Abstract
Background. We aimed to determine the antibody and T cell responses to Burkholderia pseudomallei of humans to select candidate vaccine antigens.
Methods. For antibody profiling, a protein microarray of 154 B. pseudomallei proteins was probed with plasma from 108 healthy individuals and 72 recovered patients. Blood from 20 of the healthy and 30 of the recovered individuals was also obtained for T cell assays.
Results. Twenty-seven proteins distinctively reacted with human plasma following environmental exposure or clinical melioidosis. We compared the responses according to the patient’s history of subsequent relapse, and antibody response to BPSL2765 was higher in plasma from individuals who had only 1 episode of disease than in those with recurrent melioidosis. A comparison of antibody and T cell responses to 5 B. pseudomallei proteins revealed that BimA and flagellin-induced responses were similar but that BPSS0530 could induce T cell responses in healthy controls more than in recovered patients.
Conclusions. By combining large-scale antibody microarrays and assays of T cell–mediated immunity, we identified a panel of novel B. pseudomallei proteins that show distinct patterns of reactivity in different stages of human melioidosis. These proteins may be useful candidates for development of subunit-based vaccines and in monitoring the risks of treatment failure and relapse.
The gram-negative bacillus Burkholderia pseudomallei causes human melioidosis, which ranges from a chronic localized infection to severe disseminated infection and often septicemia [1]. This disease occurs in tropical areas, particularly Southeast Asia and northern Australia, but is being increasingly reported in other tropical regions [2, 3]. B. pseudomallei can also establish persistent and asymptomatic infections that can emerge up to 62 years later as fulminant disease [4]. Individuals who recover from the disease may experience subsequent episodes of clinical illness, with 6%–13% of these cases due to reinfection but the majority a consequence of the failure to eliminate the pathogen after treatment with antimicrobials [5].
The host immune responses required to recover from melioidosis or to prevent infection in humans living in melioidosis-endemic areas are largely unknown. With use of a murine model of melioidosis, both cell-mediated and humoral immune responses have been shown to play roles in protection [6]. Cell-mediated responses involving natural killer (NK) cells and adaptive T cells producing interferon-γ (IFN-γ) play an important role in control of infection [7–9]. Our previous studies have revealed that memory CD4+, CD8+T (TEMRA), and NK cells from seropositive healthy individuals living in endemic areas or from individuals who have recovered from melioidosis are primed and produce IFN-γ in vitro in response to killed B. pseudomallei or the bacterial ABC transporter proteins, LolC, OppA, or PotF. The magnitude of these cellular responses correlated with antibody titers to killed B. pseudomallei cells detected by means of conventional indirect hemagglutination assay (IHA) [10]. However, the identity of other antigens recognized by the plasma of these individuals is not known.
High-throughput protein microarrays have previously been developed and used to map the humoral responses to individual bacterial and viral proteins [11–16]. Recently, we have devised a B. pseudomallei protein array and probed it with serum specimens from acute melioidosis patients in Northeast Thailand and Singapore. Mapping the profile of antibody responses has allowed the identification of proteins that can be used as serodiagnostic antigens for melioidosis [17]. The potential for these antigens to stimulate cell-mediated immune responses and the identification of proteins that could induce protective immune responses has not been reported.
This study aimed to identify B. pseudomallei proteins that could be candidate protective antigens. A B. pseudomallei protein array was probed with plasma from individuals who had recovered from melioidosis after receiving antibiotic therapy and from seropositive individuals living in endemic areas but with no history of melioidosis. We also sought to determine whether recurrent disease, septic disease, or localized infection influenced the antibody response profile and how these antibody responses were related to T cell responses in individuals. In the longer term, our results will support research to devise vaccines against melioidosis.
MATERIALS AND METHODS
Blood Samples
Recovered melioidosis patients and healthy control individuals were enrolled in this study and recruited by a study team based at Sappasithiprasong Hospital, Ubon Ratchathani, Northeast Thailand. Ethical permission was obtained from Ethical KKU research, no. HE470506 (Scanning the Burkholderia pseudomallei proteome for vaccine antigens). Recovered melioidosis patients were defined as individuals who had a history of clinical melioidosis (confirmed by culture positive for B. pseudomallei from clinical samples) but at the time of blood collection had completed a course of antibiotic treatment and had no sign of active melioidosis. Recurrent melioidosis infection was defined as new symptoms and signs of infection in association with a culture positive for B. pseudomallei following previous treatment and response to oral antibiotic therapy [5]. Healthy control individuals had no history of melioidosis and included seropositive individuals tested by means of IHA (titer, >40) and seronegative individuals. Plasma samples from 72 recovered melioidosis patients and 108 control individuals were used to probe B. pseudomallei protein arrays, and blood samples from 30 recovered melioidosis patients and 20 healthy control individuals were used for cell-mediated immune response assays. The details of sample demographic characteristics have been described elsewhere [10].
Antibody Detection with Use of Protein Microarray Analysis
Fabrication of the B. pseudomallei protein array and probing with plasma samples were as described elsewhere [18]. In brief, B. pseudomallei strain K96243 DNA was used as a template for the polymerase chain reaction (PCR). PCR products were cloned into a T7 expression vector by means of homologous recombination. Purified plasmids were expressed in the Escherichia coli–based rapid translation system (RTS-100) for in vitro transcription/translation (Roche) and printed without additional purification. The protein array chip used in this study comprised 154 recombinant B. pseudomallei antigens that were downselected from a larger 1205 proteome array after probing with melioidosis patient serum [17]. In addition, 4 serial dilutions of human IgG, 6 spots comprising RTS-100 reactions with nonrecombinant template vector, and 4 serial dilutions of EBNA1 protein were also printed as internal positive, negative, and serum controls, respectively. Plasma was reacted with the chips using a previously reported protocol [18]. Briefly, the 16-pad Bp chip was prewetted with 80 μL of blocking buffer (Whatman) at 25oC for 30 min. Serum samples were diluted 1:50 in blocking buffer containing 10% E. coli lysate and incubated for 2 h at 25oC with orbital shaking at 90 rpm. The arrays were washed 7 times with tris-buffered saline with 0.05% (v/v) Tween-20 for 5 min at 25oC. After adding 80 μL of goat anti–human IgG γ chain secondary antibody (Jackson Immuno) and diluting 1:100 in blocking buffer, the chips were incubated for 2 h at 25oC with orbital shaking. After washing, tertiary reagent (Streptavidin-PBXL3; Martek) diluted 1:100 in blocking buffer was added for 30–60 min with shaking. Slides were scanned using a ProScanArray HT microarray scanner and signal intensities quantified with ScanArray Express software (Perkin-Elmer). The signal intensities were normalized by using the R statistical environment (http://www.R-project.org). Normalized data were used for a Student t test (SPSS software) in differential analyses of array data. Then normalized data were retransformed into approximate raw values and presented in heat maps or plotted as graphs. In addition, the raw values were calculated for average signal intensity, standard deviation (SD), and standard error (SE) of retransformed data using Excel software (Microsoft).
Detection of Antibody to B. pseudomallei by Indirect Hemagglutination
IHA titers of all samples were determined using a passive hemagglutination assay kit (Center for Immunodiagnostic Production). Briefly, 2-fold serial dilutions of plasma samples (1:10 to 1:1280) were incubated with heat-killed B.pseudomallei–sensitized sheep erythrocytes for 2 h at 25oC, and the IHA titer was read in comparison with unsensitized sheep red blood cells [19].
Selection of B. pseudomallei Recombinant Proteins and Control Antigens for T Cell Studies
Five recombinant B. pseudomallei proteins previously shown to be immunoreactive in melioidosis patients were generated using the RTS-100 system. RTS protein was used for whole-blood assay without purification [14, 20]. Controls included phytohemagglutinin (PHA; Seromed) and IL-12 plus IL-15 (R&D Systems), which was used as a control that enhanced IFN-γ production via cytokine independently of T cell receptors. Unrelated proteins used as negative controls included recombinant proteins of Franciscella tularensis (FT1823), which were produced in the RTS system using the same method as that for B. pseudomallei proteins [21].
Whole-Blood Cultures and Assay for IFN-γ
Screening for IFN-γ production was performed by stimulating whole blood with individual B. pseudomallei proteins and measuring the IFN-γ concentration in culture supernatant by means of ELISA as previously described [10]. The experiments were conducted in the presence or absence of 0.3 μg/mL cyclosporin A (CsA; R&D Systems), a substance known to inhibit the T cell receptor-dependent pathway. Briefly, whole-blood samples were adjusted to a concentration of 9 × 105 lymphocytes/mL and incubated with medium alone or with optimal concentrations of stimulators, including 3 μg/mL of selected B. pseudomallei proteins. Controls were 1.25 μg/mL PHA, 1 ng/mL IL-12+IL15, 3 × 106 heat-killed B. pseudomallei, or 1 μg/mL F. tularensis or vaccinia virus proteins. Specific IFN-γ responses of all stimulations were subtracted from total IFN-γ when adding CsA, and the results were expressed as the mean of duplicate tests for each sample.
Statistical Analysis
The results of IFN-γ production and antibody levels were calculated using PRISM Software (GraphPad Software) and using the Mann-Whitney statistical test. Pvalues of <.05 were considered to reveal a statistically significant difference.
RESULTS
Identification of B. pseudomallei Antigens Recognized by Plasma from Healthy Control Participants and Recovered Melioidosis Patients Using a Protein Microarray
Initially, we assessed the utility of a protein microarray to profile the antibody responses to B. pseudomallei proteins in plasma taken from individuals in Thailand assigned to 2 groups: (1) 108 healthy individuals from areas where melioidosis is endemic (Northeast Thailand) but who had not previously received a diagnosis of melioidosis (“healthy control” samples) and (2) 72 recovered individuals (“recovered” samples), who had previously been treated for melioidosis, had recovered, and were currently free of any signs or symptoms of melioidosis. The array consisted of 154 recombinant B. pseudomallei polypeptide antigens previously shown to react with antibody in serum samples from melioidosis patients and control proteins [17]. Most of these polypeptides corresponded to entire B. pseudomallei proteins that were predicted from the genome sequence. Some large proteins could not be expressed in their entirety, and the immobilized antigens were polypeptides derived from these proteins.
The signals from positive control proteins (serial dilutions of human IgG, which were detected by the secondary antibody) were similar in healthy control individuals and recovered individuals, as expected. Most individuals also had signals to EBNA-1 that decreased in activity with increasing antigen dilution, which confirmed that the plasma samples contained detectable antibody. Each array also included negative control spots where DNA had been excluded from the in vitro transcription/translation system used to produce the B. pseudomallei proteins. The reactivity of these spots was low with plasma from control or recovered groups. The average “no DNA” control signal was routinely subtracted from all signals obtained from specific antigens.
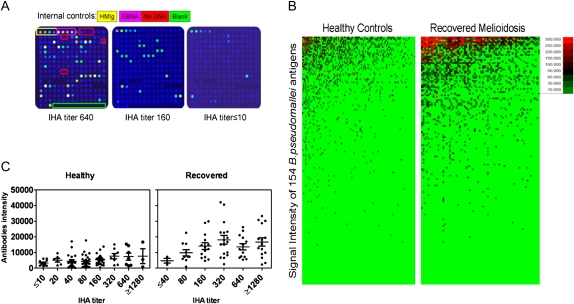
Because IHA has been widely used in endemic areas, even it has low sensitivity. Our results showed that plasma samples with high IHA titers to B. pseudomallei generally showed greater reactivity with proteins on the proteome array than did plasma samples with low IHA titers (Figure 1A). The average signal intensities from arrays probed with plasma from recovered patients (mean ± SE, 14,540 ± 2978) were significantly higher than signal intensities from arrays probed with plasma from control individuals (mean ± SE, 5041 ± 794), (Figure 1B). Using linear regression analysis, we showed that the signal intensity from arrays probed with healthy control plasma samples correlated with the antibody titer detected using conventional IHA (R2 = 0.5970; P = .0246). Although there was clearly a similar trend with samples from recovered patients, there was no correlation between array signal intensities and the IHA titers of samples from recovered patients after linear regression analysis (R2 = 0.3384; P = .2258).
Figure 1.
Antibody responses to 154 Burkholderia pseudomallei proteins on microarray and indirect hemagglutination assay (IHA) titers of healthy individuals vs recovered melioidosis patients. Microarray chips printed with 154 recombinant B. pseudomallei proteins or internal controls consisting of serial dilutions of human IgG and EBNA1 proteins were probed with plasma samples from 108 healthy individuals and 72 patients who had recovered from melioidosis and quantified by ScanArray Express software. The representative arrays probed by plasma that had different IHA titer samples are shown (A). The signal intensity of each protein was colored using Excel (Microsoft). The top reactive proteins were ranked according to the signal strength of each group. The healthy or recovered patient samples are in columns and sorted left to right by increasing average intensity to B. pseudomallei protein antigens, which are in rows (B). The average antibody intensity of each sample from healthy and recovered individuals was plotted according to antibody titer detected by means of conventional IHA assay (C).
The 20 B. pseudomallei antigens that gave the highest array signals with plasma samples from 108 healthy control individuals or 72 recovered patients were next identified (Figure 1C). In total, 27 antigens were identified, of which 13 were common to both lists (Table 1). Some of the antigens were different polypeptides derived from the same protein (BPSS1532-m1 and BPSS1532-m2, BPSS2053-s4 and BPSS2053-s9, BPSL1661-s2 and BPSL1661-s5). The finding that different polypeptides from the same protein reacted with plasma antibodies indicates that these proteins contained multiple epitopes. Some of the proteins were associated with pathogenicity, including BipB (BPSS1532), type IV pilus protein (BPSS1599), flagellin (BPSL3319), and flagella hook–associated protein (BPSL0280), whereas others, including OmpA proteins (BPSS2765) and an unknown hypothetical protein (BPSS2053), were predicted to be located on the surface. Seven proteins were more strongly recognized by plasma from recovered individuals than by plasma from control individuals. Conversely, 7 different proteins were equally recognized by plasma from both groups or more strongly recognized by plasma from healthy control individuals than by plasma from recovered individuals (Table 1).
Table 1.
Top Burkholderia pseudomallei Protein Antigens Identified by Microarray Chips Probed with Plasma Samples from Recovered Melioidosis Patients and/or Seropositive Healthy Individuals
| Locus tag | Protein name | Classification | Predicted function associated | Recovered | Healthy | |
| Top proteins of both healthy control participants and recovered melioidosis patients | ||||||
| BPSL2697 | Chaperonin GroEL | Chaperones | Pathogenicity/adaption/chaperones | 235,714 (18,662) | 46,831 (9286) | |
| BPSS1532 | Cell invasion protein (bipB) | Pathogenicity island-related functions | Pathogenicity/adaption/chaperones | 147,514 (19,384) | 34,004 (6196) | |
| BPSS1512 | Type VI secretion protein, TssM | Hypothetical protein | Unknown/hypothetical | 185,219 (20,903) | 42,267 (8240) | |
| BPSS0477 | 60 kDa chaperonin | Chaperones | Pathogenicity/adaption/chaperones | 168,271 (18,095) | 30,034 (6728) | |
| BPSL3319 | Flagellin | Chemotaxis and mobility | Pathogenicity/adaption/chaperones | 140,395 (17,474) | 48,672 (7493) | |
| BPSL2765 | Putative OmpA family lipoprotein | Membrane/exported/lipoproteins | Surfacea | 74,103 (11,987) | 20,976 (3272) | |
| BPSL2520 | Putative exported protein BPSL2520 | Membrane/exported/lipoproteins | Surfacea | 71,886 (14,942) | 8,912 (3333) | |
| BPSS1492 | Hypothetical protein Bim A | Hypothetical protein | Unknown/hypothetical | 67,895 (15,980) | 28,538 (6786) | |
| BPSL0999 | Putative OmpA family lipoprotein | Inner membrane | Surfacea | 66,679 (14,547) | 14,915 (4195) | |
| BPSL1901 | Putative membrane protein | Inner membrane | Surfacea | 46,354 (9461) | 31,068 (6991) | |
| BPSL1445 | Putative lipoprotein | Membrane/exported/lipoproteins | Surfacea | 34,941 (9806) | 10,154 (4485) | |
| BPSL3398 | ATP synthase alpha chain | ATP-proton motive force | Energy metabolismb | 27,222 (6481) | 9711 (1887) | |
| BPSS1974 | Putative lipoprotein | Membrane/exported/lipoproteins | Surfacea | 24,870 (5952) | 26,429 (5590) | |
| Top proteins of recovered melioidosis patients | ||||||
| BPSL0280 | Flagellar hook-associated protein | Chemotaxis and mobility | Pathogenicity/adaption/chaperones | 75,283 (13,077) | 1130 (521) | |
| BPSS1588 | Putative exported protein BPSS1588 | Membrane/exported/lipoproteins | Surfacea | 46,124 (11,096) | 367 (218) | |
| BPSL2522 | Outer membrane protein A (OmpA) precursor | Outer membrane | Surfacea | 40,296 (10,380) | 1685 (371) | |
| BPSS1599 | Type IV pilus biosynthesis protein (pilO) | Outer membrane | Surfacea | 39,847 (7759) | 2995 (906) | |
| BPSL2096 | Putative hydroperoxide reductase | Detoxification | Pathogenicity/adaption/chaperones | 31,387 (4269) | 2727 (837) | |
| BPSL3222 | 50S ribosomal protein L7/L12 | Ribosomal proteins—synthesis, modification | Information transferc | 29,224 (7566) | 6291 (2034) | |
| BPSS2141 | Periplasmic oligopeptide-binding protein precursor (OppA) | Carbohydrates, organic acids, and alcohols | Surfacea | 23,927 (5596) | 2133 (1321) | |
| Top protein of healthy controls | ||||||
| BPSL1465 | Peptidase | Membrane/exported/lipoproteins | Surfacea | 19,073 (2819) | 13,898 (2290) | |
| BPSL1661 | Putative hemolysin-related protein | Membrane/exported/lipoproteins | Surfacea | 10,255 (3185) | 16,491 (4924) | |
| BPSL1902 | Putative membrane protein | Inner membrane | Surfacea | 15,381 (3964) | 11,748 (2729) | |
| BPSL2063 | Putative membrane protein | Inner membrane | Surfacea | 23,599 (5191) | 22,611 (4560) | |
| BPSS0734 | Outer membrane efflux protein | Outer membrane | Surfacea | 7707 (2790) | 11,574 (3924) | |
| BPSS1434 | Putative membrane-anchored cell surface protein | Inner membrane | Surfacea | 10,840 (2676) | 19,734 (3559) | |
| BPSS2053 | Putative cell surface protein | Surface structures | Surfacea | 18,748 (4652) | 28,006 (5556) |
NOTE. Protein locus tag, name, and classification and function of B. pseudomallei proteins that were predicted on the basis of the Artemis database on the Sanger website. The mean signal intensity and standard error of the mean (SE) of individual proteins from 108 healthy control individuals and 72 recovered melioidosis patients were calculated using Excel software (Microsoft). Data are mean (SE) unless otherwise specified.
Surface consisted of outer membrane, inner membrane, and secreted and surface structures.
Information transfer consisted of transcription/translation and DNA/RNA modification.
Energy metabolism consisted of glycolysis, electrontransport, etc.
Different B. pseudomallei Antigens Are Recognized by Plasma from Melioidosis Patients with Different Forms of Disease; Antibody Profiles Change after Treatment
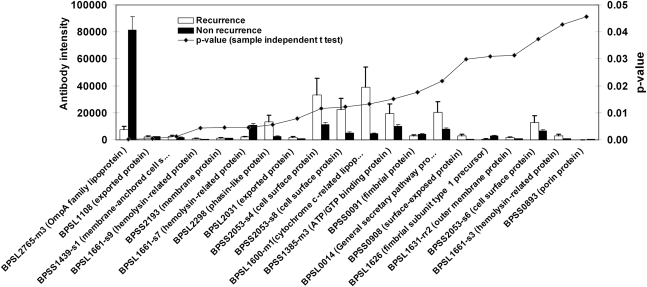
We next sought to determine whether individuals who had suffered from multiple episodes of infection developed different antibody responses from individuals who had suffered a single episode of disease. We compared antibody profiles for 7 individuals who had a history of recurrent infection after completion of antibiotic treatment and 65 patients who did not relapse during this study. A sample-independent Student t test was used to compare these groups. The results showed that 20 B. pseudomallei proteins were significantly differentially recognized by plasma from these groups (Figure 2). Notably, the average antibody response to BPSL2765 (encoding a putative OmpA family protein) was >10 times higher in plasma from individuals who had suffered a single episode of disease than in plasma from individuals with a history of recurrent melioidosis.
Figure 2.
Burkholderia pseudomallei proteins identified in serum samples from patients who had recovered from melioidosis with a history of recurrent or nonrecurrent infection. Mean of antibody intensity of plasma from patients with a history of recurrent infection (n = 7) vs nonrecurrent infection (n = 65). Data were analyzed with the sample-independent Student t test using SPSS software, and P values of each comparison were plotted on the right y-axis with signal intensities of antibodies on the left y-axis. The average intensities of 20 B. pseudomallei proteins with P < .05, ranking from low to high P values, are shown.
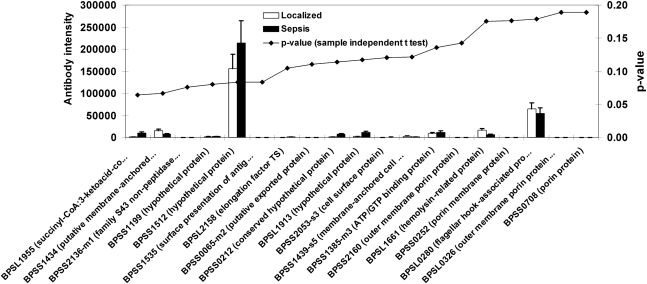
The antibody profiles of plasma from individuals who had recovered from localized melioidosis or who had recovered from melioidosis sepsis were also compared. The 20 proteins that showed the greatest difference in response between these groups (lowest P values) are shown in Figure 3. The response to B. pseudomallei antigens seemed to be higher in samples from patients with a history of sepsis than in samples from patients with localized melioidosis. However, this difference was not statistically significant (Figure 3).
Figure 3.
Burkholderia pseudomallei proteins identified by plasma from recovered melioidosis patients with a history of sepsis vs localized melioidosis. Mean antibody intensity in plasma from patients with a history of localized melioidosis (n = 21) vs patients with a history of sepsis (n = 18). Data were analyzed with the sample-independent Student t test using SPSS software, and the P value of each comparison was plotted with signal intensity. The average intensities of the top 20 B. pseudomallei proteins, ranking from low to high P values, are shown.
Finally, we determined the antibody responses in recovered melioidosis patients who attended the diabetic clinic at Sappasithiprasong Hospital (n = 30) at different times after the completion of antibiotic treatment (median, 72.5 weeks) using the Pearson correlation (Table 2). The results revealed that the responses to 10 proteins changed statistically significantly after treatment (Pearson correlation). The antibodies to 2 proteins (BPSL1901 and BPSL0326) declined after treatment (r = −0.4091 and −0.3701, respectively). In contrast, the antibodies to 8 other proteins (BPSL3222, BPSL2298, BPSL1600, BPSS0908, BPSL2827, BPSS2053, BPSS0140, and BPSL1913) increased after treatment (r-values were between 0.3573 and 0 .5572). These results suggest that antibodies to BPSL1901 and BPSL0326 had short lives (<1 year) whereas the others had longer lives (>2 years) after the development of clinical melioidosis.
Table 2.
Correlation of Antibody Intensity to 10 Burkholderia pseudomallei Proteins of Recovered Melioidosis Patients and Time after the Completion of Antibiotic Treatment Analyzed by Pearson Correlation
| No. | Locus tag | Protein name | Pearson correlation |
|
| r | P | |||
| 1 | BPSL3222 | 50S ribosomal protein L7/L12 | 0.5572 | .001 |
| 2 | BPSL2298 | Phasin-like protein | 0.5027 | .004 |
| 3 | BPSL1600 | Putative cytochrome c-related lipoprotein | 0.4991 | .004 |
| 4 | BPSS0908 | Putative surface-exposed protein | 0.4688 | .008 |
| 5 | BPSL1901 | Putative membrane protein | –0.4091 | .02 |
| 6 | BPSL2827 | Putative DnaK chaperone protein | 0.4028 | .02 |
| 7 | BPSS2053 | Putative cell surface protein | 0.389 | .03 |
| 8 | BPSS0140 | Putative sugar ABC transport system | 0.3812 | .03 |
| 9 | BPSL0326 | Putative outer membrane porin protein precursor | –0.3701 | .04 |
| 10 | BPSL1913 | Hypothetical protein BPSL1913 | 0.3573 | .048 |
Ability of Antibody-Reactive Proteins to Induce IFN-γ Production
Our previous study revealed that environmental exposure to B. pseudomallei can prime T cells to produce IFN-γ [10]. Therefore, we tested the ability of 5 B. pseudomallei proteins shown to be B cell immunogens to induce IFN-γ production by human T cells.
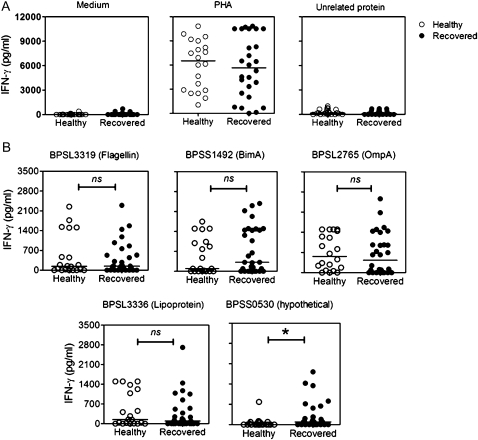
Whole-blood samples were collected from 30 recovered melioidosis patients and from 20 seropositive healthy individuals from a melioidosis-endemic area who had no history of disease. These blood samples were cocultured for 48 h with medium, PHA, a F. tularensis (FT1823) recombinant protein as a negative control, or one of the B. pseudomallei proteins. As expected, the unrelated F. tularensis protein did not induce IFN-γ production, but PHA equally induced IFN-γ production in plasma from both recovered melioidosis and seropositive healthy groups (Figure 4A). However, B. pseudomallei proteins, namely flagellin (BPSL3319), Bim A (BPSS1492), Omp A (BPSL2765), and lipoprotein (BPSL3336), but not a hypothetical protein (BPSS0530), could induce strong T cell responses in plasma from the seropositive healthy group comparable to those in plasma from recovered melioidosis patients (Figure 4B). These results suggest that seroreactive proteins are likely to evoke T cell responses in both groups.
Figure 4.
IFN-γ production in response to controls and to 5 recombinant Burkholderia pseudomallei proteins of healthy individuals and recovered melioidosis patients. Whole-blood samples of 20 healthy individuals and 30 recovered melioidosis patients were cocultured with controls; medium, phytohemagglutinin (PHA), and Franciscella tularensis FT1823 (unrelated protein) (A), and 5 selected B. pseudomallei proteins (B) for 48 h. The cultured supernatants were assayed for IFN-γ by means of ELISA. The distribution of IFN-γ levels was plotted and compared for healthy individuals vs recovered patients with use of the Mann-Whitney t test. One asterisk indicates P < .05. ns, nonsignificant.
When we analyzed both arms of the immune responses, either antibody responses and/or T cell activated specific IFN-γ responses induced by these 5 B. pseudomallei proteins (BPSL3319 flagellin, BPSL2765 OmpA, BPSS1492 BimA, BPSS0530 hypothetical protein, and BPSL3336 lipoprotein) from 26 individuals who recovered from melioidosis and 15 healthy individuals from melioidosis-endemic areas, we found that all of the plasma samples had antibody titers of >40 according to IHA, indicating the likely past exposure of these individuals to B. pseudomallei.
To analyze the patterns of antibody and T cell responses, a positive antibody response (Ab+) was defined as protein array signal intensity above the mean + 2SD of the “no DNA” control signal and a positive T cell response (T+) was defined as IFN-γ above the mean + 2SD of medium background levels. The antibody and T cell responses were grouped into 4 patterns, T+Ab+, T+Ab–, T–Ab+, and T–Ab– (Figure 5). The results showed that B. pseudomallei proteins could differentially induce antibody and T cell responses. The responses to flagellin (BPSL3319) and a hypothetical protein (BPSS0530) were broadly similar in the seropositive control and recovered groups. However, responses to OmpA (BPSL2765), BimA (BPSS1492), or a lipoprotein (BPSL3336) showed a marked bias toward T cell responses in healthy control individuals compared with recovered patients. Collectively, these results reveal distinctively reactive immune responses to B. pseudomallei proteins in humans, which suggests that different responses occur following environmental exposure or clinical melioidosis.
Figure 5.
Antibody and T cell responses to 5 Burkholderia pseudoamallei proteins of healthy individuals vs recovered melioidosis patients. The plasma and blood samples collected from 26 recovered melioidosis patients and 15 healthy control participants were tested for antibodies to B. pseudomallei proteins by microarray and IFN-γ induction by whole-blood assay. Reactive results of antibody testing were defined as a signal antibody intensity higher than the mean + 2 standard deviations (SDs) of the “no DNA” control. The positive for IFN-γ induction was defined as IFN-γ higher than the mean + 2 SDs of medium controls. Ab+ and Ab–, positive and negative for antibody response, respectively; T+ and T–, positive and negative for T cell response by IFN-γ production, respectively.
DISCUSSION
Protein microarrays have previously been used to map the antibody responses to a range of infectious diseases [18, 20, 22]. Previously, we have shown that individuals who suffered from melioidosis develop antibody responses that can be used to differentiate them from control individuals who have not experienced the disease [17]. In this study, we used a B. pseudomallei protein microarray consisting of 154 B. pseudomallei antigens previously shown to be the main protein antigens recognized by melioidosis convalescent serum [17]. The proteins immobilized on this array have previously been shown to be the most highly reactive with serum specimens from melioidosis patients in Thailand and Singapore. Therefore, although we believe that our array would have revealed antibody responses to most B. pseudomallei immunogens, it is highly likely that our study missed some antibody responses.
We first considered the possibility that proteins that are strongly recognized by plasma antibodies from individuals who have recovered from melioidosis might be potential protective antigens. In this study, we have identified 7 proteins that are recognized by recovered plasma but not by control plasma. Of these, 2 (BPSL2522 OmpA and BPSS2141 OppA) have previously been evaluated as protective antigens in a murine model of disease. Immunization with BPSL2522 (Omp3) and BPSL2765 (Omp7) provided 50% protection against a challenge with 10 times the median lethal dose of B. pseudomallei administered intraperitoneally up to 21 days after infection [23]. It should be noted that only 4 animals per group were studied in this challenge model. Therefore, additional studies with the use of enough animals to yield statistically significant results are still required.
In contrast, no protection of mice against a B. pseudomallei challenge after immunization with OppA protein has been reported [24]. The ability of the other B. pseudomallei immunoreactive proteins to elicit protective immune responses has not been examined. However, ribosomal protein L7/L12 has previously been identified as an immunodominant and protective antigen of Brucella abortus [25], and the hydroperoxide reductase family proteins of Mycobacterium avium subspecies paratuberculosis are reported to be highly immunogenic and to induce strong antibody and IFN-γ responses in experimental infection [26].
An alternative way of viewing our data is to consider the possibility that proteins that are strongly recognized by individuals in endemic areas who have not developed disease but who have substantial IHA titers might have been exposed to B. pseudomallei. Seven proteins recognized in this group but not by individuals from the same geographical area who have developed disease were identified. These proteins might therefore play a role in protecting these individuals from developing overt disease. To our knowledge, none of these proteins has been evaluated as a protective antigen. All of the proteins in this group were predicted to be surface located, and the peptidase, hemolysin-related, and outer membrane efflux proteins have been predicted to play roles in virulence [27].
Finally, we considered the possibility that protective responses might develop in individuals with localized disease or in individuals who had a single episode of disease but not in individuals who had more severe forms of disease characterized by the development of sepsis or recurrent disease. We did not identify a qualitative or quantitative difference in the antibody profiles from individuals who had localized disease or septic melioidosis. However, individuals who had experienced only 1 episode of disease showed a differential pattern of antibody response, compared with individuals who had experienced recurrent disease. Our finding that antibody levels to BPSL2765 (OmpA protein) were 10-fold higher in individuals who had only a single episode of disease suggests that this response might play a role in protection against recurrent disease. Moreover, antibodies to BPSL1901 (putative membrane protein) and BPSL0326 (putative outer membrane porin protein precursor) were short lived, suggesting a role in acute infection, and declined when patients had recovered. In contrast, 8 other proteins showed a positive correlation with time, implying that these proteins might be inducing long-lived antibodies.
Collectively, with our previous proteome array study, we are now able to identify proteins that, on the basis of antibody reactivity, are candidate protective antigens. Our findings indicate that some of these proteins are also able to induce IFN-γ production by T cells from individuals who have recovered from melioidosis or seropositive healthy individuals from melioidosis-endemic areas. Against this background, it is unclear whether candidate protective antigens should be delivered to promote humoral immunity, cellular immunity, or a combination of both. The induction of a protective immune response and B. pseudomallei clearance likely requires humoral and cell-mediated immunity [6]. The work reported here identifies proteins able to induce good humoral and cellular immune responses, and these should now be evaluated as protective antigens. Equally importantly, our work highlights the way in which additional candidates can be identified, and additional studies in animal challenge models would strengthen the relevance of these newly identified candidate antigens.
Funding
This work was supported by the Royal Golden Jubilee PhD Program of the Thai Research Fund (to D.S.) and by the US National Institutes of Health/National Institute of Allergy and Infectious Diseases (grant no. U01AI061363).
References
- 1.Leelarasamee A. Recent development in melioidosis. Curr Opin Infect Dis. 2004;17:131–6. doi: 10.1097/00001432-200404000-00011. [DOI] [PubMed] [Google Scholar]
- 2.Currie BJ, Fisher DA, Howard DM, et al. Endemic melioidosis in tropical northern Australia: a 10-year prospective study and review of the literature. Clin Infect Dis. 2000;31:981–6. doi: 10.1086/318116. [DOI] [PubMed] [Google Scholar]
- 3.Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev. 2005;18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ngauy V, Lemeshev Y, Sadkowski L, Crawford G. Cutaneous melioidosis in a man who was taken as a prisoner of war by the Japanese during World War II. J Clin Microbiol. 2005;43:970–2. doi: 10.1128/JCM.43.2.970-972.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Limmathurotsakul D, Chaowagul W, Chierakul W, et al. Risk factors for recurrent melioidosis in northeast Thailand. Clin Infect Dis. 2006;43:979–86. doi: 10.1086/507632. [DOI] [PubMed] [Google Scholar]
- 6.Healey GD, Elvin SJ, Morton M, Williamson ED. Humoral and cell-mediated adaptive immune responses are required for protection against Burkholderia pseudomallei challenge and bacterial clearance postinfection. Infect Immun. 2005;73:5945–51. doi: 10.1128/IAI.73.9.5945-5951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santanirand P, Harley VS, Dance DA, Drasar BS, Bancroft GJ. Obligatory role of gamma interferon for host survival in a murine model of infection with Burkholderia pseudomallei. Infect Immun. 1999;67:3593–600. doi: 10.1128/iai.67.7.3593-3600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haque A, Easton A, Smith D, et al. Role of T cells in innate and adaptive immunity against murine Burkholderia pseudomallei infection. J Infect Dis. 2006;193:370–9. doi: 10.1086/498983. [DOI] [PubMed] [Google Scholar]
- 9.Lertmemongkolchai G, Cai G, Hunter CA, Bancroft GJ. Bystander activation of CD8+ T cells contributes to the rapid production of IFN-gamma in response to bacterial pathogens. J Immunol. 2001;166:1097–105. doi: 10.4049/jimmunol.166.2.1097. [DOI] [PubMed] [Google Scholar]
- 10.Tippayawat P, Saenwongsa W, Mahawantung J, et al. Phenotypic and functional characterization of human memory T cell responses to Burkholderia pseudomallei. PLoS Negl Trop Dis. 2009;3:e407. doi: 10.1371/journal.pntd.0000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sundaresh S, Randall A, Unal B, et al. From protein microarrays to diagnostic antigen discovery: a study of the pathogen Francisella tularensis. Bioinformatics. 2007;23:i508–18. doi: 10.1093/bioinformatics/btm207. [DOI] [PubMed] [Google Scholar]
- 12.Xu Y, Bruno JF, Luft BJ. Profiling the humoral immune response to Borrelia burgdorferi infection with protein microarrays. Microb Pathog. 2008;45:403–7. doi: 10.1016/j.micpath.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Doolan DL, Mu Y, Unal B, et al. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics. 2008;8:4680–94. doi: 10.1002/pmic.200800194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jing L, Davies DH, Chong TM, et al. An extremely diverse CD4 response to vaccinia virus in humans is revealed by proteome-wide T-cell profiling. J Virol. 2008;82:7120–34. doi: 10.1128/JVI.00453-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benhnia MR, McCausland MM, Su HP, et al. Redundancy and plasticity of neutralizing antibody responses are cornerstone attributes of the human immune response to the smallpox vaccine. J Virol. 2008;82:3751–68. doi: 10.1128/JVI.02244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies DH, Wyatt LS, Newman FK, et al. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus ankara is comparable to that of Dryvax. J Virol. 2008;82:652–63. doi: 10.1128/JVI.01706-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felgner PL, Kayala MA, Vigil A, et al. A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross-reactive antigens. Proc Natl Acad Sci U S A. 2009;106:13499–504. doi: 10.1073/pnas.0812080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies DH, McCausland MM, Valdez C, et al. Vaccinia virus H3L envelope protein is a major target of neutralizing antibodies in humans and elicits protection against lethal challenge in mice. J Virol. 2005;79:11724–33. doi: 10.1128/JVI.79.18.11724-11733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander AD, Huxsoll DL, Warner AR, Jr, Shepler V, Dorsey A. Serological diagnosis of human melioidosis with indirect hemagglutination and complement fixation tests. Appl Microbiol. 1970;20:825–33. doi: 10.1128/am.20.5.825-833.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies DH, Liang X, Hernandez JE, et al. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci USA. 2005;102:547–52. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eyles JE, Unal B, Hartley MG, et al. Immunodominant Francisella tularensis antigens identified using proteome microarray. Proteomics. 2007;7:2172–83. doi: 10.1002/pmic.200600985. [DOI] [PubMed] [Google Scholar]
- 22.Bacarese-Hamilton T, Bistoni F, Crisanti A. Protein microarrays: from serodiagnosis to whole proteome scale analysis of the immune response against pathogenic microorganisms. Biotechniques. 2002:24–9. [PubMed] [Google Scholar]
- 23.Hara Y, Mohamed R, Nathan S. Immunogenic Burkholderia pseudomallei outer membrane proteins as potential candidate vaccine targets. PLoS One. 2009;4:e6496. doi: 10.1371/journal.pone.0006496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harland DN, Chu K, Haque A, et al. Identification of a LolC homologue in Burkholderia pseudomallei, a novel protective antigen for melioidosis. Infect Immun. 2007;75:4173–80. doi: 10.1128/IAI.00404-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo D, Ni B, Li P, et al. Protective immunity elicited by a divalent DNA vaccine encoding both the L7/L12 and Omp16 genes of Brucella abortus in BALB/c mice. Infect Immun. 2006;74:2734–41. doi: 10.1128/IAI.74.5.2734-2741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsen I, Reitan LJ, Holstad G, Wiker HG. Alkyl hydroperoxide reductases C and D are major antigens constitutively expressed by Mycobacterium avium subsp. paratuberculosis. Infect Immun. 2000;68:801–8. doi: 10.1128/iai.68.2.801-808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holden MT, Titball RW, Peacock SJ, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci USA. 2004;101:14240–5. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]