Abstract
Phototropins, a class of light-activated protein kinases, are essential for several blue light responses in plants and algae, including phototropism. These proteins contain two internal light, oxygen, and voltage sensitive (LOV) domains, which bind flavin chromophores and undergo a reversible photochemical formation of a cysteinyl-flavin adduct as part of the light sensing process. While the photodynamic properties of such photosensory domains are dictated by interactions between the chromophore and surrounding protein, more distant residues can play a significant role as well. Here we explore the role of the Phe434 residue in the photosensory response of the second LOV domain of Avena sativa phototropin 1 (AsLOV2), a model photochemical system for these LOV domains. Phe434 lies over 6 Å from the FMN chromophore in AsLOV2; nevertheless, a F434Y point mutation is likely to change several structural features of the chromophore binding site, as we demonstrate using molecular dynamics simulations. Transient absorption signals spanning 15 decades in time were compared for wildtype AsLOV2 and the F434Y mutant, showing that the latter has significantly altered photodynamics including (i) a faster intersystem crossing leading to triplet formation on a nanosecond time scale, (ii) biphasic formation of adduct state kinetics on the microsecond time scale, and (iii) greatly accelerated ground-state recovery kinetics on a second time scale. We present mechanistic models that link these spectroscopic differences to changes in the configuration of the critical cysteine residue and in the chromophore’s accessibility to solvent and oxygen according to MD trajectories and purging experiments. Taken together, these results demonstrate the importance of residues outside the chromophore-binding pocket in modulating LOV domain photodynamics.
Photosensory receptor proteins, which transduce the energy from the absorption of photons into useful physiological functions, play an essential role for most organisms to adjust their behavior and metabolism in response to the quantity and quality of light in their environment. Such proteins rely on small molecule chromophores which undergo light-dependent configurational changes to generate biological responses through biochemical signal transduction pathways (1, 2). From many perspectives, photoreceptor proteins are ideal systems to study signal perception and transduction activity, (i) since they can be triggered with short pulses of light, and (ii) since their signaling activity can be followed directly with spectroscopic techniques.
Light-sensing systems encompass multiple families of proteins including the LOV (Light-Oxygen-Voltage) domains, BLUF (Blue Light sensors Utilizing Flavin) domains, cryptochromes, photoactive yellow protein, rhodopsins, and phytochromes (1, 3–8). The phototropin, BLUF, and cryptochrome families utilize flavin chromophores that absorb light to initiate photoactivity. In cryptochromes and BLUF domains, the flavin chromophore is noncovalently bound as a flavin adenine dinucleotide (FAD) (9, 10), whereas most LOV domains bind flavin mononucleotide (FMN) chromophores. Examples of the latter domains include the tandem LOV1 and LOV2 domains within phototropins, a class of light-activated serine/threonine kinases found in plants and algae (11). These proteins mediate several classical blue light responses in plants, including phototropism (plant growth toward light sources), stomatal opening, chloroplast movement, and rapid inhibition of stem growth (12). Since their initial characterization in phototropins, LOV domains have been found in a wide variety of proteins from plants, algae, fungi and bacteria (13).
In contrast to the rapid (200 fs to 200 ps) photoisomerization mechanisms in the photoactive yellow protein (PYP) (14), rhodopsin (15), and phytochrome (8, 16–18) families (<10 ps) (16). the primary photochemistry in LOV domains involves the much slower formation of a protein-chromophore adduct (several μs) between a conserved cysteine residue and the flavin C4a carbon. While the molecular mechanisms of adduct formation remain somewhat controversial (19–21), with the latest evidence favoring a radical-pair mechanism (22), they are preceded by the fast generation (<3 ns) of a reactive triplet state populated via intersystem crossing (ISC) from the initial photoexcited singlet state (Fig. 1). The reactive triplet persists for several microseconds before generating the metastable photoadduct with a new covalent bond between FMN and a nearby cysteine residue, which is Cys450 in the model LOV system AsLOV2 (LOV2 domain of Avena sativa phototropin 1) (Fig. 2) (19). The photoadduct alters the surrounding protein structure, leading to conformational changes that trigger biological responses. For AsLOV2, adduct formation results in the unfolding of the Jα-helix that is located C-terminal to the LOV domain immediately (23, 24), generating the putative phototropin signaling state. The photoadduct recovers back to the dark-adapted singlet ground state through a light-independent mechanism on a time scale between seconds and hours, depending on the specific LOV domain (19, 25–27).
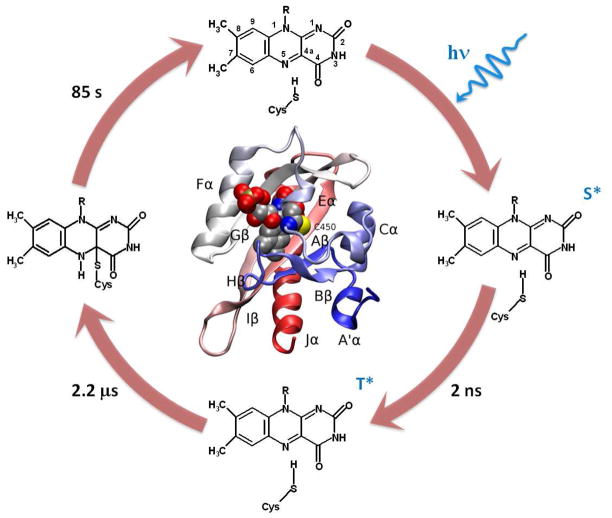
Figure 1.
Primary steps in the wildtype AsLOV2 photocycle. The dark adapted population (top) has the FMN molecule (with IUPAC atomic labels) bound to the protein via non-covalent interactions. Upon photoexcitation, FMN is promoted to an excited singlet state (right), which persists for 2 ns before generating the excited triplet state (bottom). The FMN triplet state persists for 2.2 μs before forming an adduct between the sulfur on Cys450 and the C4a on FMN (left). The adduct is stable for 85 s before completing the photocycle by recovering the dark adapted population. Inset shows a ribbon diagram of the dark-adapted AsLOV2 structure22 with secondary structure elements labeled and a space-filling view of FMN.
Figure 2.
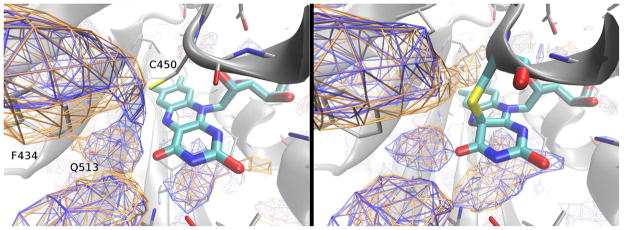
Protein structure surrounding the FMN cofactor in the wildtype AsLOV2 domain, as identified in crystal structures.22 (A) Dark structure with Cys450 in conformation I, (B) Dark structure with conformation II, (C) Light state adduct. The Ea helix contains the adduct-forming Cys450 residue, while Gln513 in the Iβ-strand makes hydrogen bonds with the O4 of the FMN isoalloxazine ring in the dark state. Phe434 on the Cα helix interacts with Gln513 through Asn414.
The flavin chromophore within a LOV domain is non-covalently bound to the protein via hydrophobic interactions complemented by a network of hydrogen bonds involving multiple amino acids (Scheme 1) (28). Each of these amino acids potentially plays a role in adjusting the photochemical properties of the receptor via their specific interactions with the FMN chromophore, surrounding protein, or both. For example, the side chain of Gln513 donates a hydrogen bond to the O4 of FMN in the dark state AsLOV2 structure (29). Although Gln513 does not directly interact with the Cys450 residue involved in the photoadduct, mutations at Gln513 can strongly affect the observed photocycle dynamics, as demonstrated by the 15-fold slower ground-state recovery kinetics of the Q513L mutant (30). Similarly, Ile427 and Asn449 stabilize the cysteinyl-flavin photoadduct via their van der Waals contacts with Cys450 (29). as argued by the faster observed adduct decay kinetics observed in an I427V mutant AsLOV2 that removes this favorable interaction (31).
Scheme 1.
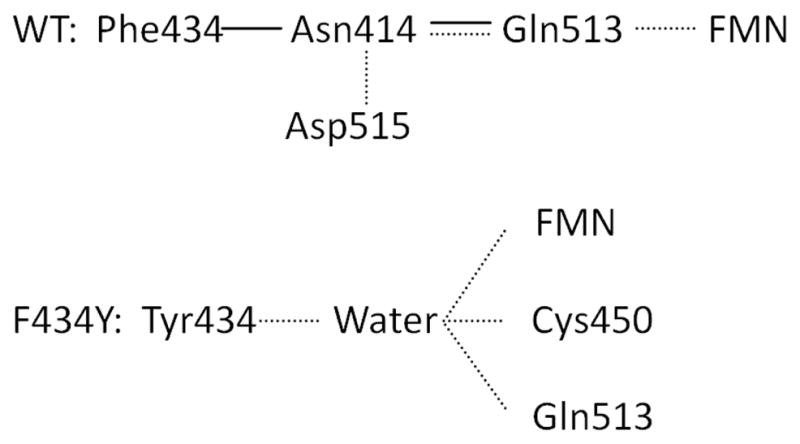
Phe434 in wild type and Tyr434 in mutant are contact with other amino acid residues which contributes hydrogen binding to FMN. Solid line: vdW; Dashed line: hydrogen bond. In the light-induced state a structural change induces a breakage of hydrogen bond between Asn414 and Asp515.
While Ile427, Asn449 and Gln513 have been identified as important for LOV photocycling by either physical proximity (28). or random mutagenesis (31), as have certain other residues by mechanism-based approaches (32), there is a need for further rationally-predicted mutations that alter LOV domain photochemistry. Molecular dynamics (MD) simulations provide a useful computational route to such predictions, via their ability to explore conformations that are functionally critical, but difficult to characterize by X-ray crystallography or solution NMR spectroscopy. An example of this is provided by recent MD simulations of wildtype AsLOV2 which suggest that formation of the cysteinyl-flavin photoadduct forces the Gln513 side chain to rotate away from the flavin and break the hydrogen bond to the FMN O4 atom observed in the dark state (Fig. 2A) (33). This alteration of the Gln513 conformation tilts the Iβ strand and destabilizes the interface with the Jα helix by introducing a bulge into the binding surface and by changing the conformational distribution of the Hβ-Iβ loop (Fig. 1, inset) (33, 34).
Given the nature of the structural changes to Gln513 observed in the light-induced state, it was hypothesized that the introduction of a hydrogen bond donating residue in the FMN binding pocket “above” the plane of the isoalloxazine ring system (in the direction of Cys450) would result in a flipped rotamer of Gln513 that would persist in the dark state and yield a constitutively active LOV2 domain. Based on structural analysis, three mutations were selected to achieve this goal: V416T, F434Y and L453T. All three mutants were investigated with MD simulations in both the dark and light states and subsequently overexpressed in E. coli and purified (Supporting Information). While the V416T variant appeared to be unperturbed from wildtype based on similarities in NMR and visible absorbance spectra, and the L453T mutant was insufficiently stable for biophysical characterization (data not shown), aspects of the F434Y-containing protein warranted further investigation. Notably, simulations of the F434Y mutant showed altered Gln513 behavior and the rotameric distribution of the photoreactive Cys450 residue that suggested that this variant might demonstrate significant changes in photodynamic properties. It should be noted that while classical molecular dynamics simulations cannot treat the changes in electronic structure involved in the photoreaction itself, they can provide crucial data on the environment of the flavin chromophore in both the dark and light states as demonstrated here.
Presented here is a combined simulation/experimental study that explores the influence of the F434Y mutation on the photodynamics of the AsLOV2 domain extending over 15 decades in time. Atomistic MD simulations provide detailed information regarding chromophore binding pocket dynamics and energetics, including the conformational kinetics of nearby residues. We also resolve water and oxygen accessibilities to the binding pocket. Experimentally, the formation and decay kinetics of both the triplet state of the photoexcited FMN cofactor and the photoadduct species were resolved with a combination of absorption spectroscopy techniques sensitive to ultrafast (fs-ns), fast (μs) and slow (s) photodynamics.
Materials and Methods
The details of sample preparation, experimental techniques and computational approaches are described in depth in the Supporting Information and are briefly summarized here. The ultrafast (fs to ns) photodynamics in both the dark and light states for each AsLOV2 variant studied were resolved using atomistic molecular dynamics simulations with three 200 ns trajectories in each case for the F434Y mutant that are compared to five 200 ns trajectories in each state from a previous study of wildtype AsLOV2 (33). Solution NMR experiments were performed on Varian Inova 600 and 800 MHz spectrometers at 25°C. The transient absorption signals resolving the ultrafast dynamics were measured with a amplified mode-locked Ti:Sapphire laser spectrometer, generating both 50 fs, 400 nm excitation pulses and broadband visible probe pulses (35). The photodynamics extending from 10 ns to 20 μs were collected with an ancillary flash photolysis setup using 50 fs, 400 nm laser pulses for excitation and a 640 nm CW laser diode as the probe.
Results
Molecular dynamics simulations
A number of experimental and computational studies have identified motion of Gln513 as a key step in activation of the LOV domain after the photoreaction (20, 24, 28, 34–40). While it has generally been assumed that the rotation of Gln513 that allows hydrogen bonding with the newly protonated N5 nitrogen is the key structural change for activation, recent MD simulations of wildtype AsLOV2 (using the same structures as in the present study) suggested that this hydrogen bond is only transiently populated (present in <10% of timesteps) (33). Similar results had been previously obtained for a LOV2 domain from Adiantum capillus-veneris phy3 (34), and notably, a high-resolution crystal structure of AsLOV2 failed to observe formation of this hydrogen bond upon illumination (29). Instead, the primary structural change observed in the MD simulations (33). of the light-induced state is one in which Gln513 rotates away from FMN altogether, and in many cases is replaced by a water interacting directly with FMN at O4 and/or N5. This light-induced rotation of Gln513 was correlated with broader structural changes such as tilting of the Iβ strand that contribute to Jα dissociation.
In addition to the observed changes in the conformation of Gln513 (Supporting Information), the F434Y mutation introduces a water binding site adjacent to FMN that is similar to light-induced structural changes reported elsewhere (33). Volumetric maps of the water occupancies observed in both the wildtype and F434Y mutant simulations are compared in Fig. 3. Between all four states (i.e., F434Y and wildtype under both light and dark conditions), a total of three different major water binding sites are resolved: (1) one adjacent to Asn492 and distal to FMN, between the Iβ strand and Cα helix (which will be referred to as the antechamber site), (2) one positioned to interact with the O4 atom of FMN, a similar site interacting with the FMN N5 atom, and (3) one adjacent to Cys450 distal to FMN as observed in the crystal structure (29). Site 1 corresponds to a previously identified void in several LOV domain crystal structures proposed to allow imidazole occupancy near the chromophore (site 2 of reference (41).), but the others have not been identified previously and appear to emerge only in some non-crystallographic conformations sampled during MD simulations. The occupancies of these three sites throughout the simulations reported here, and their correlations with the conformation of Q513, are shown in Tables SI1 and SI2. Very little water binding in the sites interacting directly with FMN is apparent in the wildtype dark state simulations, and when present such waters do not cause Q513 to lose contact with O4. However, the photoadduct state allows water binding to both the O4 and N5 sites, displacing Gln513 as discussed above; as seen in Tables SI1 and SI2, the occupancy of conformations in which Gln513 can form a hydrogen bond with the O4 atom of FMN drops from 0.7 to 0.25 between the dark and light state (33). As expected, the F434Y mutation significantly alters the pattern of water binding in the vicinity of FMN (Scheme 1); in the F434Y dark state, strong occupancy of the N5 site is observed, although the location of water binding is slightly above the FMN plane (vs. in the plane in the wildtype light state). This is because the water molecules bound here are generally hydrogen bonded with both Tyr434 and FMN. The F434Y light state shows a stronger water occupancy of the O4 site and weaker occupancy of the N5 site, but is otherwise similar to the wildtype light state. These water binding sites strongly influence the behavior of Gln513, as displacement of Gln513 away from FMN occurs primarily in the presence of water molecules bound to the O4 and N5 sites. When in the flipped conformation, Gln513 can interact with both the water that displaces it and the antechamber water.
Figure 3.
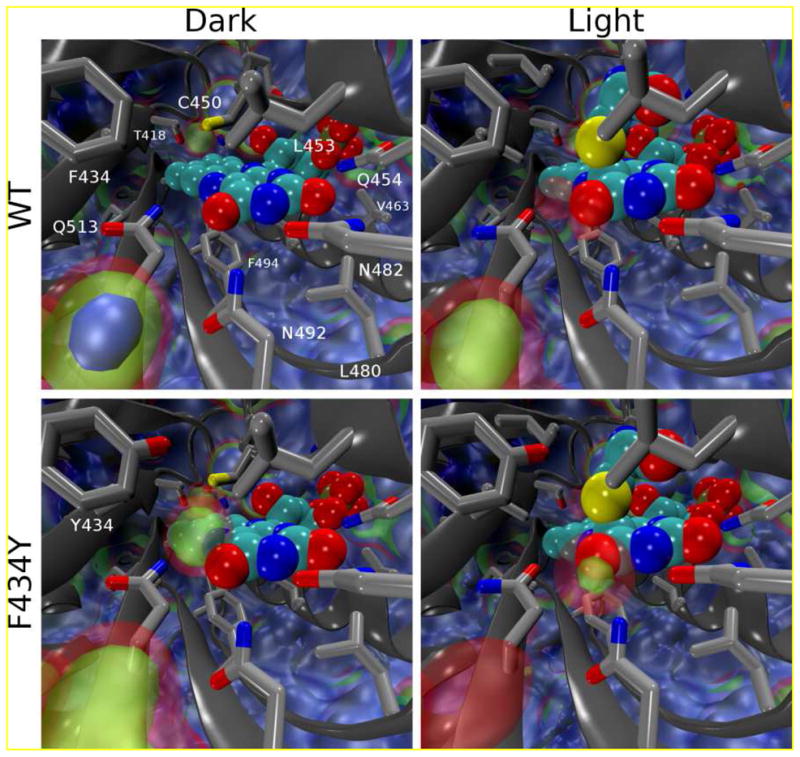
Volumetric maps showing water occupancy in and around the FMN binding site from MD simulations. The value of each volume element (voxel) corresponds to the average number of water oxygens occupying it; the voxels span 0.5 Å along each dimension and are arranged in a grid along arbitrary orthogonal axes. The median value for bulk water was found to be 0.475 waters/voxel; blue, green, and red isosurfaces indicate occupancies of 1x, 0.5x, and 0.25x the density of bulk water, respectively. Clockwise from top left, occupancies averaged over: D_WT1-5, L_WT1-5, L_F434Y1-3, D_F434Y1-3.
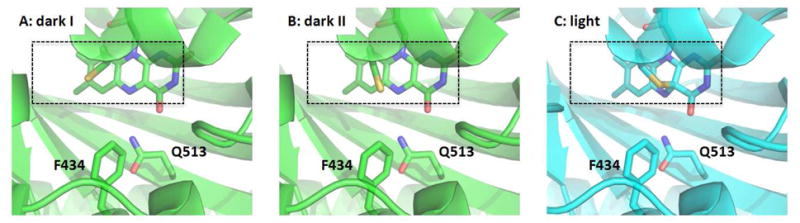
As observed in the crystal structures of CrLOV1 (Chlamydomonas reinhardtii phototropin 1 LOV1) (37). and AsLOV2 (29), the MD simulations of wildtype AsLOV2 resolved two conformations of Cys450, differing by rotation about the χ1 angle (Figure 2C,D). Similar cysteine inhomogeneity can be observed in FTIR measurements (42, 43). In conformation I, Cys450 is rotated to a position with its sulfur atom directly above the C4a atom of FMN, whereas in conformation II, the cysteine is rotated away from the C4a atom and toward a water binding site. The occupancies of conformations I and II extracted from the CrLOV1 and AsLOV2 crystal structures are strongly biased towards conformation II (0.3:0.7 for CrLOV1 (37) and 0.1:0.9 for AsLOV2) (29). However, our MD simulations of wildtype AsLOV2 in solution yield different occupancies of 0.74 (± 0.03) and 0.25 (± 0.03) for the conformation I and II states, respectively (treating each non-overlapping 50 ns segment of trajectory as a statistically independent measurement). The F434Y mutation significantly alters the energetics and kinetics of Cys450 rotation to favor conformer II with occupancies of 0.29 ± 0.05 and 0.71 ± 0.05 for conformer I and II, respectively (Table 1).
Table 1.
Rate constants in the kinetic simulations of wildtype and F434Y for the conformer exchange.
| Parameter | WT (A) | F434Y (B) | |
|---|---|---|---|
| MD simulation conformer time constants* | k I→II | (2400 ps)−1 | (950 ps) −1 |
| k II→I | (900 ps) −1 | (2400 ps) −1 | |
| MD simulation conformer occupancies | conformer I | 74.5% | 28.8% |
| conformer II | 25.5% | 71.1% | |
| conformer time constants | k I→II | (1200 ps) −1 | (1100 ps) −1 |
| k II→I | (900 ps) −1 | (1875 ps) −1 | |
| conformer Occupancies | conformer I | 57 % | 37 % |
| conformer II | 43 % | 63 % | |
| S2→S1 time constants | Ikvib,IC | (1 ps) −1 | (0.3 ps) −1 |
| IIkvib,IC | (0.5 ps) −1 | (0.3 ps) −1 | |
| radiative time constants | Ikrad | (17 ns) −1 | (17 ns) −1 |
| IIkrad | (17 ns) −1 | (17 ns) −1 | |
| IC time constants | IkIC | (20 ns) −1 | (1.68 ns) −1 |
| IIkIC | (21 ns) −1 | (2.15 ns) −1 | |
| ISC time constants | IkISC | (1.5 ns) −1 | (1.5 ns) −1 |
| IIk,ISC | (3.0 ns) −1 | (3.0 ns) −1 | |
| triplet quantum yield, φ | φI,ISC | 87% | 41% |
| S-FMN distances | conformer I S-FMN mean distance | 3.74 Å | 3.80 Å |
| conformer II S-FMN mean distance | 4.54 Å | 4.84 Å | |
Estimated conformer equilibration time constants are underestimated by 30% to 90% due to the use of a Langevin thermostat (shaded parameters).
These MD simulations demonstrate that interconversion between C450 conformers is rapid, occurring on nanosecond time scales in both the wildtype and F434Y structures. The distribution of dwell times in one conformation prior to switching in the wildtype dark state can be fitted with a single exponential decay with rate constants of (2400 ps)−1 and (890 ps)−1 for the I → II and II → I transitions, respectively. Coincidentally, the rates for the same transitions in the F434Y dark state are almost exactly reversed at (950 ps)−1 and (2400 ps)−1 for the I → II and II → I transition, respectively. Introducing the F434Y mutation shifts the free energy difference between the rotamers by ~1 kcal/mol to more strongly favor conformation II, presumably due to steric hindrance by the Tyr434 phenolic oxygen and the introduction of a water binding site that competes with conformer I. Due to the formation of a covalent bond between Cys450 and the C4a atom of FMN, conformation II is not populated in any of the light state simulations.
Structural effects of F434Y mutation as identified by solution NMR
To establish the effects of the F434Y point mutation on the AsLOV2 structure, we compared multidimensional NMR spectra acquired on uniformly 15N-labeled samples of the wildtype and F434Y forms of the protein. An overlay of 2D 15N/1H HSQC spectra of both proteins (Fig. SI2a) shows similar overall patterns of crosspeaks, with only a subset of sites being affected. This is qualitatively consistent with only minor perturbations of the overall LOV domain structure. More detailed analysis using the minimum chemical shift difference method (44), which conservatively estimates changes between a fully assigned reference protein (i.e., wildtype AsLOV2) and the unassigned mutant, shows that structural effects are primarily focused to the region around the F434Y point mutation (Fig. SI2b,c). Several sites near the FMN chromophore are affected but a limited number of locations overall are perturbed. Taken together, these data are consistent with the F434Y protein maintaining a similar global structure as the wildtype protein, with minor conformational changes near Tyr434, Cys 450 and the isoalloxazine ring.
Ultrafast Triplet Formation Dynamics (<10 ns)
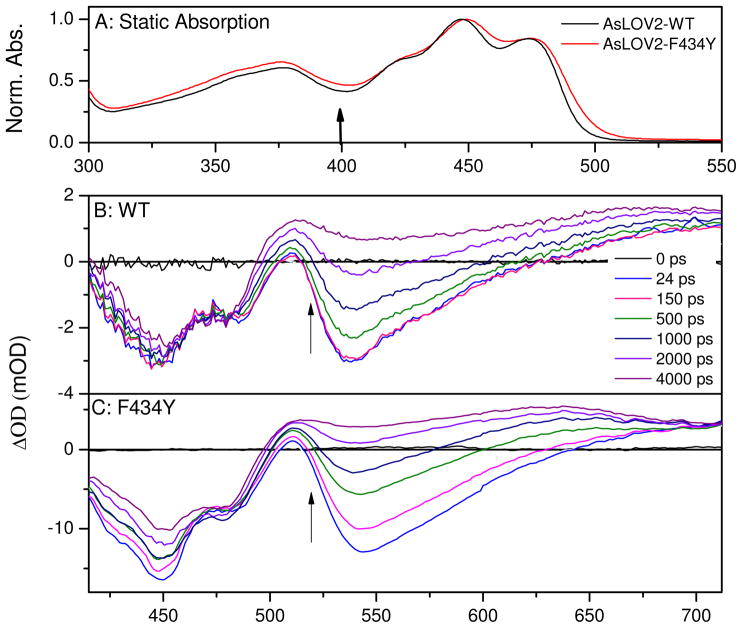
The static absorption spectra of both wildtype AsLOV2 and the F434Y mutant are very similar (Fig. 4A), with vibronic peaks at 422, 447, and 474 nm associated with the S0→S1 transition. A higher energy S0→S2 transition overlaps the S0→S1 transition and peaks near 365 nm (45). The measured fs-μs photodynamics for both systems were initiated via 400 nm ultrafast excitation. Although this introduces excess vibrational energy into the system vs. lower energy excitation, it has previously been established that excitation wavelengths between 360 to 470 nm produce similar photocycles and quantum yields (46, 47).
Figure 4.
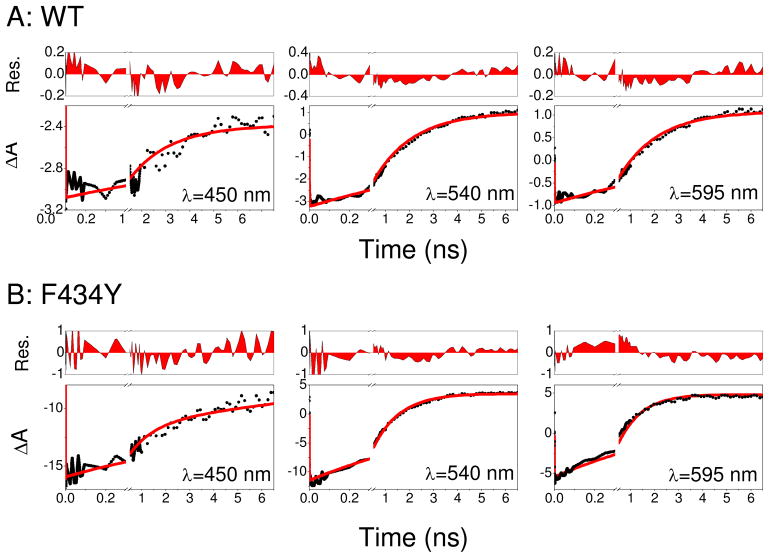
(A) Normalized absorption spectra of wildtype AsLOV2 (black lines) and F434Y mutant (red line), in 50 mM sodium phosphate buffer (pH 6.0) with 100 mM NaCl. Arrow indicates the excitation wavelength for the transient experiments. Ultrafast transient absorbance spectra of (B) wildtype and (C) F434Y. Arrows indicate transient trend of data as the singlet S1 population evolves to the triplet population. Note: the F434Y mutant exhibits greater transient loss of bleach (<500 nm) signal than the wildtype signals.
The sub-ns photoinduced signals in F434Y and wildtype protein exhibit similar spectroscopic features (Fig. 4B and C). Immediately upon 400 nm photoexcitation, a negative ground-state bleach (GSB) between 400 and 500 nm is observed, concomitant with a negative stimulated emission (SE) band near 540 nm and positive excited-state absorption (ESA) bands at 510 nm and above 625 nm. Since the transient spectra in Figure 4 resolve both the bleach and excited-state signals simultaneously, a complete model with ground-state recovery kinetics can be constructed. In the F434Y sample, the sub-picosecond signals exhibit a fast (~2 ps) blue-shift of the SE band that is ascribed to vibrational cooling of the S1 population (Fig. 4C) (48). Transient signals induced with lower energy excitation (470 nm) do not exhibit this sub-picosecond behavior (27, 41). The 1 ps spectrum is similar to the excited singlet spectrum previously identified for oxidized FMN (48) and the 7 ns spectrum strongly resembles the known triplet state spectrum of flavin with a broad absorption band extending from 490 nm into the red (49). Apart from this fast vibrational relaxation, the observed dynamics agree well with previous ultrafast measurements on wildtype AsLOV2 by Kennis and co-workers (27) with the initial photo-excited singlet state generating an excited triplet state on a nanosecond time scale. The addition of imidazole, used to accelerate the ground-state recovery kinetics, does not affect the ultrafast ISC kinetics in either sample (data not shown) (31, 32, 41).
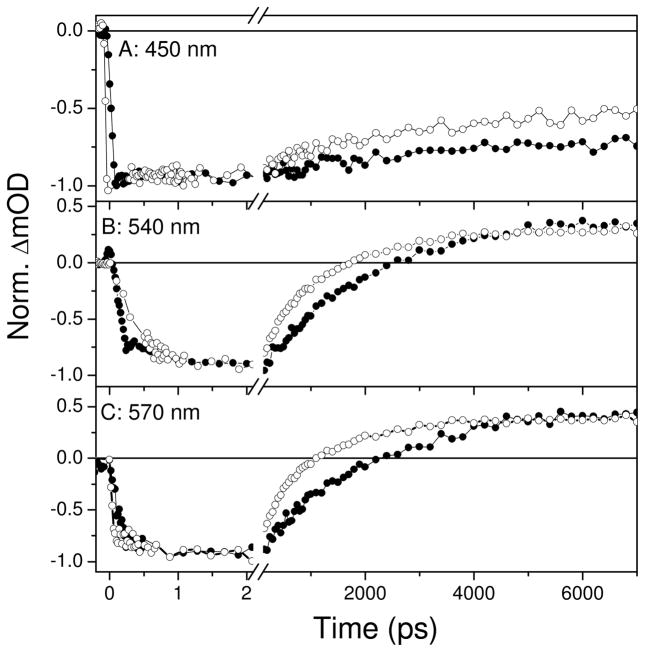
The differences between wildtype AsLOV2 and F434Y signals are more apparent when viewed temporally, and differ in both time scale and functional form (Figs. 5 and SI3). The singlet excited state (with its characteristic SE and ESA bands) decays distinctly faster for F434Y faster than for wildtype AsLOV2. Global multi-wavelength fits to the wildtype AsLOV2 data demonstrate that a single exponential decay with τ = 2.09 ns (Fig. SI3) is sufficient to describe the data well, in agreement with previous measurements of Kennis and co-workers (27). In contrast, the F434Y mutant photodynamics required a bi-exponential decay model with time constants of 320 ps (34 %) and 1.4 ns (66 %) to describe the data (Fig. SI3B).
Figure 5.
Ultrafast transient absorption kinetic traces probed at select wavelengths of wildtype AsLOV2 (filled circles) and F434Y mutant (open circles) during the formation of the triplet state via intersystem crossing. λpr=450 nm (A); 540 nm (B); 570 nm (C).
Adduct Formation Dynamics (10 ns – 5 μs)
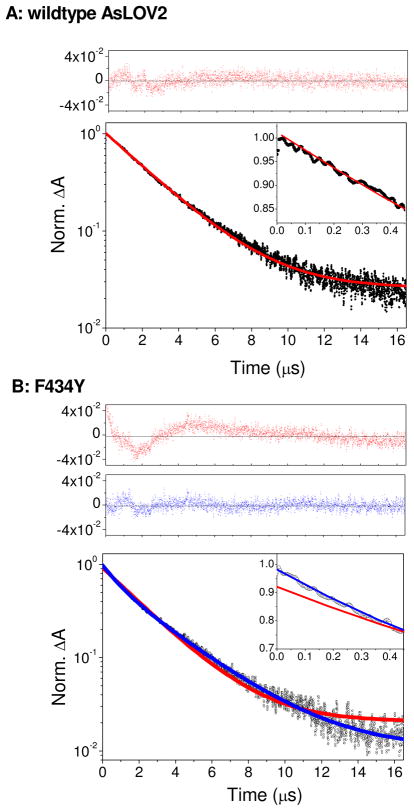
Although the triplet state formation kinetics is clearly resolved in the sub-10 ns signals, the subsequent decay and adduct formation dynamics between Cys450 and FMN evolve on a microsecond time scale that cannot be directly monitored with the ultrafast apparatus. To characterize the adduct formation dynamics that quench the triplet population, the time-resolved signals of the triplet state were probed at 640 nm, near the peak of the triplet absorption spectrum (Fig. 4B,C). The adduct absorbs very weakly at this wavelength (19). The parameters extracted from fitting these data to a sum of exponentials are comparable (Table SI4) to other LOV studies (31). The triplet state decay for the wildtype protein can be fit with a single exponential decay with 2.49 μs time constant (Fig. 6A). In contrast, the F434Y signals require a bi-exponential model to achieve a comparable quality of fit (Fig. 6B), with time constants of 3.02 μs (66%) and 0.93 μs (34%). As with the ultrafast triplet formation kinetics, the addition of imidazole to either sample does not affect the observed triplet decay kinetics (data not shown).
Figure 6.
Triplet state decay kinetics of wildtype AsLOV2 (A) and F434Y (B) at 640 nm near the peak of triplet state absorption (Fig. 4B,C). Excitation pulses were centered at 400 nm with an energy of 160 nJ. Experimental data (circles) were normalized at the peak signal (time=0). Red lines indicate a single-exponential fit, whereas blue lines show a bi-exponential fit. Data and fits at early times are compared in insets. Residuals of fits are contrasted at top panels as indicated. The transient data were best fit with the following functions A(t)=0.98·e−t/2.50+0.025 and A(t)=0.33·e−t/0.93+0.64·e−t/3.02+0.010 for wildtype and F434Y, respectively, with t in units of μs. The small (1%) static amplitude represents the weak absorption of the adduct photoproduct.
Adduct Decay Dynamics (500 ms – 100 sec)
The photoadduct recovery kinetics in wildtype AsLOV2 exhibits a mono-exponential decay with a time constant of 85 s under the conditions of the protein sample. Although this is a longer timescale than previously observed for AsLOV2 (e.g., 45 s observed in Reference 40), different sample conditions were used here. The F434Y mutant also exhibits mono-exponential photoadduct recovery kinetics, but with an appreciably faster time constant of 7.8 s (at 25°C in pH 6.0 buffer, see Fig. SI7). The F434Y mutation also induced a greater sensitivity (observed as an acceleration) of the adduct decay kinetics to residual imidazole in the buffer than for wildtype (data not shown), indicative of the greater solvent access to the FMN cofactor for the F434Y mutant identified in the MD simulations.
Discussion
Intersystem Crossing Dynamics (100 fs to 10 ns)
The ultrafast kinetics of the studied LOV domains shown in Figures 4 and 5 are strongly dominated by the high yield (>50%) ISC dynamics that generate the reactive triplet populations. For wildtype AsLOV2, these signals can be well described with a single 2 ns exponential model in agreement with Kennis and co-workers (27). In contrast, fitting the F434Y signals requires multiple exponential components (Fig. SI3), which is often interpreted as a signature of an underlying inhomogeneity. The coexistence of two conformations of Cys450 offers a convenient and often-used justification for the multi-exponential μs kinetics in LOV domains. For wildtype AsLOV2, the occupancies of conformations I and II in the x-ray crystal structures are estimated at 10% and 90%, respectively (29). The MD simulations on wildtype AsLOV2 demonstrate that the two conformations are in a dynamic equilibrium with Cys450 flipping from one conformer to the other on a nanosecond time scale (33, 42). A static phenomenological model consisting of two separate populations exhibiting differing ISC kinetics can be constructed to characterize the F434Y signals (Fig. SI4C). This model, however, does not sufficiently explain both the bi-exponential kinetics of the F434Y mutant and the mono-exponential kinetics in wildtype AsLOV2 (27), given that the MD simulations demonstrate that inter-conformation population flow occurs on time scales comparable to those of the ISC dynamics for both wildtype and F434Y mutant AsLOV2.
Rather, the underlying ultrafast signals for both proteins require a dynamic multi-state model that incorporates the interchange of conformers identified in the MD simulations. Underlying this “dynamic conformer model” in Figure 7 is that both cysteine conformations differ not only in orientation, but also in the average distance between the sulfur atom of Cys450 and the FMN co-factor (Table 1). From the MD simulations, these distances average 3.80 Å (conformer I)/4.84 Å (conformer II) for F434Y, and 3.74 Å (conformer I)/4.54 Å (conformer II) for wildtype LOV2 structures. It is known that the addition of sulfur atoms to riboflavin solutions in DMSO enhances ISC kinetics (50) and that Cys→Ser mutations which remove the sulfur atom of the conserved cysteine residue in CrLOV1 (Cys57 in CrLOV1 and Cys450 in AsLOV2) significantly slows ISC dynamics (from τF = 2.9 ns to τF = 4.6 ns) (50, 51). The heavy atom mechanism affecting ISC dynamics is a spin-orbit coupling that scales exponentially with the distance between the sulfur and the FMN chromophore (52–54). The ISC rates are thus strongly modulated by the conformer occupations and the underlying Cys450 flipping dynamics (55, 56).
Figure 7.
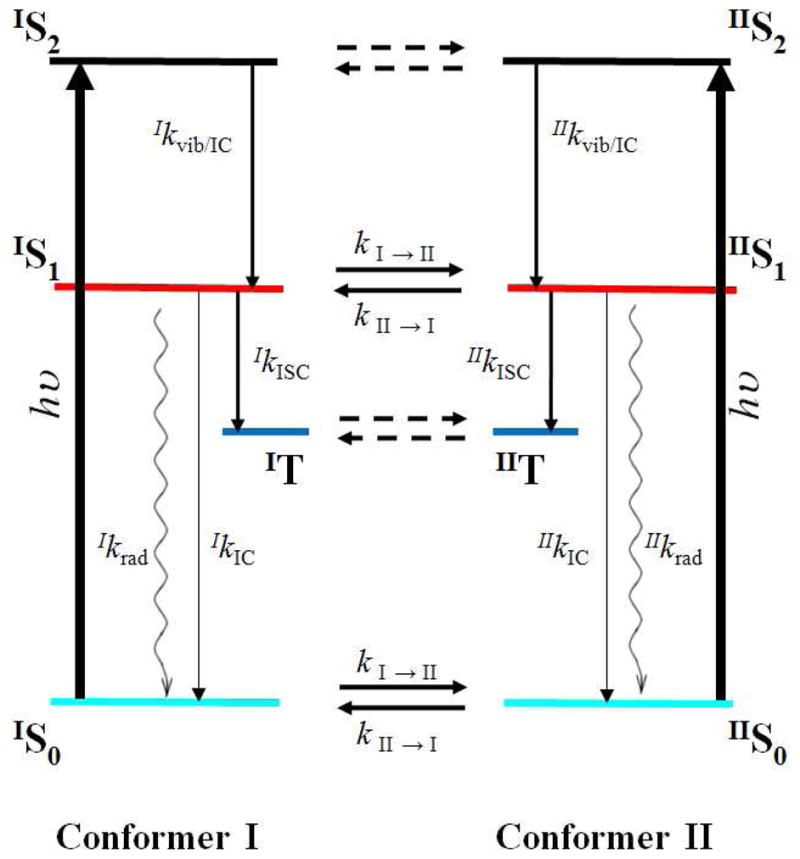
Dynamic conformer model used to fit both wildtype and F434Y mutant ultrafast dynamics. The connectivity consists of two photoexcited populations (conformer I and conformer II) representing different conformer states (and distances to the FMN chromophore) of the Cys450 residue. Each population is indicated via right superscripts. 400 nm photoexcitation pulses (thick vertical black lines) excite both the S2 and higher lying vibrational-excited S1 (black lines) in each conformer population. Evolution via vibrational relaxation and S2 to S1 occurs via internal conversion. The S1 population persists for ns and decays via either intersystem crossing to the triplet state (blue line), radiative emission (wavy lines) or internal conversion to the ground state populations (cyan curve). Horizontal lines represent inter-conformer dynamics. The modeling explicitly included the inter-conformer conversion of the S0 and S1 populations (which were locked to same kinetics); the dashed lines presumably exist, but do not affect the signals and were ignored for convenience. Initial occupations of each conformer population result exclusively from the inter-conformer time constants. States are color coded to spectra and concentration profiles in Figure SI6.
The dynamic conformer model in Figure 7 separates the photodynamics into contributions from the two conformers with different intrinsic ISC rates, with population flow occurring between them. Although more complicated than a static inhomogeneous model (Fig. SI4C), a number of constraints can be introduced to limit the range of estimated parameters to describe the transient data: (1) the Species Associated Difference Spectrum (SADS) (57, 58). that describes the spectrum for each contributing population in the dynamics is assumed to be insensitive to the C450 conformation (IS1 = IIS1); (2) the initial occupancies of the conformers are determined exclusively by the isomerization time constants, where the occupancy ratio is equal to the ratio of isomerization time constants; (3) the ISC time constants for each conformer flow is independent of the sample (i.e., IkISC for wildtype AsLOV2 is the same as IkISC for the F434Y mutant); (4) the radiative, ISC, and internal conversion (kIC) time constants are accurately described by simultaneously fitting the ground-state bleach recovery and the excited-state signal, and (5) reasonable agreement with MD simulations of conformer time constants including both time scales and trends.
Subject to the constraints described above, the dynamic conformer model accurately describes both the wildtype AsLOV2 and the F434Y mutant ultrafast signals (Fig. 8) with the parameters outlined in Table 1. The concentration profiles for the contributing populations and their SADS are compared in Fig. SI6 (57, 58). Although intrinsically multi-exponential in decay character, the predicted dynamics for the wildtype AsLOV2 sample is primarily mono-exponential due to the trade off of occupancies and intrinsic ISC timescales described below. In agreement with the MD simulations, the combination of the greater conformer I occupancy (57%) vs. conformer II (43%) with the faster inter-conformer kinetics of conformer II to conformer I (900 ps) vs. the reverse (1200 ps) generates a near mono-exponential behavior. In contrast, the F434Y mutation alters the conformer occupancies to favor the conformer II (37% vs. 63%), with a slower inter-conformer time scale (conformer I to conformer II), which generates biexponential ISC kinetics.
Figure 8.
Select decay kinetic traces of wildtype AsLOV2 (A) and F434Y (B) ultrafast signals. Solid lines indicate fits obtained from the dynamic inhomogeneous model outlined in Fig. 7. Inter-conformer kinetics parameters were obtained via MD simulations. The S2, S1, and T populations for each conformer were locked to each other as were the same intersystem crossing time constants for each conformer in both samples (Table 1). Releasing these temporal constraints results in slightly better fits (Supporting Information), but loses information regarding the connectivity between the C450 isomerization dynamics and observed ISC dynamics observed in both samples. Residuals are shown in top panels.
While the time constant for rotation of Cys450 from the II to the I conformation in MD simulations is approximately 2.5-fold too slow (k−1I→II = 2400 ps and k−1II→I = 900 ps) to match the experimentally observed slow component of the F434Y ISC kinetics (2400 ps), the Langevin thermostat in the MD simulations is known to overestimate true rates of barrier crossing (59). Indeed, in 200 ns simulations of both the wildtype and F434Y dark states in the NVE ensemble (i.e., with no thermostat), all Cys450 rotation rates except for the wildtype kI→II were estimated to be faster, by factors of 1.3–1.9 (data not shown). This accounts for the small discrepancy with experimental time scales used in the global modeling and the MD simulations.
As indicated by the bleach recovery dynamics in Fig. 5A, the F434Y mutation also induces faster IC population kinetics. This may originate from the presence of the water molecule identified in the MD simulations (Fig. 2) that can hydrogen bond to the FMN to facilitate a faster quenching mechanism (60). The peak of the static absorption spectrum of F434Y is red-shifted compared to the wildtype (Fig. 4A). This spectral shift may reflect a local reordering of hydrogen bonding around the FMN with the hydroxyl group of the newly introduced tyrosine forming a new hydrogen bond to some amino residue in the AsLOV2 domain (Scheme 1) (61). It has been observed that the lack of hydrogen bonding between A. capillus-veneris phy3 LOV2-Q1029L and FMN induces a similar blue-shifted absorption spectrum in the dark (36). The molecular dynamics simulations indicate that Tyr434 accepts a new hydrogen bond to a water molecule (Fig. 3), which mediates interactions between Cys450 and the O4 atom of FMN. Even though the ISC rate constants are distinctly faster for the F434Y mutant, which increases the triplet yield and overall photosensitive efficiency of the protein, an increase in IC timescales, which reduces the triplet yield, counters this increase perfectly to generate a near identical yield for both wildtype and F434Y proteins. An alternative mechanism underlying this faster IC for the F434Y mutant involves a competing electron transfer process from the introduced tyrosine to FMN that would quench the singlet excited state. A similar mechanism has been proposed for activating BLUF domains (46, 62) whereby a conserved Tyr donates an electron to the photoexcited flavin. However, the transient data does not support the growth of a secondary reduced quinoid species overlapping the singlet and triplet populations, which indicates that if electron transfer mechanism were functioning in the LOV2 domains, that it does not result in stable intermediates that would rapidly recombine to recover the ground state.
Adduct Formation Dynamics (100 ns – 5 μs)
Many naturally occurring LOV domains (e.g., AsLOV2 and Bacillus subtilis YtvA LOV) exhibit single exponential μs photoadduct formation kinetics, whereas others (e.g., CrLOV1) exhibit bi-exponential kinetics (Table SI3). The single-exponential decay kinetics of the wildtype AsLOV2 triplet state (Fig. 6A) suggests that either a single photoadduct population exists or that an inhomogeneity is “equilibrated out” on a faster time scale to generate an effective homogeneous system with a rapid population flow between the two populations (e.g., conformers). The F434Y mutation of AsLOV2 switches the protein from exhibiting single- to exhibiting double-exponential triplet decay kinetics, with one phase approximately 2.5 times faster (0.93 μs) and a second phase slightly slower (3.02 μs) than wildtype AsLOV2. The nature of the inhomogeneity responsible for the bi-exponential triplet decay kinetics in the F434Y mutant is not resolved in the data presented here and is outside the time scales of MD simulations. Since the dynamic equilibration of the Cys450 that manipulates the ISC kinetics on the ns-time scale will “average out” on the slower μs time scale, the contributions of various local interactions to the FMN cofactor such as hydrogen bonding and van der Waals interactions on cysteinyl-FMN adduct formation kinetics must be considered. The fast kinetic phase could arise either because the introduced Tyr434 residue alters the conformational distributions of the flavin and Cys450 (Scheme 1) to accelerate adduct formation, or because Tyr434 (or the binding site water bound adjacent to it) directly participates in adduct formation (e.g., by acting as acid/base catalyst to deprotonate Cys450 and protonate the N5 nitrogen of the flavin). Since the dark state MD simulations show that water binding and unbinding into the FMN pocket occur on time scales of tens of nanoseconds, which is orders of magnitude too fast to explain the multiple kinetic phases observed on μs time scale, this eliminates the presence or absence of water as the cause of the two kinetic phases in the mutant signals unless the exchange of water into the binding site is radically slower in the triplet state (not simulated).
It is more likely that a slow, large-scale structural change exists in the F434Y mutant that gives rise to a configuration (e.g., due to the relative orientation of Tyr434 and the flavin) that more rapidly forms the photoadduct. Chemical shift NMR analysis (Fig. SI2) indicates that in the F434Y mutant, sites near Tyr434, Cys450 and the O1/N2/O3 portion of the isoalloxazine ring undergo small changes while the rest of the protein remains globally the same as wildtype.
Adduct Decay Dynamics (500 ms – 100 sec)
Based on observations that the I427V mutation accelerates adduct decay kinetics by destabilizing the adduct through the removal of favorable contacts (31), free energy perturbation (FEP) calculations were performed (Supporting Information) to quantify the change in photoadduct stability for the F434Y mutant compared to wildtype protein. Energetic destabilization of the photoadduct in the F434Y mutant is unlikely to be a factor for the rapid recovery timescale, as FEP calculations indicate that the photoadduct is more stable in the F434Y mutant than in the wildtype, by 1.0 ± 0.3 kcal/mol (with error bars based on block averaging of 10 blocks and given at two standard deviations). The molecular nature of the increased adduct stability for F434Y mutant is unresolved, but the removed constraints of the unfolded Jα-helix force a structural perturbation in the Iβ-strand to induce a small movement of Tyr434 on the Cα helix through flipping of Gln513 and Asp517. The water molecule which donates the new hydrogen bond to Tyr434 (Scheme 1) shifts the conformation properties of Cys450 and this structural change in F434Y alters the dihedral distribution of Cys450 and the conformation of Gln513 (33). The resulting changes in the C4a-distal rotamer and water occupancy to two different conformers might induce β-strand loosening, resulting in faster adduct formation in the light adapted state and faster adduct relaxation in the dark state (Fig. 6 and SI7).
To test alternative causes of the accelerated photoadduct decay in the F434Y mutant, the solvent (Fig. 3) and oxygen (Fig. 9) accessibility of the binding site were explored. Implicit ligand sampling calculations based on the equilibrium MD trajectories indicate the presence of an expanded oxygen-accessible region immediately adjacent to the cysteinyl-flavin bond in the F434Y photoadduct. The estimated free energy for oxygen binding in this region is approximately 2.5 kT more favorable for F434Y than the wildtype protein. The obtained solvent accessibility maps (Fig. 3) indicate a significantly enhanced water binding site adjacent to the O4 atom of the light adapted photoadduct in the F434Y mutant. This raises the possibility of a water-catalyzed decay mechanism (the same site is also presumably responsible for the enhanced imidazole sensitivity of the F434Y mutant) for recovery dynamics, which is, in part, supported by D2O/H2O kinetic isotope effects observed in the recovery of wt-AsLOV2 by Bogomolni and co-workers that identifies a proton transfer reaction involved in initiating the recovery dynamics (19, 63). Both the oxygen and water accessibility analysis was performed on trajectories which began, and remained, in the Jα-associated conformation, due to the very long (by MD standards) timescales for Jα dissociation. Hence, the water and oxygen accessibility may thus be further altered by the dissociation of Jα, in the light state. In biased MD simulations in which Jα was completely dissociated from the beta sheet core, no substantial changes to the FMN binding site were observed besides rotation of Q513 as noted above (P. Freddolino, unpublished data), suggesting that the non-dissociated simulations provide a good approximation for the vicinity of the FMN binding site in the Jα dissociated, light-adapted state.
Figure 9.
Oxygen accessibility of AsLOV2 from MD simulations, calculated using implicit ligand sampling. Left: Dark state oxygen accessibility for wildtype (blue) and F434Y (orange) AsLOV2. An isosurface is shown at a free energy change equivalent to that for moving oxygen from vacuum into bulk TIP3 water (1.97 kcal/mol) (64). Right: Light state oxygen accessibility for wildtype (blue) and F434Y (orange) AsLOV2; isosurfaces are defined as for the left panel.
To evaluate the hypothesis that increased accessibility of molecular oxygen to the FMN binding pocket in the F434Y mutant is, at least in part, responsible for the accelerated recovery timescale of the mutant vs. wild type, the recovery kinetics measured in samples exposed to oxygen in the air was contrasted with samples purged with argon (Figure SI8). A clear slowing of the recovery timescales is observed in the purged, but freshly exposed to the air, sample vs. the fully exposed samples. Furthermore, when the argon purged sample is exposed to air for over 5 minutes, near identical kinetics to the non-purged sample can be observed, indicative of the sensitivity of O2 to the recovery mechanism. In agreement with our MD simulations, the recovery kinetics of wild type samples exhibits a significantly smaller sensitivity to oxygen (not shown) due to a lower exposure of the cysteinyl-FMN adduct to O2.
Clearly, exposure to molecular oxygen facilitates adduct disruption and results in an oxidation of the FMN to reform the dark-adapted population. Although molecular oxygen is known to break thioether bonds to generate oxidized sulfoxide species (65, 66), no persistent degradation of the F434Y was observed in our measurements. This suggests that O2 catalyzes the adduct bond breakage, but does not incorporate directly into the FMN or the cysteine residue, which would essentially inactivate the protein photoactivity. Alternatively, the isoalloxazine ring of the FMN is established as an excellent electron acceptor capable of many interesting redox chemistries, which can involve one to two electron oxidation processes (67). Flavins can form transient adducts with molecular oxygen observed in many flavin-dependent flavoenzymes (68, 69), including oxidases and dehydrogenases. These new peroxy adducts may potentially displace the FMN-S bond, but also result in the incorporation of oxygen atoms into substrates species (e.g., cysteine in AsLOV2) like the chemistry of direct attack of the thioether bonds. It is unclear of the exact mechanism underlying the observed oxygen-catalyzed adduct breakage in F434Y, and is a topic of further study. It should also be noted that although the purging of O2 significantly slows the recovery kinetics, the observed timescales in purged F434Y samples are still considerably faster than observed in wild type AsLOV2 (Figure SI7), so other unresolved factors (e.g., acid/base catalysis by Y434) must also contribute to the faster photocycle recovery.
Concluding Comments
LOV domains are of particular interest at present for their suitability both for basic science and engineering applications, many of which are based on approaches to confer photosensitivity to target proteins via genetic fusion of LOV domains onto exogenous targets (5). While LOV domains have several natural advantages compared to other sensory domains for such engineering, including their small size and use of common FMN and FAD chromophores, certain properties may not always be ideal for each application. As such, versions of AsLOV2 and other LOV domains containing point mutations which confer “improved” characteristics (e.g. dynamic range) (70) are valuable both as research reagents and vehicles to test mechanistic hypotheses. Unfortunately, identifying new mutations with beneficial properties is challenging – rational design is slow, and genetic selection strategies are highly dependent on assay conditions to identify a novel phenotype. Our work demonstrates the merits of a combined computational/experimental approach to identify a novel mutant in a residue outside the chromophore-binding pocket to generate an AsLOV2 variant with new photochemical properties. Future use of such an approach, perhaps accelerated through the use of coarse-grained computational and rapid experimental methods, will be essential for identifying additional mutations useful for biophysical study and applications.
Supplementary Material
Acknowledgments
We are thankful to Kimberly Shahi for technical assistance and Radio Paradise for auditory stimulation. The Lagarias Lab (UCD) is acknowledged for use of their UV-VIS instrumental setup.
Abbreviations
- LOV
light, oxygen, and voltage
- AsLOV2
second LOV domain of Avena sativa phototropin 1
- BLUF
Blue Light sensors Utilizing Flavin
- FAD
flavin adenine dinucleotide
- FMN
flavin mononucleotide
- PYP
photoactive yellow protein
- ISC
intersystem crossing
- MD
Molecular dynamics
- CrLOV1
Chlamydomonas reinhardtii light, oxygen, and voltage domain one
- GSB
ground-state bleach
- SE
stimulated emission
- ESA
excited-state absorption
- SADS
Species Associated Difference Spectrum
- FEP
free energy perturbation
Footnotes
This work was supported with a Career Development Award (CDA0016/2007-C) from the Human Frontiers Science Organization (DSL); NIH grants P41-RR05969 (KS), MCB0744057 (KS) R01 GM081875 (KHG), T32 GM008297 (AN); LRAC MCA93S028 (KS); and grant I-1424 from the Robert A. Welch Foundation (KHG).
Supporting Information Available: Details of the following topics are available free of charge via the Internet at http://pubs.acs.org: (1) Sample expression and purification, (2) Molecular Dynamics simulation methods, (3) MD simulations of GLN513 dynamics, (4) Solution NMR signals and (5) chemical shift difference maps, (6) Time-resolved methods, (7) global analysis details underlying dispersed transient data and (8) adduct recovery signals.
References
- 1.Briggs WR, Spudich JL. Handbook of Photosensory Receptors. Wiley-VCH Verlag GmbH & Co; KGaA: 2005. [Google Scholar]
- 2.Hoersch D, Otto H, Cusanovich MA, Heyn MP. Time-resolved spectroscopy of dye-labeled photoactive yellow protein suggests a pathway of light-induced structural changes in the N-terminal cap. Phys Chem Chem Phys. 2009;11:5437–5444. doi: 10.1039/b821345c. [DOI] [PubMed] [Google Scholar]
- 3.Moglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moglich A, Yang X, Ayers RA, Moffat K. Structure and function of plant photoreceptors. Annu Rev Plant Biol. 2010;61:21–47. doi: 10.1146/annurev-arplant-042809-112259. [DOI] [PubMed] [Google Scholar]
- 5.Zoltowski BD, Gardner KH. Tripping the Light Fantastic: Blue-Light Photoreceptors as Examples of Environmentally Modulated Protein-Protein Interactions. Biochemistry. 2011;50:4–16. doi: 10.1021/bi101665s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Horst MA, Hellingwerf KJ. Photoreceptor proteins, “star actors of modern times”: a review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc Chem Res. 2004;37:13–20. doi: 10.1021/ar020219d. [DOI] [PubMed] [Google Scholar]
- 7.Losi A. Flavin-based Blue-Light photosensors: a photobiophysics update. Photochem Photobiol. 2007;83:1283–1300. doi: 10.1111/j.1751-1097.2007.00196.x. [DOI] [PubMed] [Google Scholar]
- 8.Rockwell N, YSS, Lagarias J. Phytochrome structure and signaling mechanisms. Ann Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuda S, Bauer CE. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell. 2002;110:613–623. doi: 10.1016/s0092-8674(02)00876-0. [DOI] [PubMed] [Google Scholar]
- 10.Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, Dutton PL, Cashmore AR. Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science. 1995;269:968–970. doi: 10.1126/science.7638620. [DOI] [PubMed] [Google Scholar]
- 11.Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR. Arabidopsis NPH1: A protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 12.Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci U S A. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briggs WR, Christie JM. Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci. 2002;7:204–210. doi: 10.1016/s1360-1385(02)02245-8. [DOI] [PubMed] [Google Scholar]
- 14.Larsen DS, van Stokkum IH, Vengris M, van Der Horst MA, de Weerd FL, Hellingwerf KJ, van Grondelle R. Incoherent manipulation of the photoactive yellow protein photocycle with dispersed pump-dump-probe spectroscopy. Biophys J. 2004;87:1858–1872. doi: 10.1529/biophysj.104.043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuda M, Nelson B, Chang CH, Govindjee R, Ebrey TG. Characterization of the chromophore of the third rhodopsin-like pigment of Halobacterium halobium and its photoproduct. Biophys J. 1985;47:721–724. doi: 10.1016/S0006-3495(85)83969-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadori H, Yoshihara K, Tokutomi S. Primary process of phytochrome-initial step of photomorphogenesis in green plants. J Am Chem Soc. 1992;114:10958–10959. [Google Scholar]
- 17.Andel F, Hasson KC, Gai F, Anfinrud PA, Mathies RA. Femtosecond time-resolved spectroscopy of the primary photochemistry of phytochrome. Biospec. 1997;3:421–433. [Google Scholar]
- 18.Muller MG, Lindner I, Martin I, Gartner W, Holzwarth AR. Femtosecond kinetics of photoconversion of the higher plant photoreceptor phytochrome carrying native and modified chromophores. Biophys J. 2008;94:4370–4382. doi: 10.1529/biophysj.106.091652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swartz TE, Corchnoy SB, Christie JM, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J Biol Chem. 2001;276:36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- 20.Crosson S, Moffat K. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell. 2002;14:1067–1075. doi: 10.1105/tpc.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schleicher E, Kowalczyk RM, Kay CW, Hegemann P, Bacher A, Fischer M, Bittl R, Richter G, Weber S. On the reaction mechanism of adduct formation in LOV domains of the plant blue-light receptor phototropin. J Am Chem Soc. 2004;126:11067–11076. doi: 10.1021/ja049553q. [DOI] [PubMed] [Google Scholar]
- 22.Alexandre MTA, Domratcheva T, Bonetti C, van Wilderen L, van Grondelle R, Groot ML, Hellingwerf KJ, Kennis JTM. Primary Reactions of the LOV2 Domain of Phototropin Studied with Ultrafast Mid-infrared Spectroscopy and Quantum Chemistry. Biophys J. 2009;97:227–237. doi: 10.1016/j.bpj.2009.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 24.Harper SM, Christie JM, Gardner KH. Disruption of the LOV-Jalpha helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 25.Kottke T, Heberle J, Hehn D, Dick B, Hegemann P. Phot-LOV1: photocycle of a blue-light receptor domain from the green alga Chlamydomonas reinhardtii. Biophys J. 2003;84:1192–1201. doi: 10.1016/S0006-3495(03)74933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Losi A, Polverini E, Quest B, Gartner W. First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys J. 2002;82:2627–2634. doi: 10.1016/S0006-3495(02)75604-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kennis JT, Crosson S, Gauden M, van Stokkum IH, Moffat K, van Grondelle R. Primary reactions of the LOV2 domain of phototropin, a plant blue-light photoreceptor. Biochemistry. 2003;42:3385–3392. doi: 10.1021/bi034022k. [DOI] [PubMed] [Google Scholar]
- 28.Crosson S, Moffat K. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc Natl Acad Sci U S A. 2001;98:2995–3000. doi: 10.1073/pnas.051520298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halavaty AS, Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochem. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 30.Nash AI, Ko WH, Harper SM, Gardner KH. A conserved glutamine plays a central role in LOV domain signal transmission and its duration. Biochem. 2008;47:13842–13849. doi: 10.1021/bi801430e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christie JM, Corchnoy SB, Swartz TE, Hokenson M, Han IS, Briggs WR, Bogomolni RA. Steric interactions stabilize the signaling state of the LOV2 domain of phototropin 1. Biochem. 2007;46:9310–9319. doi: 10.1021/bi700852w. [DOI] [PubMed] [Google Scholar]
- 32.Zoltowski BD, Vaccaro B, Crane BR. Mechanism-based tuning of a LOV domain photoreceptor. Nat Chem Biol. 2009;5:827–834. doi: 10.1038/nchembio.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freddolino PL, Nash AI, Harper SM, Gardner KH, Schulten K. Photoreaction-induced conformational changes in A. sativa Phot1-LOV2: A molecular dynamics study. manuscript in preparation. [Google Scholar]
- 34.Freddolino PL, Dittrich M, Schulten K. Dynamic switching mechanisms in LOV1 and LOV2 domains of plant phototropins. Biophys J. 2006;91:3630–3639. doi: 10.1529/biophysj.106.088609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dittrich M, Freddolino PL, Schulten K. When light falls in LOV: a quantum mechanical/molecular mechanical study of photoexcitation in Phot-LOV1 of Chlamydomonas reinhardtii. J Phys Chem B. 2005;109:13006–13013. doi: 10.1021/jp050943o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nozaki D, Iwata T, Ishikawa T, Todo T, Tokutomi S, Kandori H. Role of Gln1029 in the photoactivation processes of the LOV2 domain in adiantum phytochrome3. Biochem. 2004;43:8373–8379. doi: 10.1021/bi0494727. [DOI] [PubMed] [Google Scholar]
- 37.Fedorov R, Schlichting I, Hartmann E, Domratcheva T, Fuhrmann M, Hegemann P. Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii. Biophys J. 2003;84:2474–2482. doi: 10.1016/S0006-3495(03)75052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Losi A, Ghiraldelli E, Jansen S, Gartner W. Mutational effects on protein structural changes and interdomain interactions in the blue-light sensing LOV protein YtvA. Photochem Photobiol. 2005;81:1145–1152. doi: 10.1562/2005-05-25-RA-541. [DOI] [PubMed] [Google Scholar]
- 39.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 40.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexandre MT, Arents JC, van Grondelle R, Hellingwerf KJ, Kennis JT. A base-catalyzed mechanism for dark state recovery in the Avena sativa phototropin-1 LOV2 domain. Biochem. 2007;46:3129–3137. doi: 10.1021/bi062074e. [DOI] [PubMed] [Google Scholar]
- 42.Iwata T, Yamamoto A, Tokutomi S, Kandori H. Hydration and temperature similarly affect light-induced protein structural changes in the chromophoric domain of phototropin. Biochem. 2007;46:7016–7021. doi: 10.1021/bi7003087. [DOI] [PubMed] [Google Scholar]
- 43.Laan W, Bednarz T, Heberle J, Hellingwerf KJ. Chromophore composition of a heterologously expressed BLUF-domain. Photoche Photobio Sci. 2004;3:1011–1016. doi: 10.1039/b410923f. [DOI] [PubMed] [Google Scholar]
- 44.Farmer BT, Constantine KL, Goldfarb V, Friedrichs MS, Wittekind M, Yanchunas J, Robertson JG, Mueller L. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nat Struct Biol. 1996;3:995–997. doi: 10.1038/nsb1296-995. [DOI] [PubMed] [Google Scholar]
- 45.Mueller F. Chemistry and biochemistry of flavoenzymes. Boca Raton: CRC Press; 1991. [Google Scholar]
- 46.Gauden M, van Stokkum IHM, Key JM, Luhrs DC, Van Grondelle R, Hegemann P, Kennis JTM. Hydrogen-bond switching through a radical pair mechanism in a flavin-binding photoreceptor. Natl Acad Sci USA. 2006;103:10895–10900. doi: 10.1073/pnas.0600720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolf MMN, Schumann C, Gross R, Domratcheva T, Diller R. Ultrafast Infrared Spectroscopy of Riboflavin: Dynamics, Electronic Structure, and Vibrational Mode Analysis. J Phys Chem B. 2008;112:13424–13432. doi: 10.1021/jp804231c. [DOI] [PubMed] [Google Scholar]
- 48.Stanley RJ, MacFarlane AW. Ultrafast excited state dynamics of oxidized flavins: Direct observations of quenching by purines. J Phys Chem A. 2000;104:6899–6906. [Google Scholar]
- 49.Heelis PF, Parsons BJ, Phillips GO, Mckellar JF. The Flavin Sensitized Photo-Oxidation of Ascorbic-Acid - a Continuous and Flash-Photolysis Study. Photochem Photobio. 1981;33:7–13. [Google Scholar]
- 50.Holzer W, Penzkofer A, Fuhrmann M, Hegemann P. Spectroscopic characterization of flavin mononucleotide bound to the LOV1 domain of Phot1 from Chlamydomonas reinhardtii. Photochem Photobiol. 2002;75:479–487. doi: 10.1562/0031-8655(2002)075<0479:scofmb>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 51.Salzmann S, Silva MR, Thiel W, Marian CM. Influence of the LOV Domain on Low-Lying Excited States of Flavin: A Combined Quantum-Mechanics/Molecular-Mechanics Investigation. J Phys Chem B. 2009;113:15610–15618. doi: 10.1021/jp905599k. [DOI] [PubMed] [Google Scholar]
- 52.Berberan-Santos MN. External heavy-atom effect on fluorescence kinetics. Phys Chem Comm. 2000;5:18–23. [Google Scholar]
- 53.Chandra AK, Turro NJ, Lyons AL, Stone P. Intra-Molecular External Heavy-Atom Effect in Bromonorbornenes, Benzonorbornenes And Naphthonorbornenes. J Am Chem Soc. 1978;100:4964–4968. [Google Scholar]
- 54.Alexandre MTA, van Grondelle R, Hellingwerf KJ, Robert B, Kennis JTM. Perturbation of the ground-state electronic structure of FMN by the conserved cysteine in phototropin LOV2 domains. Phys Chem Chem Phys. 2008;10:6693–6702. doi: 10.1039/b810040c. [DOI] [PubMed] [Google Scholar]
- 55.Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Autrey T, Devadoss C, Sauerwein B, Franz JA, Schuster GB. Solvent Cage Recombination of 4-Benzoylphenylthiyl Radicals - Fast Intersystem Crossing of Triplet Sulfur-Centered Radical Pairs. J Phys Chem. 1995;99:869–871. [Google Scholar]
- 57.van Stokkum IH, Larsen DS, van Grondelle R. Global and target analysis of time-resolved spectra. Biochim Biophys Acta 1657. 2004:82–104. doi: 10.1016/j.bbabio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 58.Holzwarth AR. Biophysical Techniques in Photosynthesis. Dordrecht, The Netherlands. 1996;Chapter 5:75–92. [Google Scholar]
- 59.Zagrovic B, Pande V. Solvent viscosity dependence of the folding rate of a small protein: Distributed computing study. J Comp Chem. 2003;24:1432–1436. doi: 10.1002/jcc.10297. [DOI] [PubMed] [Google Scholar]
- 60.Zirak P, Penzkofer A, Mathes T, Hegemann P. Photo-dynamics of roseoflavin and riboflavin in aqueous and organic solvents. Chem Phys. 2009;358:111–122. [Google Scholar]
- 61.Alexandre MTA, van Grondelle R, Hellingwerf KJ, Kennis JTM. Conformational Heterogeneity and Propagation of Structural Changes in the LOV2/J alpha Domain from Avena sativa Phototropin 1 as Recorded by Temperature-Dependent FTIR Spectroscopy. Biophys J. 2009;97:238–247. doi: 10.1016/j.bpj.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bonetti C, Mathes T, van Stokkum IHM, Mullen KM, Groot ML, van Grondelle R, Hegemann P, Kennis JTM. Hydrogen Bond Switching among Flavin and Amino Acid Side Chains in the BLUF Photoreceptor Observed by Ultrafast Infrared Spectroscopy. Biophys J. 2008;95:4790–4802. doi: 10.1529/biophysj.108.139246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Corchnoy SB, Swartz TE, Szundi I, Lewis JW, Briggs WR, Bogomolni RA. Proton transfers in the photocycle of the LOV2 domain of phototropin 1. Biophys J. 2003;84:398A–398A. doi: 10.1074/jbc.M209119200. [DOI] [PubMed] [Google Scholar]
- 64.Cohen J, Arkhipov A, Braun R, Schulten K. Imaging the migration pathways for O-2, CO, NO, and Xe inside myoglobin. Biophys J. 2006;91:1844–1857. doi: 10.1529/biophysj.106.085746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Foote CS, Peters JW. Chemistry of singlet oxygen: Reactive Intermediate in Sulfide Photooxidation. J Am Chem Soc. 1971;93:3795. [Google Scholar]
- 66.Schoneich C, Aced A, Asmus KD. Mechanism Of Oxidation Of Aliphatic Thioethers To Sulfoxides By Hydroxyl Radicals - The Importance Of Molecular-Oxygen. J Am Chem Soc. 1993;115:11376–11383. [Google Scholar]
- 67.Massey V. The chemical and biological versatility of riboflavin. Biochem Soc Trans. 2000;28:283–296. [PubMed] [Google Scholar]
- 68.Chlumsky LJ, Sturgess AW, Nieves E, Jorns MS. Identification of the covalent flavin attachment site in sarcosine oxidase. Biochem. 1998;37:2089–2095. doi: 10.1021/bi972705s. [DOI] [PubMed] [Google Scholar]
- 69.Steenkamp DJ, Kenney WC, Singer TP. Novel Type Of Covalently Bound Coenzyme In Trimethylamine Dehydrogenase. J Bio Chem. 1978;253:2812–2817. [PubMed] [Google Scholar]
- 70.Strickland D, Yao XL, Gawlak G, Rosen MK, Gardner KH, Sosnick TR. Rationally improving LOV domain-based photoswitches. Nat Meth. 2010;7:623–U618. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.