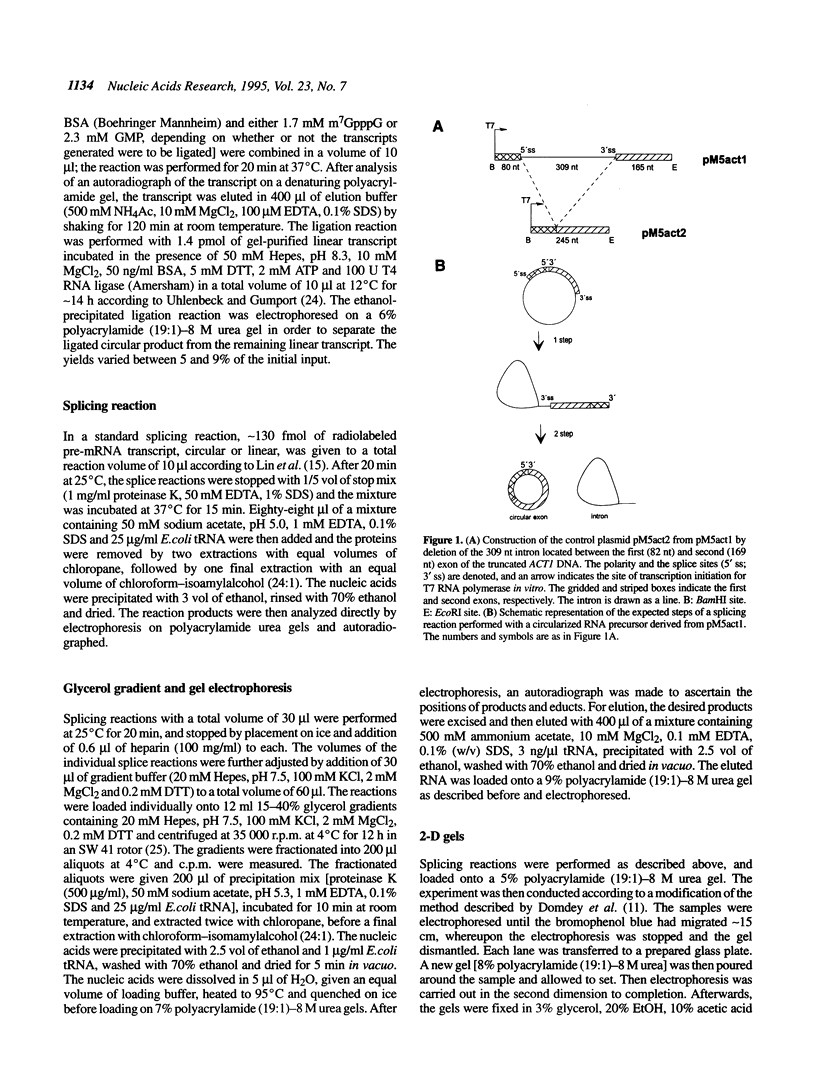
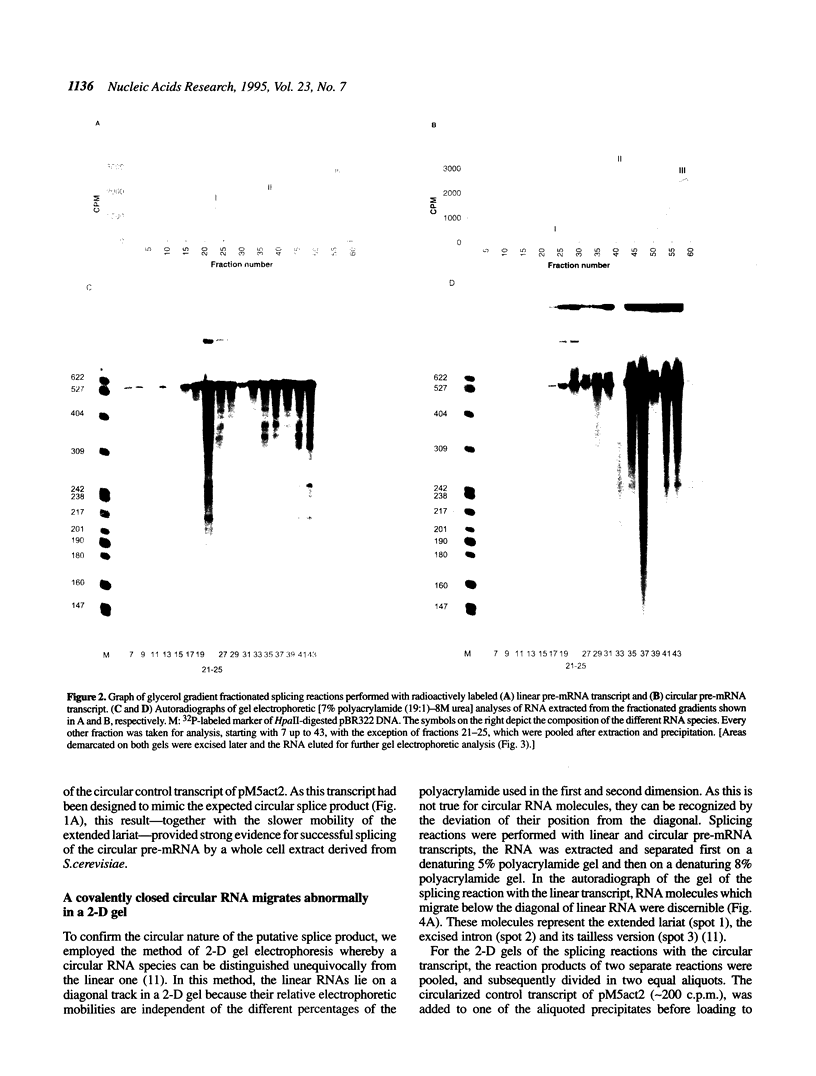
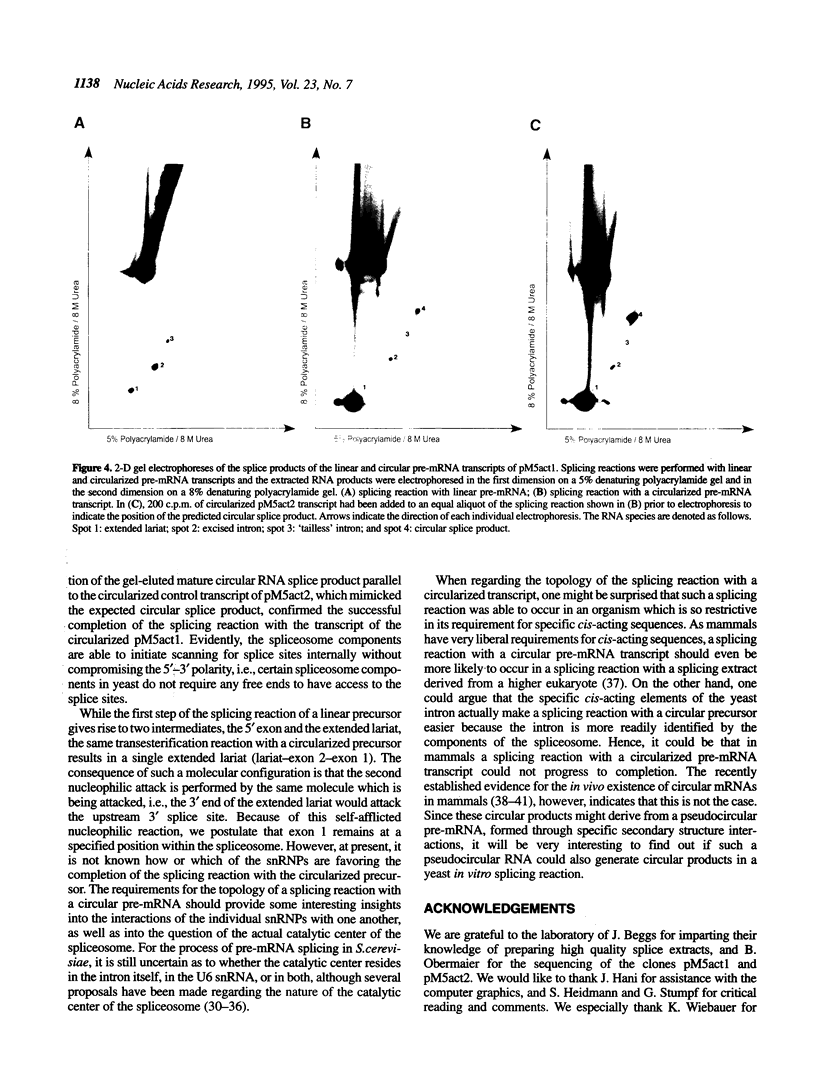
Abstract
We have tested the fate of a circularized synthetic pre-mRNA transcript in a whole cell splicing extract of Saccharomyces cerevisiae. Our results demonstrate that this circular precursor RNA is able to induce spliceosome formation in vitro and that the products of the following splicing reaction are the lariat-shaped intron, and a mature circular mRNA. Thus, it would appear that free 5' and/or 3' ends are not obligatory for a splicing reaction to occur, although we find its efficiency to be strongly influenced by the presence or lack of free ends. To our knowledge, this is the first demonstration that a circular pre-mRNA molecule is recognized as a suitable substrate by an eukaryotic mRNA splicing apparatus.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brody E., Abelson J. The "spliceosome": yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985 May 24;228(4702):963–967. doi: 10.1126/science.3890181. [DOI] [PubMed] [Google Scholar]
- Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993 Jun 4;73(5):1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- Cocquerelle C., Daubersies P., Majérus M. A., Kerckaert J. P., Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992 Mar;11(3):1095–1098. doi: 10.1002/j.1460-2075.1992.tb05148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerelle C., Mascrez B., Hétuin D., Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993 Jan;7(1):155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- Domdey H., Apostol B., Lin R. J., Newman A., Brody E., Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984 Dec;39(3 Pt 2):611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- Duchêne M., Löw A., Schweizer A., Domdey H. Molecular consequences of truncations of the first exon for in vitro splicing of yeast actin pre-mRNA. Nucleic Acids Res. 1988 Aug 11;16(15):7233–7239. doi: 10.1093/nar/16.15.7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery I., Sonenberg N. Cap-dependent RNA splicing in a HeLa nuclear extract. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7590–7594. doi: 10.1073/pnas.82.22.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E., Ares M., Jr Synthesis of circular RNA in bacteria and yeast using RNA cyclase ribozymes derived from a group I intron of phage T4. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3117–3121. doi: 10.1073/pnas.91.8.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz H. J., Hohlmaier J., Kramer W., Ohmayer A., Wippler J. Oligonucleotide-directed construction of mutations: a gapped duplex DNA procedure without enzymatic reactions in vitro. Nucleic Acids Res. 1988 Jul 25;16(14B):6987–6999. doi: 10.1093/nar/16.14.6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski P. J., Padgett R. A., Sharp P. A. Messenger RNA splicing in vitro: an excised intervening sequence and a potential intermediate. Cell. 1984 Jun;37(2):415–427. doi: 10.1016/0092-8674(84)90372-6. [DOI] [PubMed] [Google Scholar]
- Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991 Jul 12;253(5016):157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- Hodges P. E., Beggs J. D. RNA splicing. U2 fulfils a commitment. Curr Biol. 1994 Mar 1;4(3):264–267. doi: 10.1016/s0960-9822(00)00061-0. [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Lewis J., McGuigan C., Jankowska M., Darzynkiewicz E., Mattaj I. W. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994 Aug 26;78(4):657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- Kandels-Lewis S., Séraphin B. Involvement of U6 snRNA in 5' splice site selection. Science. 1993 Dec 24;262(5142):2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Lin R. J. Pre-mRNA splicing within an assembled yeast spliceosome requires an RNA-dependent ATPase and ATP hydrolysis. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):888–892. doi: 10.1073/pnas.90.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinz F. J., Gallwitz D. Size and position of intervening sequences are critical for the splicing efficiency of pre-mRNA in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1985 Jun 11;13(11):3791–3804. doi: 10.1093/nar/13.11.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Padgett R. A., Sharp P. A. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984 Oct;38(3):731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhrer K., Domdey H. Splicing and spliceosome formation of the yeast MATa1 transcript require a minimum distance from the 5' splice site to the internal branch acceptor site. Nucleic Acids Res. 1988 Oct 25;16(20):9457–9475. doi: 10.1093/nar/16.20.9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhrer K., Vogel K., Domdey H. A yeast tRNA precursor containing a pre-mRNA intron is spliced via the pre-mRNA splicing mechanism. EMBO J. 1990 Mar;9(3):705–709. doi: 10.1002/j.1460-2075.1990.tb08163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Lesser C. F., Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993 Dec 24;262(5142):1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- Lin R. J., Newman A. J., Cheng S. C., Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985 Nov 25;260(27):14780–14792. [PubMed] [Google Scholar]
- Lossky M., Anderson G. J., Jackson S. P., Beggs J. Identification of a yeast snRNP protein and detection of snRNP-snRNP interactions. Cell. 1987 Dec 24;51(6):1019–1026. doi: 10.1016/0092-8674(87)90588-5. [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992 Nov 27;71(5):803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Guthrie C. Randomization-selection analysis of snRNAs in vivo: evidence for a tertiary interaction in the spliceosome. Genes Dev. 1994 May 1;8(9):1071–1086. doi: 10.1101/gad.8.9.1071. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R., Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro J. M., Cho K. R., Fearon E. R., Kern S. E., Ruppert J. M., Oliner J. D., Kinzler K. W., Vogelstein B. Scrambled exons. Cell. 1991 Feb 8;64(3):607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- Parker R., Guthrie C. A point mutation in the conserved hexanucleotide at a yeast 5' splice junction uncouples recognition, cleavage, and ligation. Cell. 1985 May;41(1):107–118. doi: 10.1016/0092-8674(85)90065-0. [DOI] [PubMed] [Google Scholar]
- Pause A., Méthot N., Sonenberg N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol Cell Biol. 1993 Nov;13(11):6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikielny C. W., Rosbash M. mRNA splicing efficiency in yeast and the contribution of nonconserved sequences. Cell. 1985 May;41(1):119–126. doi: 10.1016/0092-8674(85)90066-2. [DOI] [PubMed] [Google Scholar]
- Plumpton M., McGarvey M., Beggs J. D. A dominant negative mutation in the conserved RNA helicase motif 'SAT' causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 1994 Feb 15;13(4):879–887. doi: 10.1002/j.1460-2075.1994.tb06331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Sawa H., Abelson J. Evidence for a base-pairing interaction between U6 small nuclear RNA and 5' splice site during the splicing reaction in yeast. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11269–11273. doi: 10.1073/pnas.89.23.11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer E. J., Steitz J. A. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993 Dec 24;262(5142):1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- Teigelkamp S., McGarvey M., Plumpton M., Beggs J. D. The splicing factor PRP2, a putative RNA helicase, interacts directly with pre-mRNA. EMBO J. 1994 Feb 15;13(4):888–897. doi: 10.1002/j.1460-2075.1994.tb06332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson-Jäger S., Domdey H. Yeast pre-mRNA splicing requires a minimum distance between the 5' splice site and the internal branch acceptor site. Mol Cell Biol. 1987 Nov;7(11):4010–4016. doi: 10.1128/mcb.7.11.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]