Abstract
The crystal structure of the first endolytic peptidoglycan lytic transglycosylase MltE from Escherichia coli is reported herein. The degradative activity of this enzyme initiates the process of cell wall recycling, which is an integral event in the bacterial existence. The structure sheds light on how MltE recognizes its substrate, the cell wall peptidoglycan. It also explains the ability of this endolytic enzyme to cleave in the middle of the peptidoglycan chains. Furthermore, the structure reveals how the enzyme is sequestered on the inner leaf let of the outer membrane.
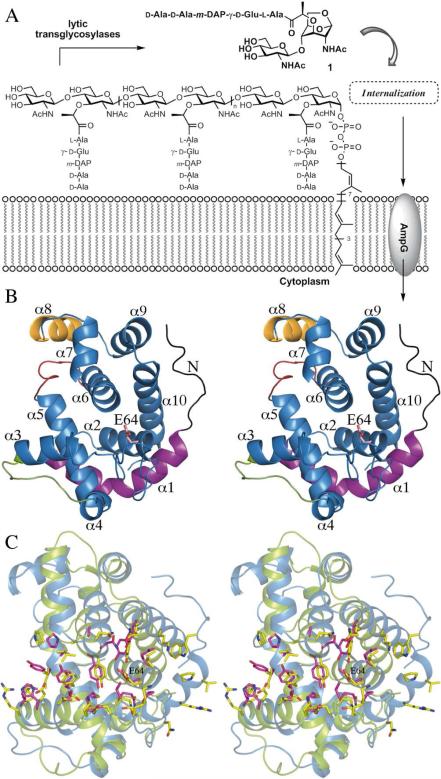
The cell wall is recycled during the course of the normal doubling of bacterial cultures as well as in response to damage, such as might be inflicted by antibiotics (1,2). The major component of cell wall is the peptidoglycan (PG), the neighboring strands of which undergo cross linking to assemble the functional cell wall. PG is comprised of alternating β-1,4-linked N-acetylmuramic acid (MurNAc) and N-acetylglucosamine (GlcNAc) residues. A unique peptide stem, the site of cross-linking, is appended to lactyl groups of the MurNAc residues (1). The reaction of lytic transglycosylases (LTs) is unusual, in that the β-1,4-glycosidic bond between MurNAc and GlcNAc of PG is cleaved in an initial step that is believed to go through an oxocarbonium species, which in turn entraps the C6 hydroxyl moiety of MurNAc. This substrate assisted catalysis leads to the formation of metabolite 1, which produces a non reducing 1,6-anhydromuramyl residue (Figure 1A). Metabolite 1 is taken up by the permease AmpG (3), and once in the cytoplasm, it undergoes a series of reactions that ultimately leads to the formation of lipid II, which is translocated to the surface of the bacterium for de novo synthesis of PG. Of the seven known E. coli LTs, six are anchored to the outer membrane (MltA, MltB, MltC, MltD, MltE and MltF) and one is soluble (Slt70) (4). A comparison of genes encoding LTs allowed their classification into four families (5). The LT family 1 includes Slt70, MltC, MltD, MltE and MltF, whereas MltA and MltB represent families 2 and 3, respectively (6, 7). Most of LTs appear to act as exolytic enzymes, releasing metabolite 1 from the ends of the glycan strands. The only exception in E. coli would appear to be MltE, which is proposed to be an endolytic enzyme. This is the ability to fragment the peptidoglycan strand in the middle, which ultimately would lead to the formation of 1 by the action of other LTs on products of the reaction of MltE (8). Structural research on LTs has so far resulted in the X ray structures of the 70 kDa soluble Slt70 (9), the 36 kDa soluble Slt35 of Escherichia coli (7, 10) and the 38 kDa MltA (6, 11, 12). Slt70 structure is built up of three distinct domains (named U-, L- and C-domains respectively), which are all rich in α-helices. The N-terminal domain is packed in a U-shaped conformation and connected to a L-domain forming a closed ring with a large central opening (11). The C-terminal domain is the catalytic one and is packed on top of this ring, interacting with both L-and U-domains. This C-domain has a globular structure and presents a fold similar to the goose-type lysozyme (11). Structural studies of Slt70 with peptidoglycan fragments (9) revealed the presence of six saccharide binding sites in the active-site groove. Slt35 is a fully active, soluble form of the 40 kDa membrane anchored lytic transglycosylase B (MltB). The structure of Slt35 of LT family 3 shows three domains named the α, β, and core domains. The core domain resembles the fold of goose-type lysozyme and the catalytic domain of Slt70 (7). It contains Glu162, which coincides with the catalytic Glu478 of Slt70 and Glu73 of goose type lysozyme after superposition. Both Slt35 and Slt70 are exo-muramidases that require the peptide side chains in peptidoglycan for activity (13, 14). It is believed that the N-terminal, doughnut-shaped domain of Slt70 together with the presence of a specific binding site for 1,6-anhydromuropeptide product and the “exo loop” of Slt35 (residues 99−108) impose this exo-muramidase activity (7, 15). MltA (LT family 2), whose crystal structures from E. coli and N. gonorrhoeae have been reported (11, 12), presents a catalytic domain completely different from that of lysozyme. It comprises two main domains resembling the catalytic domain of endoglucanase V (16). For LT family 1 only the three dimensional structure of soluble exolytic Slt70 has been reported. We report herein the first high-resolution (2.0 Å) crystal structure for the endolytic MltE.
Figure 1.
(A) PG fragmentation by LTs leads to the formation of metabolite 1, which is transported across plasma membrane to initiate its recycling. (B) 3D structure of MltE. Main structural differences with Slt70 are colored in black (N-loop), in magenta (α1 helix), in red (the loop connecting the α5 and α6) and in yellow (α8). The catalytic Glu64 is labeled. (C) Structural superimposition of MltE (blue ribbons) and catalytic domain of Slt70 (green ribbons). Side-chains building active sites are represented as sticks (in yellow for MltE and in pink for Slt70).
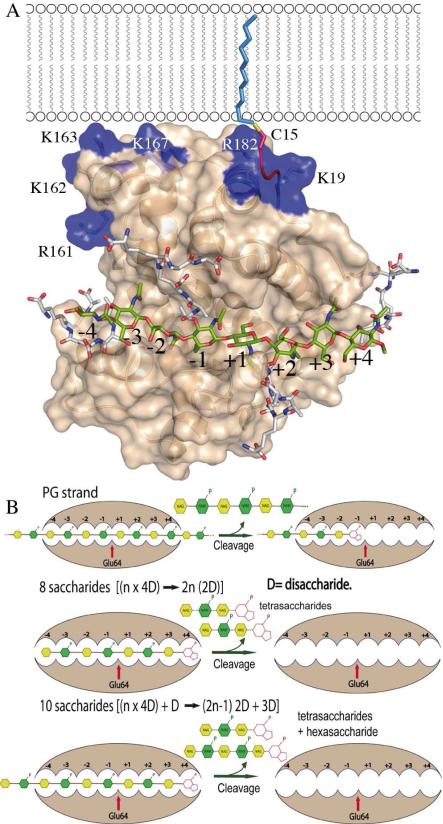
Despite significant (35%) sequence identity between MltE and the catalytic module of Slt70, molecular replacement method failed in providing a viable solution. Multisolution phasing through search of small fragments derived from the catalytic module of Slt70 (15) combined with density modi fication was successfully applied using the ARCIMBOLDO program (17). Two monomers were built in the asymmetric unit (r.m.s.d of 0.19 Å for all Cα atoms), giving good electron density for all residues, which refined up to 2.0 Å resolution. MltE has a globular structure with ten α-helices (Figure 1B). Its overall fold is similar to that of goose-type lysozyme. Utilizing the Dali server (18), the closest structural models of MltE are the catalytic module of Slt70 (15) (r.m.s.d of 3.1 Å for 161 Cα atoms; see Fig. S1A), the G- type lysozyme (19) (r.m.s.d of 2.6 Å for 144 Cα atoms) and the Slt35 lytic transglycosylase (7) (r.m.s.d of 2.5 Å for 127 Cα atoms). Despite the overall structural homology, there exist significant differences in both size and secondary structure distribution between the membrane-bound endolytic transglycosylase MltE and the catalytic domain of Slt70 (Fig. 1C and Fig. S1). This is relevant as high sequence homology is observed in Slt70s (15) and MltEs (Fig. S1C). Among structural differ ences, three of them are critical to explain specific features in MltE: the presence of a N-terminal loop (residues 19-30) and the long α1 helix (residues 30-50) that is markedly longer than the corresponding helix in Slt70 and the presence in MltE of a new α helix (α8) presenting a basic patch of surface on one side of the protein (Arg161, Lys162, Lys163, Lys167). While the first two differences are important for the endolytic character of MltE, the orientation of the N-terminal region and the new α8 have implications for interactions with the outer membrane by MltE (see below). As revealed in the MltE crystal structure, the long N-terminal loop (residues 19-30), not present in the Slt70 structure, is strongly stabilized through interactions with the core of the protein (see Table S2) and provides a rigid scaffolding that connects the point of membrane anchoring (acy lated-Cys15, discussed below; Fig. 2) with the active site. In fact Trp27, a residue within the N-terminal loop, is part of the active site (Fig. 1C and Fig. S2). As indicated earlier, MltE is the only LT of E. coli with peptidoglycan endolytic activity. As such, the structure reveals it to have an extended active site that binds at least eight saccharides of the peptidoglycan chain (vide infra) as obtained by molecular dynamics (MD) simulations. The substrate binding cleft of lysozymes and other glycosyl hydrolases accommodate several saccharide units at subsites designated as positions -i (the non-reducing end through +j (in the other direction). The saccharide units flanking) the scissile glycosidic bond are designated as positions -1 and +1. The ellipsoid shape of MltE with its two lobes is linked by the long α5 helix. The active site is sequestered in a deep groove that spans the two lobes within the -4 to +4 saccharide-binding subsites (Fig. 2) vs. the -4 to +2 saccharide-binding subsites reported for Slt70 (9). The peptide-binding site at -1 is formed by a shallow groove between α7 and the loop connecting α9 and α10 (SI) similarly to that found for St70 (15). The site at -3 is formed by the groove between α3 and α5. Another point of distinction concerns the α1 helix. This helix (residues 30-50) is longer than the corresponding helix in Slt70 (20 vs. 8 residues, respectively), and provides an extension of the substrate-binding site up to subsite +4. In addition, the loop connecting α5 with α6 creates a groove rich in acidic amino acids at the backside of the substrate-binding site (Fig. 1B and Fig. S3) whose function is likely glycan stabilization of a neighboring cross-linked PG strands. The last significantly distinct feature is the presence of an additional α-helix (α8). This helix comprises a cluster of basic residues (Fig. 2) that forms, together with Arg182 (from α9) and Lys19 (from the N-terminal loop) a membrane-interacting surface near the acylated Cys15 (Fig. 2A).
Figure 2.
(A) MltE could accommodate saccharides at sites -4 to +4 (and also the associated peptides attached to the MurNAc moieties), consistent with the solution NMR structure for the peptidoglycan (shown in capped sticks). Docked saccharide rings and peptide-stems from PG are colored differently (carbon atoms in green for saccharides and in white for peptides). N-lipidated Cys15, the membrane anchor, is shown as a red coil. Some residues within the basic patch are labeled. (B) Scheme shows the proposed mechanism for endotransglycosylase MltE. Since MltE has eight sugar binding sites and there are no other structural domains, MltE can cleave in the middle of the polymeric substrate. Four disaccharides (4D) fill the active site and experience cleavage. For strands having [n × 4D] saccharides the final products will be tetrasaccharides. For substrates having [(n × 4D)+ D] saccharides (as a polysaccharide with 10 monosaccharide units) the final products will be (2n-1) tetrasaccharides plus one hexasaccharide. This scheme is consistent with the experimental observations (8).
Glu64 is the catalytic residue at subsite -1 based on its equivalence in position to the catalytic glutamic acid in Slt70 (Fig. 1C). The acidic form of Glu64 protonates the glycosidic oxygen that departs from the scissile bond in the substrate to give rise to the intermediary oxocarbonium ion. The nowdeprotonated Glu64 would promote the C6-hydroxyl in the formation of the 1,6-anhydromuramyl moiety of the product (16). The three-dimensional structure of MltE and MD simulations reveal key insights into its unique endolytic activity, beyond the aforementioned expansive active site that spans from -4 to +4 subsites for binding to the polymeric substrate. The active-site surface accommodates the three-fold symmetric right-handed helical structure of the peptidoglycan, as determined previously by NMR (20) (Fig. S3). It has binding sites for the extended peptide stems that would be appended to the MurNAc moieties, in agreement with the peptide stem requirement for the activity of some LTs (15). The sites could accommodate the peptides regardless of whether they are cross-linked to a neighboring peptidoglycan or not. Potential steric encumbrance by such cross-linking would not appear to make a difference. This, in large measure is likely due to MltE being the only single-domain LT in E. coli. The second step reaction of the enzyme, which leads to the formation of the 1,6-anhydroMurNAc bicyclo ring, leads to the switch of all substituents from the equatorial to the axial positions. The conformational influence of this switch is profound, but especially so as far as the peptide moiety is concerned. It has been suggested that the Slt70 active site actually has a surface for binding of the peptide in the 1,6-anhydroMurNAc at the subsite +2, but a corresponding peptide-binding site is not seen in the structure of MltE (Fig. S4 and Fig. S5). This subsite in Slt70 has been postulated as essential to direct the product 1 from the subsite -1 to +2 after one turnover event, which allows processivity in the function of the exolytic enzymes such as Slt70(15) (Fig. S6). The absence of this subsite in MltE abrogates this opportunity, whereas the presence of eight saccharide-binding subsites supports the reported catalytic outcome for a tetra and hexasaccharide 1,6-anhydroMurNAc variants of the peptidoglycan as products (Fig. 2B). In this vein, activity assays indicated that tetrasaccharide analog is not a substrate for MltE (data not shown).
Removal of the signal peptide in MltE by signal peptidase is followed by N-acylation of Cys15 by a fatty acid (8). The lipid penetrates the inner leaflet of the outer membrane. Whereas the contribution to anchoring by this lipid penetration might be quite significant, the structure of MltE suggests that an electropositive surface near the N-terminus might actually provide the draw to the membrane head groups and also could contribute to substrate orientation during in vivo catalysis. A cluster of positively charged residues is located on this electropositive surface (Fig. 2 and Fig. S3). The presence of basic patches has been associated with membrane sensing and binding in BAR domains (21). The relevance of this finding in the catalytic mechanism of LTs awaits additional structural information on other en-zymes.
Supplementary Material
Acknowledgment
This work was supported by grants bfu2008-01711 and eu-cp223111. The work in the USA was supported by the National Institutes of Health. LIL is a Pew Latin American Fellow in the Biomedical Sciences, supported by The Pew Charitable Trusts. KM acknowledges the Deutsche Forschungsgemeinschaft DFG.
Footnotes
Supporting Information Available: Experimental procedures for structural determination and molecular dynamics calculations. The crystallographic coordinates are deposited in the Protein Data Bank (PDB code 2y8p). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Vollmer W, Joris B, Charlier P, Foster S. FEMS Microbiol Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 2.Suvorov M, Fisher JF, Mobashery S. Practical Handbook of Microbiology. 2008:153–183. [Google Scholar]
- 3.Cheng Q, Park JT. J Bacteriol. 2002;184:6434–36. doi: 10.1128/JB.184.23.6434-6436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheurwater E, Reid CW, Clarke AJ. Int J Biochem Cell Biol. 2008;40:586–591. doi: 10.1016/j.biocel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn NT, Clarke AJ. J Mol Evol. 2001;52:78–84. doi: 10.1007/s002390010136. [DOI] [PubMed] [Google Scholar]
- 6.van Straaten KE, Barends TR, Dijkstra BW, Thunnissen AM. J Biol Chem. 2007;282:21197–21205. doi: 10.1074/jbc.M701818200. [DOI] [PubMed] [Google Scholar]
- 7.van Asselt EJ, Dijkstra AJ, Kalk KH, Takacs B, Keck W, Dijkstra BW. Structure. 1999;7:1167–1180. doi: 10.1016/s0969-2126(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 8.Kraft AR, Templin MF, Holtje JV. J Bacteriol. 1998;180:3441–3447. doi: 10.1128/jb.180.13.3441-3447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thunnissen A-MWH, Dijkstra AJ, Kalk KH, Rozeboom HJ, Engel H, Keck W, Dijkstra BW. Nature. 1994;367:750–753. doi: 10.1038/367750a0. [DOI] [PubMed] [Google Scholar]
- 10.van Asselt EJ, Perrakis A, Kalk KH, Lamzin VS, Dijkstra BW. Acta Crystallogr. 1998;D 54:58–73. doi: 10.1107/s0907444997010330. [DOI] [PubMed] [Google Scholar]
- 11.van Straaten KE, Dijkstra BW, Vollmer W, Thunnissen AM. J. Mol. Biol. 2005;352:1068–1080. doi: 10.1016/j.jmb.2005.07.067. [DOI] [PubMed] [Google Scholar]
- 12.Powell AJ, Liu ZJ, Nicholas RA, Davies C. J. Mol. Biol. 2006;359:122–36. doi: 10.1016/j.jmb.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 13.Beachey EH, Keck W, de-Pedro MA, Schwarz U. Eur. J. Biochem. 1981;116:355–358. doi: 10.1111/j.1432-1033.1981.tb05342.x. [DOI] [PubMed] [Google Scholar]
- 14.Romeis T, Vollmer W, Höltje J-V. FEMS Microbiol. Lett. 1993;111:141–146. doi: 10.1111/j.1574-6968.1993.tb06376.x. [DOI] [PubMed] [Google Scholar]
- 15.van Asselt EJ, Thunnissen AM, Dijkstra BW. J Mol Biol. 1999;291:877–98. doi: 10.1006/jmbi.1999.3013. [DOI] [PubMed] [Google Scholar]
- 16.Thunnissen AM, Rozeboom HJ, Kalk KH, Dijkstra BW. Biochemistry. 1995;34:12729–12737. doi: 10.1021/bi00039a032. [DOI] [PubMed] [Google Scholar]
- 17.Rodríguez DD, Grosse C, Himmel S, González C, M de Ilarduya I, Becker S, Sheldrick GM, Usón I. Nature Meth. 2009;6:651–654. doi: 10.1038/nmeth.1365. [DOI] [PubMed] [Google Scholar]
- 18.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucl. Acids Res. 2010;38:545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helland R, Larsen RL, Finstad S, Kyomuhendo P, Larsen AN. Cell. Mol. Life Sci. 2009;66:2585–2598. doi: 10.1007/s00018-009-0063-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meroueh SO, Bencze KZ, Hesek D, Lee M, Fisher JF, Stemmler TL, Mobashery S. Proc Natl Acad Sci U S A. 2006;103:4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reider A, Barker SL, Mishra SK, Im YJ, Maldonado-Baez L, Hurley JH, Traub LM, Wendland B. EMBO J. 2009;28:3103–3116. doi: 10.1038/emboj.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.