobery TW, Dubey SA, Anderson K, et al. A comparison of standard immunogenicity assays for monitoring HIV type 1 gag-specific T cell responses in Ad5 HIV Type 1 gag vaccinated human subjects. AIDS Res Hum Retroviruses. 2006;22:1081–90. doi: 10.1089/aid.2006.22.1081. [DOI] [PubMed] [Google Scholar]
- 29.Trigona WL, Clair JH, Persaud N, et al. Intracellular staining for HIV-specific IFN-gamma production: statistical analyses establish reproducibility and criteria for distinguishing positive responses. J Interferon Cytokine Res. 2003;23:369–77. doi: 10.1089/107999003322226023. [DOI] [PubMed] [Google Scholar]
- 30.Bhattacharya T, Daniels M, Heckerman D, et al. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315:1583–6. doi: 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- 31.Kiepiela P, Ngumbela K, Thobakgale C, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 32.Crawford H, Lumm W, Leslie A, et al. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J Exp Med. 2009;206:909–21. doi: 10.1084/jem.20081984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frahm N, McElrath MJ. AIDS Vaccine Conference. Paris, France: 19–22 October 2009. Recent immunologic findings from the Step and related HIV vaccine clinical trials. [Google Scholar]
- 34.Liu Y, McNevin J, Zhao H, et al. Evolution of human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitopes: fitness-balanced escape. J Virol. 2007;81:12179–88. doi: 10.1128/JVI.01277-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goepfert PA, Lumm W, Farmer P, et al. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J Exp Med. 2008;205:1009–17. doi: 10.1084/jem.20072457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Middleton D, Menchaca L, Rood H, Komerofsky R. New allele frequency database: doi: 10.1034/j.1399-0039.2003.00062.x. http://www.allelefrequencies.net. Tissue Antigens 2003; 61:403–7. [DOI] [PubMed] [Google Scholar]
- 37.Maiers M, Gragert L, Klitz W. High-resolution HLA alleles and haplotypes in the United States population. Hum Immunol. 2007;68:779–88. doi: 10.1016/j.humimm.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Gloster SE, Newton P, Cornforth D, et al. Association of strong virus-specific CD4 T cell responses with efficient natural control of primary HIV-1 infection. AIDS. 2004;18:749–55. doi: 10.1097/00002030-200403260-00005. [DOI] [PubMed] [Google Scholar]
- 39.Boritz E, Palmer BE, Wilson CC. Human immunodeficiency virus type 1 (HIV-1)-specific CD4+ T cells that proliferate in vitro detected in samples from most viremic subjects and inversely associated with plasma HIV-1 levels. J Virol. 2004;78:12638–46. doi: 10.1128/JVI.78.22.12638-12646.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lichterfeld M, Kaufmann DE, Yu XG, et al. Loss of HIV-1-specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1-specific CD4+ T cells. J Exp Med. 2004;200:701–12. doi: 10.1084/jem.20041270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carlson JM, Brumme ZL. HIV evolution in response to HLA-restricted CTL selection pressures: a population-based perspective. Microbes Infect. 2008;10:455–61. doi: 10.1016/j.micinf.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 42.Martinez-Picado J, Prado JG, Fry EE, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol. 2006;80:3617–23. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brumme ZL, Tao I, Szeto S, et al. Human leukocyte antigen-specific polymorphisms in HIV-1 Gag and their association with viral load in chronic untreated infection. AIDS. 2008;22: 1277–86. doi: 10.1097/QAD.0b013e3283021a8c. [DOI] [PubMed] [Google Scholar]