Abstract
Background. Annual vaccination is the primary means for preventing influenza. However, great interindividual variability exists in vaccine responses, the cellular events that take place in vivo after vaccination are poorly understood, and appropriate biomarkers for vaccine responsiveness have not been developed.
Methods. We immunized a cohort of healthy male adults with a licensed trivalent influenza vaccine and performed a timed assessment of global gene expression before and after vaccination. We analyzed the relationship between gene expression patterns and the humoral immune response to vaccination.
Results. Marked up regulation of expression of genes involved in interferon signaling, positive IL-6 regulation, and antigen processing and presentation, were detected within 24 hours of immunization. The late vaccine response showed a transcriptional pattern suggestive of increased protein biosynthesis and cellular proliferation. Integrative analyses revealed a 494-gene expression signature—including STAT1, CD74, and E2F2—which strongly correlates with the magnitude of the antibody response. High vaccine responder status correlates with increased early expression of interferon signaling and antigen processing and presentation genes.
Conclusions. The results highlight the role of a systems biology approach in understanding the molecular events that take place in vivo after influenza vaccination and in the development of better predictors of vaccine responsiveness.
Influenza viruses circulate worldwide, causing an estimated 250,000–500,000 deaths each year [1]. In the United States alone, yearly epidemics affect an estimated 2–5% of the population [2] and are responsible for an average of 200,000 hospitalizations [3] and 36,000 deaths [4]. Since 1977, influenza A (H3N2), influenza A (H1N1), and influenza B viruses have been responsible for the majority of documented infections. Trivalent inactivated vaccines, updated yearly based on the currently circulating strains, are the primary tools used for preventing influenza [5, 6]. Recent systematic reviews have highlighted the need for better vaccines [7–9]. It is clear that the protection offered by the currently available vaccines is incomplete, and that large interindividual variation exists [10]. Furthermore, the cellular events that take place in vivo after vaccination and the correlates of vaccine-induced protection are incompletely understood. Genome-wide transcriptional analysis has recently been introduced as a useful tool to study the mechanisms of viral infection [11, 12] and host responses to vaccination [13–15]. However, an integrated, systematic evaluation of the transcriptional response to influenza vaccination using multiple time points and incorporating antibody response data has not been performed to date.
We immunized a cohort of 119 healthy male adults, ages 18–40, with a licensed trivalent influenza vaccine and assessed the genome-wide gene expression patterns in peripheral blood cells before and on days 1, 3, and 14 after vaccination. Antibody titers were measured before and on days 14 and 28 after vaccination. We then studied the correlation between changes in gene expression and the antibody response following influenza vaccination.
METHODS
Study Subjects
Healthy volunteers ages 18–40 years were enrolled (n = 119). Given the systematic differences in gene expression between sexes and among ethnic groups [16, 17], this initial cohort was limited to male individuals of self-reported Caucasian ancestry. Individuals who were known to have received an influenza vaccine in the previous three years were not included. Enrollment, vaccination, and sample collection were conducted at a university campus. The protocol was approved by the institutional review boards of all participating institutions. Informed consent was obtained from each subject prior to enrollment.
Vaccine
Participants were immunized on day 0 with the 2008–2009 trivalent influenza vaccine [A/Brisbane/59/2007(H1N1), A/Brisbane/10/2007(H3N2), B/Florida/4/2006; Sanofi-Pasteur].
RNA Samples
Peripheral blood samples for RNA purification (n = 461) were obtained before and on days 1, 3, and 14 after immunization. To minimize changes in gene expression induced by sample handling and processing, whole blood samples (2.5 mL) were collected in PAXgene RNA stabilization tubes (QIAGEN) and frozen at −80°C. RNA purification was performed using the PAXgene Blood RNA system (QIAGEN). Spectrophotometry (NanoDrop-1000 Spectrophotometer, Thermo Fisher Scientific) and microfluidic electrophoresis (Experion Automated Electrophoresis System, Bio-Rad Laboratories) were used for quality control.
Microarray Analysis
In vitro transcription was performed using Ambion Illumina TotalPrep RNA Amplification Kits (Applied Biosystems/Ambion). cRNA was hybridized onto Illumina Human HT-12v3 Expression BeadChips (Illumina), following the manufacturer’s protocol. All samples for each individual were processed simultaneously. The arrays included over 25,000 well-characterized genes (including splice variants) and nonannotated gene candidates. Standard quality control thresholds were applied after preprocessing of signal intensity data, and failed microarrays were removed [18, 19]. We also excluded arrays from individuals for which data was not available at all time points. The final set of 368 arrays represented 92 individuals at four time points. We required a detection P value of <.05 in at least 65% of the samples for a transcript to be considered detected. There were 12,795 detected transcripts in our data. To identify genes with levels of expression that changed after vaccination, we used the expression values of each transcript at the four time points in a linear mixed-effects analysis of variance (ANOVA) model, using day as a fixed effect and person as a random effect.
Serum Samples and Antibody Titer Measurements
Whole blood (10 mL) was collected in Vacutainer Serum Separator Tubes (Beckton-Dickinson). Serum was separated by centrifugation prior to storage at −20°C. Hemagglutination inhibition (HAI) tests were performed as previously described [20], except for a starting serum sample dilution of 1:4 and the use of turkey red blood cells (RBCs). HAI test antigens were allantoic fluid harvests from infected embryonated hen’s eggs (whole virus antigens). Neutralizing antibody tests were performed as previously described [21] except that hamster serum was not included. Test strains were the same as those used in the vaccine.
Data Processing and Statistical Analysis
Initial quality control of the microarray signal intensity data was performed using the lumi Bioconductor package [18] in the R programming language [22]. The same software was used for variance stabilization transformation [19] and robust spline normalization of the expression data. Regression and ANOVA were carried out in R.
Pathway Enrichment and Content Analysis
The Gene Ontology (GO) vocabulary was obtained from the GO Web site (http://www.geneontology.org, 2009 build). Sequence information for reporters in the microarrays was converted to nuID annotation [18]. The corresponding Entrez identifiers were mapped to the GO data structure. Using our own ontology analysis system, OntologyTraverser [23], we tabulated the genes annotated at or below each GO node for the entire array. We used a hypergeometric sampling model to examine the statistical representation of each GO node for genes in our gene sets. To compare sets, we took differences between the standardized scores determined for each gene set. Gene lists were simultaneously analyzed using Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems) and DAVID Ontology [24, 25] (http://www.david.abcc.ncifcrf.gov) to confirm significantly associated pathways.
Calculation of the Titer Response Index
We performed regression analyses for each vaccine component in each of the two antibody titer measurements (Figure S1, supplementary data). Because the results for the two antibody titer data sets (HAI and neutralization) were strongly correlated, we combined the information from the two assays for downstream TRI calculations. We related the change in titer between pre- and postvaccination measurements (response variable) to the prevaccination titer (explanatory variable) using a simple linear model with an additional term to account for spurious variation attributable to recruitment dates (Figure S1, supplementary data). We next determined the residuals from the above linear regressions and used them as the input values for the individual response scores. The residuals are the distance of each individual point from the regression fit (Figure S2, supplementary data). These residuals reflect how the antibody titer rise of an individual deviates from the expected rise given the typical response of individuals within the cohort with a given prevaccination titer level. To increase the accuracy of combining the titer scores across vaccine components, we standardized the residuals by dividing by the residual standard deviation for each component; we then computed four outcome scores within each titer response measurement for each individual. These four summaries are the mean standardized residual across vaccine components, median standardized residual across vaccine components, mean rank (across all individuals, across vaccine components), and median rank. These scores were correlated (Figure S2, supplementary data). The scores were then standardized again (mean = 0, variance = 1 across all individuals), and scores within each individual were averaged to obtain the titer response index (TRI). The TRI, therefore, is a summary of the aggregate residuals of all antigen components in the vaccine with both HAI and neutralization antibody titers incorporated. The TRI is designed specifically for carrying out the correlation analyses between gene expression and antibody response.
Correlation of the Gene Expression and Antibody Titer Data
In this regression model, we treated the expression data for all detected transcripts as the outcome, and the day and TRI as explanatory variables. We computed false discovery rate (FDR) q values (Benjamini-Hochberg method) for the partial F statistics of titer score against expression. We used an FDR cutoff of 0.05 and a TRI slope absolute magnitude cutoff of 0.1.
Cross-validation
We sequentially excluded the values for each individual and used the expression data and titer scores on the remaining individuals to determine the correlation between the TRI and the expression values. We then attempted to predict the TRI value of the individual that was excluded from the model selection and fitting process using the expression values for the target individual and the model fit from the rest of the cohort. To make the process more robust and to balance the effect of each gene, we standardized the expression data to have a mean of zero and variance of 1 among the set of genes and individuals considered. We used a subset of genes with known functions in antigen presentation, interferon response, and peroxidase activity processes (Table 1). We identified day 1 and day 3 after vaccination as the days with the strongest correlation. We then fit the TRI to expression relation within each of these days. The cross-validation prediction for each individual was determined as the maximum absolute prediction value obtained from either day 1 or day 3.
Table 1.
Subset of Transcripts Used for Prediction of the TRI
| lumi ID | Accession No. | Gene | Upregulated in |
| ix1KUCgkUiUOgfjpXA | NM_007315 | STAT1 | High responders |
| 0tbjDbteANdkRHp66I | NM_006084 | IRF9 | High responders |
| Ki3QtbOwNI3Vd0VdUU | NM_001080547 | SPI1 | High responders |
| xVb14TRJJdQRCnUIXI | NM_001025158 | CD74 | High responders |
| lBLUXe7cU4VX10R4Xs | NM_005516 | HLA-E | High responders |
| fcZTqE3D0iggrU7f6o | NM_006573 | TNFSF13B | High responders |
| Qa6_OmX0CK4J7aHECo | NM_005809 | PRDX2 | Low responders |
| KWIca7nvIK9635HCK0 | NM_006793 | PRDX3 | Low responders |
| TWl6.Zetu384kU7CXU | NM_004091 | E2F2 | Low responders |
| cw3oT2KD6We0efvv5k | NM_000314 | PTEN | Low responders |
| r7J67r56D73z6X5PHk | NM_002211 | ITGB1 | Low responders |
RESULTS
Maximum Changes in Gene Expression Occur in the Initial 24 Hours After Vaccination
We identified expression changes in 4740 transcripts (Dataset 1) after imposing a FDR cutoff of 0.01 on the P value for the day effect. Due to the assay’s design, some reporters mapped to different regions or alternatively spliced transcripts of the same gene. The reporters corresponding to differentially expressed transcripts in our data set uniquely mapped to 3854 RefSeq-annotated genes and 242 nonannotated gene candidates. The maximum expression change occurred on day 1, while day 14 had the greatest number of genes with maximum expression values. Interindividual differences in expression are reflected in the magnitude of person-effects in the ANOVA model (Dataset 1). The top 1% differentially expressed genes included interferon-inducible genes (eg, IFIT1, MX1, and IRF9), the signal transducer and activator of transcription (STAT) gene family, genes with ribosomal functions (eg, RPL7, RGS18, and RPS27), and genes with unclear functions (eg, WARS and the family of guanylate-binding proteins).
Three Distinct Patterns of Gene Expression Are Observed After Influenza Vaccination
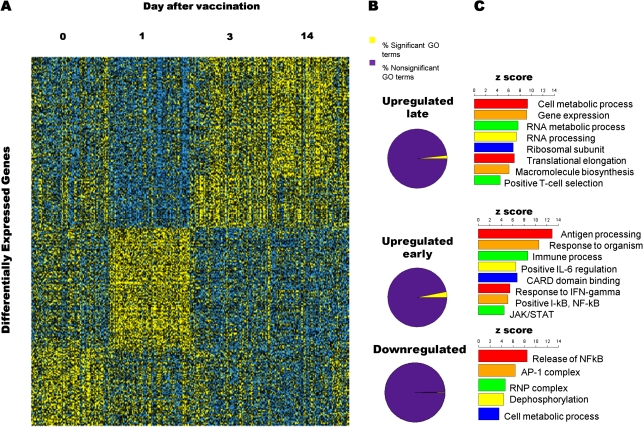
A visual summary of the dynamic changes in the transcriptional response to vaccination over time shows three major gene expression patterns: downregulation, early upregulation, and late upregulation (Figure 1A). Genes with late upregulation of expression were not significantly different between the last two time points. We evaluated the gene content of each of the above patterns of expression using three methodologies: association of Gene Ontology (GO) terms (http://www.geneontology.org) using our own R software package [16], the DAVID bioinformatics database [17, 18], and Ingenuity Pathway Analysis software. These results are summarized in Figure 1B, and complete gene lists are provided in Datasets 2–8. As expected, the early phase of transcriptional activation was marked by upregulation of genes involved in multiple immune processes. The data suggest early expression of genes whose products act on host viral sensing via Toll-like receptors TLR7 and TLR8. Overall, there were significantly increased transcript levels for genes participating in antiviral defense response (eg, GO:0009615), cellular activation (eg, GO:0001775), and cellular differentiation (eg, GO:0045582). Activation of antigen processing and presentation is implied by the increased expression of major histocompatibility complex (MHC) Class I genes (eg, GO:002474). Associations were most significant for the interferon response pathway (GO:0005062, GO:0034341, and GO:0034340) when compared with other cytokines. Of the signal transduction pathways, the Janus kinase/signal transducers and activators of transcription (JAK/STAT) (GO:007259) and nuclear factor kappa beta (NFκβ) (GO:0043122) axes were particularly enriched. In contrast, the late vaccine response was characterized by increased detection of transcripts for genes involved in intracellular processes suggestive of active cellular proliferation, including macromolecule biosynthesis (eg, GO:0009059), cellular protein metabolism (eg, GO:0044267), RNA metabolic processing (eg, GO:0016070), and regulation of antiapoptosis (GO:0045767).
Figure 1.
The human transcriptional response to trivalent influenza vaccination shows three distinct patterns of gene expression. (A) Heatmap illustrating the patterns of expression for all differentially expressed genes. Yellow indicates a higher value. Three distinct patterns of expression are evident: early upregulation, late upregulation and downregulation. (B) Proportions of significant and nonsignificant Gene Ontology (GO) categories for each of the three expression patterns. (C) Selection of the most important GO terms for each expression pattern. Higher z scores indicate that a GO term is more likely to be enriched in a given set of differentially expressed genes.
A Classification Model for Antibody Responsiveness
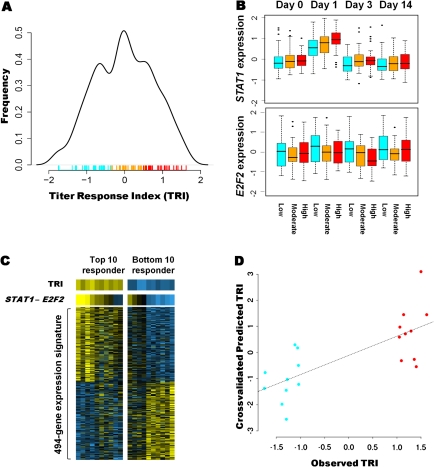
Antibody titers were measured by both hemagglutination inhibition (HAI) and neutralization assays. The titer results from these assays were strongly correlated, and the titers generally plateaued after day 14. We constructed a classification model for vaccine responsiveness (the Titer Response Index, TRI) based on the aggregate titer changes for the three antibodies. The TRI robustly aggregates both the individual’s response across the vaccine components and the individual’s response relative to the rest of the cohort. Interestingly, the distribution of the TRI scores is trimodal, facilitating an unbiased classification of individuals as high, intermediate, and low vaccine responders (Figure 2A), depending on where the individual’s TRI falls under this distribution. The TRI and rise in HAI titer for the 92 individuals show a significant positive relationship (r2 = 0.4585).
Figure 2.
Early patterns of gene expression correlate with responsiveness to trivalent influenza vaccine. (A) Probability density estimate of the titer response index (TRI) in our cohort. A trimodal distribution for the vaccine response is observed. The hash marks below the curve are the TRI values for each individual, and the colors correspond to low, intermediate, and high responders. (B) STAT1 and E2F2 expression in the three responder groups at each time point. STAT1 expression increases after vaccination, most prominently on day 1 and in the high-responder group. E2F2 expression is downregulated after vaccination, most prominently on day 3 and with greater downregulation in the high-responder group. (C) Heatmap of the expression signature of vaccine responsiveness in the top 10 (left) and bottom 10 (right) responders. The expression values for each gene were standardized at each time point, and the day 1 and day 3 values were averaged. Yellow indicates a higher value. Each column represents an individual. The top row illustrates the TRI for each individual. The difference between STAT1 and E2F2 expression alone can differentiate individuals at the two extremes of the response spectrum, as illustrated in the second row. The bottom rows display the expression patterns for the 494 genes in the gene expression signature. (D) Cross-validation prediction data, illustrating how expression values can be used to predict the TRI.
A Distinct Gene Expression Signature Correlates with the Magnitude of the Antibody Response to Influenza Vaccination
To assess whether specific patterns of gene expression correlated with the magnitude of the antibody response, we studied the relationship between gene expression and the TRI in all subjects. We found that the abundance level of 494 transcripts significantly correlated with the TRI (Dataset 8). These mapped to 481 known genes and 13 nonannotated gene candidates. Two opposing expression trends influenced the magnitude of the antibody response, and the maximum effects were observed at specific time points as illustrated by the genes STAT1 and E2F2 in Figure 2B. A visual heatmap display of the entire gene expression signature in those individuals with the 10 highest and 10 lowest TRI scores is given in Figure 2C (bottom). Remarkably, the difference between STAT1 and E2F2 expression alone generates a gradient that corresponds clearly to the TRI at the two extremes of the response spectrum (Figure 2C, top).
We then performed a cross-validated linear prediction procedure utilizing the expression values of a subset of differentially expressed genes (Table 1) to predict the antibody response scores of those individuals with the highest and lowest antibody responses as depicted in Figure 2C. We show that the residual expression values on days 1 and 3 are predictive of the observed TRI for each individual and, more importantly, that the cross-validated predicted TRI recategorized the highest and lowest responders into the appropriate groups (Figure 2D). The prediction performed particularly well in the individuals who mounted the least immune response.
Early Upregulation of Interferon Response and Antigen Presentation Pathways is Associated With a Higher Antibody Response
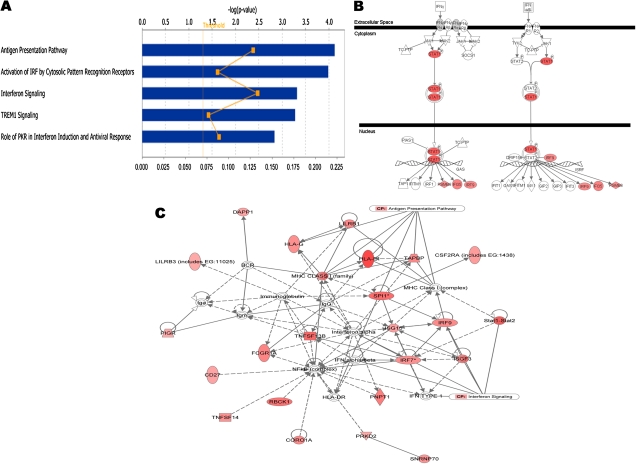
Content analysis of the genes that were upregulated in the high-responder group showed enrichment of GO categories involved in cellular immune responses, most notably the interferon response and antigen presentation pathways (Figure 3A and Datasets 9, 10). Genes within the interferon response pathway were highly upregulated in the high-responder group (Figure 3B). A functional interaction network for this subset of genes is presented in Figure 3C. In contrast, the majority of upregulated genes and enriched functional pathways in low responders were not specifically related to cell-mediated immune responses (Figure S3 and Datasets 11, 12).
Figure 3.
Interferon signaling and antigen presentation genes are preferentially upregulated in individuals with high vaccine responsiveness. (A) Top enriched pathways in the high responder group. The negative log P value, along the x-axis, increases as a pathway is more significantly associated. The ratio indicates the proportion of upregulated genes relative to all the genes present in a pathway. (B) Genes that act in the interferon pathway and which are preferentially upregulated in the high vaccine responders are shown in red. (C) Top functional network for genes that are preferentially upregulated in the high responder group, including genes that interact directly (solid lines) and indirectly (broken lines). Functionally, the network corresponds to antigen presentation, cellular growth and proliferation, and hematological system development and functions. Shades of red are used to display the level of expression for each gene in the high-responder relative to the low-responder groups.
DISCUSSION
This study illustrates how a systems biology approach can be applied in a clinical scenario to understand the complex molecular events that take place in vivo after trivalent influenza vaccination, and to develop better molecular biomarkers for vaccine responsiveness. The implications of the findings, therefore, extend from vaccine biology to vaccine development and clinical vaccinology.
The data support a model in which genes involved in interferon signaling and antigen presentation pathways are strongly upregulated in the initial 24 hours after vaccination, and the expression pattern of early-activation genes correlates strongly with the magnitude of the antibody response measured 14 and 28 days after vaccination. The simultaneous assessment of transcript abundance for over 25,000 genes and gene candidates offers an unbiased, genome-wide view of the transcriptional events that take place after vaccination. By performing this analysis before and at three time points after vaccination, we have gained a deeper understanding of these events and their timing than had been possible before. The three patterns of coexpression that became evident from our analysis are quite distinct and suggest a previously undescribed biphasic transcriptional response to vaccination. The observation of maximum expression changes in the initial 24 hours after vaccination and the correlation between early expression and the magnitude of the antibody response are important new contributions to the understanding of the vaccination response sequence. Content analysis suggests a central role in the early vaccine response for genes whose products are involved in viral sensing via TLR7 and TLR8, MHC Class I presentation, interferon signaling, IL-6, and the NFκβ and JAK/STAT signaling pathways. These genes are also known to be activated in innate cells (ie, macrophages and dendritic cells) during viral infections [26]. Days 3 and 14 show similar patterns of expression, more suggestive of increased RNA processing and protein synthesis. For inactivated influenza vaccines, therefore, these findings suggest that the first 24 hours after vaccination are of great biological importance.
As the field of clinical vaccinology moves into an era of high-throughput experimental data generation and systems-level biology, it is imperative to give serious statistical consideration to the limitations of current methods of assessment of the magnitude of immune responses. With trivalent influenza vaccines, which in the case of influenza are of high clinical importance, three factors are known to complicate the quantitative analysis of antibody titer data in clinical studies. First, by young adulthood, most individuals have been exposed to influenza antigens through infection or prior vaccination. Indeed, despite our exclusion of individuals who had been vaccinated over the previous three years, most of the study participants had measurable prevaccination antibody titers. Second, individuals with higher prevaccination titers have smaller differences between pre- and postvaccination titers. Accordingly, we observed in our data an approximately linear inverse correlation between prevaccination titers and the rise in titer. For this reason, a simple calculation of the titer change (the titer delta) is inadequate as a way to classify the vaccine responsiveness of individuals. Finally, influenza vaccination currently involves the concomitant administration of three antigens and, while the immune response takes place simultaneously and separations are somewhat artificial, there is inter- and intraindividual variation in the response to each. These observations are depicted in Figure S2. We, therefore, developed the TRI (Titer Response Index) as a classification method for these data. The TRI accounts for an individual’s prevaccination antibody titers, their titers for the three antigens, and the magnitude of their response relative to other individuals in the population. The observed trimodal distribution clearly deviates from what would be expected by chance alone, permitting a statistically significant classification of trivalent influenza vaccine recipients on the basis of responsiveness.
Compared to previous studies, our study has a larger sample size, which provides sufficient power to detect significant correlations between transcript abundance and responsiveness. By assessing the relationship between the early patterns of gene expression and the magnitude of the antibody response, we have discovered 494 transcriptional biomarkers that strongly correlate with the humoral immune response to trivalent influenza vaccines. Our content and network analyses of this predictive gene expression signature again point clearly toward the interferon and antigen presentation pathways. Genes in these pathways, as well as other genes with levels of expression that are clearly different between the highest and lowest vaccine responders in our study (CD74, HLA-E, E2F2, and PTEN) are potential targets for functional studies linking the early molecular events that follow vaccination with the subsequent adaptive immune response. Genomic regions that control expression of these genes could be important for studies seeking to explain interindividual variation in vaccine responsiveness at the DNA level.
A full predictive analysis using the 92 individuals for which expression data was analyzed would be ideal for validating these correlated transcripts. However, our power to perform cross-validation on individuals in the intermediate response range is affected by the expression differences that result from interindividual variation. Fortunately, these subjects appear to develop protective immune responses after vaccination and are less crucial to the interpretation of the data. Since individuals who did not mount a response to the trivalent influenza vaccine are the most clinically interesting and, given that the vaccine response of the entire cohort follows a trimodal distribution, we sampled from the extreme ends of this distribution in order to maximize the power of the cross-validation analyses. Specific patterns of gene expression characterize individuals at the two extremes of the antibody response spectrum, and cross-validation illustrates their predictive value. The fact that a simple gradient of STAT1 to E2F2 expression can predict an individual’s response status at the extremes of the spectrum, and the fact that cross-validation could be performed using a small subset of representative genes, underscore the magnitude of the differences and suggest that simplified predictors based on early gene expression patterns are possible. Further validation of our gene expression-based prediction model in another and preferably larger group of volunteers is warranted. Ideally, this would include individuals of both sexes and different ethnicities.
Early molecular predictors of vaccine responsiveness can be useful in the development and comparison of new vaccines and adjuvants. They can also play a role in studies of vaccine response among different subgroups (children, the elderly, or immunocompromised patients, for example) and open the door for studies of individualized vaccine regimens. Past and recent influenza pandemics highlight the need for studies geared toward rapid implementation of clinical and translational research findings.
DATASETS
The following datasets will be available for download from our laboratory’s Web site (www.bcm.edu/cnrc/faculty/cnrcbelmont):
Dataset 1. List of Differentially Expressed Transcripts.
Dataset 2. Gene Ontology Results for Downregulated Genes.
Dataset 3. Ingenuity Pathway Analysis Results for Downregulated Genes.
Dataset 4. Gene Ontology (GO) Results for Early Upregulated Genes.
Dataset 5. Ingenuity Pathway Analysis Results for Early Upregulated Genes.
Dataset 6. Gene Ontology (GO) Results for Early Upregulated Genes.
Dataset 7. Ingenuity Pathway Analysis Results for Late Upregulated Genes.
Dataset 8. Gene Expression Signature for Influenza Vaccine Responses.
Dataset 9. DAVID Gene Ontology (GO) Results for Highly Expressed Genes in High Responders.
Dataset 10. Ingenuity Pathway Analysis Results for Highly Expressed Genes in High Responders.
Dataset 11. DAVID Gene Ontology (GO) Results for Highly Expressed Genes in Low Responders.
Dataset 12. Ingenuity Pathway Analysis Results for Highly Expressed Genes in Low Responders.
Supplementary Data
Supplementary data are available at http://jid.oxfordjournals.org online.
Funding
This work is supported by a contract from the U.S. National Institutes of Health (NO1-AI-030039). K.L.B. is supported by a Ruth L. Kirschtein NRSA Pre-doctoral Training Grant (F31AI071372-03).
References
- 1.World Health Organization. Seasonal influenza. http://www.who.int/mediacentre/factsheets/fs211/en/. Accessed April 2009. [Google Scholar]
- 2.Centers for Disease Control Prevention. Influenza: the disease. http://www.cdc.gov/flu/weekly/. Accessed 4 February 2011. [Google Scholar]
- 3.Thompson WW, Shay DK, Weintraub E, et al. Influenza-associated hospitalizations in the United States. JAMA. 2004;292:1333–40. doi: 10.1001/jama.292.11.1333. [DOI] [PubMed] [Google Scholar]
- 4.Thompson WW, Shay DK, Weintraub E, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–86. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 5.Couch RB. Seasonal inactivated influenza virus vaccines. Vaccine. 2008;26(Suppl 4):D5–9. doi: 10.1016/j.vaccine.2008.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keitel WA, Couch RB. Inactivated influenza vaccines. In: Potter CW, editor. Influenza. New York: Elsevier; 2002. pp. 145–77. [Google Scholar]
- 7.Jefferson T, Rivetti A, Harnden A, Di Pietrantonj C, Demicheli V. Vaccines for preventing influenza in healthy children. Cochrane Database Syst Rev. 2008 doi: 10.1002/14651858.CD004879.pub3. CD004879. [DOI] [PubMed] [Google Scholar]
- 8.Jefferson TO, Rivetti D, Di Pietrantonj C, Rivetti A, Demicheli V. Vaccines for preventing influenza in healthy adults. Cochrane Database Syst Rev. 2007 doi: 10.1002/14651858.CD001269.pub3. CD001269. [DOI] [PubMed] [Google Scholar]
- 9.Rivetti D, Jefferson T, Thomas R, et al. Vaccines for preventing influenza in the elderly. Cochrane Database Syst Rev. 2006;3 doi: 10.1002/14651858.CD004876.pub2. CD004876. [DOI] [PubMed] [Google Scholar]
- 10.Thomas C, Moridani M. Interindividual variations in the efficacy and toxicity of vaccines. Toxicology. 2010;278:204–10. doi: 10.1016/j.tox.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Shapira SD, Gat-Viks I, Shum BO, et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–67. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaas AK, Chen M, Varkey J, et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe. 2009;6:207–17. doi: 10.1016/j.chom.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haining WN, Wherry EJ. Integrating genomic signatures for immunologic discovery. Immunity. 2010;32:152–61. doi: 10.1016/j.immuni.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Zhu W, Higgs BW, Morehouse C, et al. A whole genome transcriptional analysis of the early immune response induced by live attenuated and inactivated influenza vaccines in young children. Vaccine. 2010;28:2865–76. doi: 10.1016/j.vaccine.2010.01.060. [DOI] [PubMed] [Google Scholar]
- 15.Querec TD, Akondy RS, Lee EK, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–25. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitney AR, Diehn M, Popper SJ, et al. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci USA. 2003;100:1896–901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Idaghdour Y, Storey JD, Jadallah SJ, Gibson G. A genome-wide gene expression signature of environmental geography in leukocytes of Moroccan Amazighs. PLoS Genet. 2008;4:e1000052. doi: 10.1371/journal.pgen.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du P, Kibbe WA, Lin SM. Lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–8. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 19.Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dowdle WR, Kendal AP, Noble GR. Influenza viruses. In: Lenette EH, Schmidt NJ, editors. Diagnostic procedures for viral, rickettsial and chlamydial infections. Washington, DC: American Public Health Association; 1979. pp. 603–5. [Google Scholar]
- 21.Frank AL, Puck J, Hughes BJ, Cate TR. Microneutralization test for influenza A and B and parainfluenza 1 and 2 viruses that uses continuous cell lines and fresh serum enhancement. J Clin Microbiol. 1980;12:426–32. doi: 10.1128/jcm.12.3.426-432.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R Development Core Team. R: a language environment for statistical computing. (2.9 ed) Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 23.Young A, Whitehouse N, Cho J, Shaw C. OntologyTraverser: an R package for GO analysis. Bioinformatics. 2005;21:275–6. doi: 10.1093/bioinformatics/bth495. [DOI] [PubMed] [Google Scholar]
- 24.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 26.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–20. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]