Abstract
The binding of transcription factors to specific DNA target sequences is the fundamental basis of gene regulatory networks. Chromatin immunoprecipitation combined with DNA tiling arrays or high-throughput sequencing—ChIP-chip and ChIP-seq—has produced many recent studies that detail the binding sites of various transcription factors. Surprisingly, data from a variety of model organisms and tissues have demonstrated that transcription factors vary greatly in their number of genomic binding sites, and that binding events can significantly exceed the number of known or possible direct gene targets. Thus, our current understanding of transcription factor function must expand to encompass what role, if any, binding might play outside of direct transcriptional target regulation. Here, we discuss the biological significance of genome-wide binding of transcription factors and present models that can account for this phenomenon.
Regulatory networks and the core model of gene regulation
The complex interactions between multiple transcription factors and gene targets across various tissues, cellular contexts, and time points are termed `transcriptional regulatory networks' (Box 1). It has been stated that a truly thorough understanding of such interactions should theoretically explain how an organism is `computed' from its DNA [1]. The core model of gene regulation posits that transcription factors recruit a polymerase complex to the transcriptional start site [2]. Transcription factors initiate this by binding at nearby or distant DNA sequences and directly interacting with components of the polymerase complex or with complexes that indirectly mediate the polymerase interaction. In eukaryotes, the latter may include chromatin remodelers or modifiers that facilitate access or increase protein-protein affinities via histone modifications [3,4]. The simplest view of the core model would suggest that factor binding directly correlates with transcriptional regulation. However, numerous examples of the separate regulation of factor binding and transcriptional activation suggest otherwise [5–7]. For example, recent studies indicate that the sequence of the DNA binding site can induce conformational changes in the bound transcription factor that permits transcriptional regulation by subsets of a transcription factor family that can bind to similar sites [8,9].
Defining the relationship between transcription factor binding and target regulation across the entire genome of various species has become an attainable goal with the recent explosion in advanced computing and information processing tools. These advances have resulted in some remarkable progress in reconstructing and predicting regulatory networks [10]. The advent of ChIP-chip (chromatin immunoprecipitation coupled to microarray hybridization) and ChIP-seq (chromatin immunoprecipitation coupled to high-throughput sequencing) have now allowed for determination of the precise, genome-wide distribution of transcription factor binding sites. The results of numerous studies employing these techniques have been at times predictable and at other times surprising. While some studies have shown the expected correlation between factor binding and gene regulation, others have observed binding events that vastly exceed the number of expected gene targets (Table 1). Given these findings, it is timely to reconsider the relationship between transcription factors and gene regulation and the role, if any, that widespread transcription factor binding may play outside of direct gene target regulation.
Table 1.
Numbers of transcription factor bound sites from selected ChIP-chip and ChIP-seq experiments
| Transcription factor | Species | Technique | Reported numbers of bound sites | Reference |
|---|---|---|---|---|
| Ste12 | S. cerevisiae | ChIP-chip | 65/57a | [12] |
| PHA-4 | C. elegans | ChIP-seq | 4350/4808b | [15] |
| Twist | D. melanogaster | ChIP-chip | 2096 | [16] |
| Twist | D. melanogaster | ChIP-chip | 3000 | [21] |
| NRSF | Human | ChIP-seq | 1946 | [18] |
| TAL1 | Mouse | ChIP-seq | 2994 | [31] |
| TAL1 | Human | ChIP-seq | 6315 | [29] |
| PXR | Mouse | ChIP-seq | 3812/6446c | [19] |
| CaRF | Mouse | ChIP-seq | 176 | [20] |
| STAT1 | Human | ChIP-seq | 11004/41582d | [27] |
| GATA1 | Mouse | ChIP-seq | 15360 | [28] |
| CTCF | Human | ChIP-chip | 13804 | [48] |
| CTCF | Human | ChIP-seq | 20262 | [49] |
| CTCF | Mouse | ChIP-seq | 39609 | [38] |
| MyoD | Mouse | ChIP-seq | 25956/59267e | [34] |
Binding sites are those specifically identified in either mating or filamentous growth conditions, respectively,
Binding sites are in embryos, and L1 larvae, respectively,
Binding sites are listed for basal conditions, and conditions in which a synthetic activator of PXR was used, respectively,
Binding sites are listed for conditions of non-stimulated and interferon-γ-stimulated cells, respectively,
Binding sites are listed at two different statistical cutoffs (false discovery rates of 10−7 and 0.018, respectively)
Transcription Factor Binding and Direct Gene Regulation
Several genome-wide transcription factor binding studies in various model organisms have supported a relatively direct connection between factor binding and gene regulation. One of the first genome-wide assessments of transcription factor binding in yeast reported transcription factor binding in promoter regions, in spite of the presence of binding motifs in both coding and intergenic regions [11]. Another report evaluating over 100 tagged factors in yeast identified more than 4,000 promoter-transcription factor interactions and described numerous regulatory circuits [10]. The subset of circuits that comprised feed-forward networks (Figure 1a) alone was extensive, involving 39 factors, 49 distinct networks, and greater than 10% of all bound areas. This study emphasized both the importance of regulatory networks in controlling gene expression, as well as the ability of ChIP studies to uncover such networks.
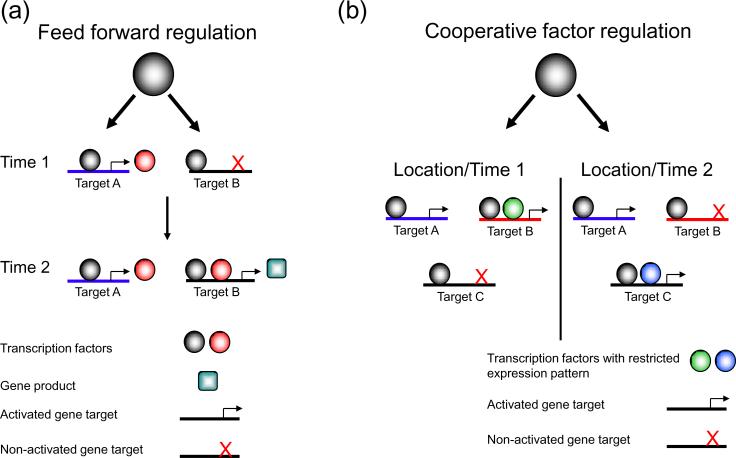
Figure 1. Examples of regulatory motifs used to control transcription.
A variety of mechanisms, or regulatory motifs, are used to control the expression of specific gene targets over unique spacial (eg. specific tissue types) and/or temporal contexts. (A) Feed-forward regulation permits temporal control of the targets of a single transcription factor. A transcription factor, represented by the grey circle, binds to multiple DNA targets (blue and black targets), but only activates one of them (Time 1). The gene target that it activates (red circle) can then also bind to one of the same gene targets as the original factor (black), and together they activate transcription (Time 2). (B) The use of cooperative factors permits transcription factors to be expressed widely, but discriminately activate gene targets. A single transcription factor, again represented by the grey circle, binds to multiple gene targets, activating one (the blue line) consistently, regardless of the cellular context (either tissue type or time). Other targets that it binds to in both cases (black and red targets), are activated only if they are also bound by another factor (compare activation of black and red targets between location/time 1 and 2), expressed specifically in that cellular condition.
A later study looking at an individual transcription factor in yeast, with roles in both filamentous growth and mating behavior, also found that DNA binding tightly correlated with function. Under cellular conditions that activated either growth or mating functions individually, Ste12 was found to occupy approximately 60 unique binding sites that were located in the promoters of genes with appropriate corresponding functions [12]. This binding was noted to be dependent on another transcription factor for the process of filamentation, an example of the importance of cooperative factor binding (Figure 1b) in mediating transcription factor activity.
The forkhead box A homolog PHA-4 regulates organogenesis of the pharynx in Caenohabditis elegans, and provides an example of factor binding correlating closely with direct gene target effects in a multicellular organism. Initial studies demonstrated that expression of its targets correlated with PHA-4 binding sites in promoter regions, and that the timing of target expression correlated with binding affinity between transcription factor and its target sequence [13]. Follow-up studies refined this model, providing evidence for other factors that cooperated with PHA-4 binding to modulate timing of target expression [14]. Taken together, the data suggested that pharyngeal organ development is regulated by a combination of PHA-4 binding affinity and cooperating factors to temporally regulate gene expression. It also suggested that it should be possible to predict the time of expression of a putative PHA-4 target gene solely from analysis of its DNA sequence.
Recent ChIP-seq data for PHA-4 has been in agreement with this assessment. The great majority (>90%) of the bound sites identified in either embryos or larvae can be designated as `gene-associated' using a distance cutoff of 2 kb or less between a bound site and nearest gene [15]. Overlapping the binding with gene expression data (high-throughput sequencing of RNA), most (87%) of the associated genes were expressed when PHA-4 binding was present, and a decrease in factor binding was associated with a reduction in expression for most (60%) presumptive targets, suggesting that binding of the factor activated the expression of those genes.
Studies in Drosophila melanogaster have identified the importance of cis-regulatory modules (CRMs), which are short DNA sequences (~300–500 nucleotides in length) that integrate multiple input signals to control gene expression. For example, the binding of Mef2, an important factor in mesodermal development, changes temporally during the course of muscle development [16]. At the time points evaluated, different factor motifs were noted at Mef2 binding regions, suggesting a cooperative factor mechanism used to temporally regulate the expression of various Mef2 targets. Further complexity in regulation is also suggested by a study comparing the binding profiles of Mef2 and lameduck (Lmd) [17]. Mutants of Mef2 and Lmd show a similar defect in myoblast fusion, suggesting similar or overlapping biological roles; however, while their DNA binding profiles overlap significantly, the effect of binding is widely variable. Depending on the enhancer target, co-binding can lead to additive, synergistic, or repressive effects, as demonstrated in reporter assays using eight different characterized enhancers. For example, co-expression of Lmd and Mef2 activates the blow enhancer while expression of Lmd counteracts the positive effect of Mef2 on the CG9416 enhancer. While these results reveal the potential complexity of regulatory networks, a relatively direct relationship can still be inferred between DNA binding and target gene effects.
The close relationship between DNA binding and gene target effect has also been observed in mammalian systems. In one of the first studies to use ChIP-seq, the binding of the zinc-finger protein neuron-restrictive silencer factor (NRSF) was mapped to only ~2000 sites in the human genome [18]. It was found that a few hundred potential target genes showed relatively `low' gene expression compared to average cellular transcript expression when a NRSF peak was located nearby (≤1 kb), suggesting that NRSF was exerting its transcriptionally repressive effects at those genes when bound nearby. Studies of other factors, such as pregnane X receptor (PXR) [19] and calcium-response factor (CaRF) [20], have also demonstrated a direct correlation of factor binding with gene regulation in mammalian cells.
Transcription Factor Binding in Excess of Known Direct Targets
In contrast to the model of direct gene regulation, several studies have demonstrated transcription factor binding at a large number of sites, many of which cannot be clearly connected with target gene regulation. In Drosophila, several ChIP-chip studies using whole genome tiling arrays have been performed for developmental transcription factors [21,22]. These studies have identified a large number of binding regions, on the order of several thousands, for individual factors in the developing embryo, indicating a greater amount of DNA binding by developmental factors than had been anticipated. For example, over 2,000 binding regions were observed for Twist in the Drosophila genome in two separate studies utilizing distinct microarray designs [21,23], vastly exceeding the number of known Twist targets and including many intronic and intergenic sites. Also unexpectedly, Twist binding overlaps significantly with both Dorsal and Snail binding sites, and many of these sites possess highly conserved motifs. Their conservation suggests they are likely to be functional sites, but their significance is still unclear.
While widespread binding of early developmental transcription factors is perhaps not entirely surprising [24], the unexpected finding has been the identification of numerous binding sites of unclear function, including for other factors as well. Studies of the binding and gene regulation of Myc and other proteins of the dMax family in Drosophila and human cells have shown extensive binding across the genome, but that binding did not necessarily correlate with transcriptional regulation of the nearby target genes [25,26].
In an early ChIP-seq study examining the interferon-γ (IFN-γ) responsive transcription factor STAT1 in human cells, a strikingly large number of bound sites was observed [27]. In unstimulated cells, over 10,000 binding sites were identified, and this increased more than four-fold after stimulation with IFN-γ. In both conditions, approximately 50% of the total sites were intragenic and 25% intergenic. While there was a strong overlap with sites of known STAT1 activity, the majority of binding sites were not located adjacent to STAT1 regulated genes, suggesting that many, or most, bound sites were not directly regulating a nearby gene target. The authors suggested that many of the STAT1 sites might correspond to weaker, less favored binding sites, or possibly functional sites with STAT1 bound in only a subset of the total cell population.
As another example of widespread binding, the hematopoietic factor GATA1 was reported to have over 15,000 DNA binding sites in a mouse erythroblast line [28]. GATA1-factor binding is apparently necessary for the binding of another hematopoietic factor, the basic helix-loop-helix (bHLH) factor TAL1, to an adjacent E-box motif, the consensus binding site for bHLH factors. There is a strong association of TAL1 binding with erythroid gene regulation [29–31], with over 2000 genes, most of which (90%) were categorized as related to erythroid development, having TAL1 binding within putative regulatory elements in one study, and over half of TAL1-regulated genes containing TAL1 bound within a proximal or distal regulatory element in another study [29]. In this case, the widespread binding of GATA1 might be identifying the sites that can be bound by TAL1, and possibly other factors at different times or in different cells, to execute cell-type specific programs of gene expression.
The myogenic bHLH factor MyoD is another transcription factor that offers potential insight into genome-wide binding. MyoD directly regulates genes expressed during skeletal muscle differentiation [32] and orchestrates a temporal pattern of gene expression through a feed-forward circuit [33]. ChIP-seq on MyoD in skeletal muscle cells identified approximately 30,000–60,000 MyoD binding sites [34]. As anticipated, genes regulated by MyoD during myogenesis had associated MyoD binding sites. However, almost 75% of all genes were associated with a MyoD binding site and about 25% of the MyoD sites were in intergenic regions. Therefore, the majority of MyoD binding events were not directly associated with gene regulation. Although regional transcription was not detected at these intergenic sites, MyoD binding was demonstrated to induce local chromatin modifications, specifically acetylation of histone H4 that is generally associated with active and/or accessible regions of the genome.
Together with the studies discussed above, these findings demonstrate that some transcription factors have binding events that are vastly in excess of the genes that they directly regulate. The remainder of this review will discuss the possible significance of these large number of transcription factor binding events that are not directly related to gene transcription. One proposed explanation for large-scale genome-wide transcription factor binding is the presence of `non-functional' binding sites that serve no biological purpose [22]. Alternatively, it has been proposed that transcription factors may bind to many low affinity sites in the genome and contribute to gene expression at levels that are low but sufficient to allow evolutionary conservation, an idea proposed from a large scale ChIP-chip study in yeast [35]. Presuming that these sites are functional, other possibilities include roles in affecting the functional concentration of factors, induction of chromatin looping, changing chromatin and nuclear structure, or the evolution of new transcriptional regulatory networks.
Site Accessibility Model
It has been suggested that binding sites occurring outside of areas directly involved in gene regulation may be `non-specific,' or random. However, these intergenic sites contain the factor-specific binding motifs and have been validated both experimentally and statistically, the latter by passing very strict statistical cutoffs [27,34]. Thus, it seems more appropriate to conclude that the observed genome-wide binding of some transcription factors is a biologically specific event; however, the biological role at many of the sites remains largely undetermined.
Based on the binding of the lac repressor to bacterial DNA, it was suggested that genome-wide binding at non-regulatory sites might function to maintain an optimum amount of available transcription factor in the nucleus [36]. In this model, some of the transcription factor binding sites that are located in intergenic regions or repetitive elements might serve that function, helping to fine-tune gene expression by limiting the concentration of unbound factors and preventing binding to sites that need to be regulated by co-factor occupancy and cooperative binding. In this model, the genome-wide binding serves as a reservoir for factors, sequestering them in a manner analogous to other biological buffering systems.
Some studies provide support for this model. For example, in the Drosophila studies that show binding at thousands of sites in the genome in addition to binding at regulated genes [22,37], higher-affinity binding occurred at regulated genes, and lower-affinity binding occurred in regions not regulated by the factors. This is consistent with the model that accessible DNA serves as a low-affinity reservoir for transcription factors and that these sites are not directly regulating regional gene transcription.
Other studies provide additional support for the notion that transcription factors will bind to any available sites genome-wide. ChIP-seq of 15 transcription factors and regulators involved in mouse embryonic stem (ES) cell biology demonstrated binding for multiple factors at the same 3,583 sites in both promoter and intergenic regions [38]. Similarly, in Drosophila several of the patterning factors exhibit notable overlap in their binding sites, although there is variability in the degree of overlap. And while analyses of binding site sequences demonstrate, in general, factor specificity for preferred DNA-binding motifs previously identified in vitro, many regions also exist which lack consensus binding motifs [22]. Therefore, some genome-wide binding might reflect factor interaction with accessible DNA regions that have not been specifically selected for a role in regional gene transcription.
Although likely correct in many instances, this model does not explain why there is an order of magnitude, or more, difference in genome-wide binding for factors with equivalently complex binding motifs. As noted above, MyoD has ~30,000–60,000 binding sites whereas TAL1 is reported to have ~3,000–6,000 sites in erythroid cells [29–31,34]. Both are bHLH factors that dimerize with an E-protein and recognize the core CANNTG E-box motif. The substantial difference in their genome-wide binding, however, suggests that sequence complexity is not the only determinant of binding. One possibility is that some factors are more constrained by site accessibility than others. MyoD can initiate chromatin remodeling at inaccessible sites and can bind independently of other factors, whereas the related bHLH factor Myogenin is more constrained to bind to accessible sites [33,34,39,40] and the TAL1 bHLH factor might require GATA1 or other factors to bind [29]. This suggests that the difference in the number of MyoD and TAL1 binding sites might, at least in part, reflect their relative ability to make new sites accessible for binding and to bind independently of other factors.
Chromosome Looping and Changes in Nuclear Architecture
Another, non-exclusive, model is that intergenic binding sites regulate gene transcription at a distance. Chromatin looping provides a mechanism for transcriptional control by bringing regulatory elements into proximity with target genes. Chromosome conformation capture studies indicate that the interaction of the distant locus control region (LCR) with the beta globin gene is required for high-level transcription. Interestingly, this interaction is dependent on GATA1 acting as an anchor [41]. Given that GATA1 binds to over 15,000 sites, it is plausible that some proportion of these may affect transcription by inducing chromatin loops. In agreement with this idea, the LCR is necessary for globin genes to associate with transcriptionally-engaged PolII sites [42], while other experiments demonstrated the association of hundreds of specific genomic loci with the murine globin genes in `transcription factories' [43]. In another specific example of chromatin looping leading to gene regulation, a Wnt-responsive enhancer downstream of the Myc gene has been shown to loop to cooperate with a 5' enhancer in a beta-catenin/TCF dependent fashion to regulate Myc expression [44]. These studies suggest that genome-wide binding might establish productive long-range interactions, either by looping to bring distant enhancers together with promoters, or in more complex interactions such as the co-regulation found in transcription factories.
Genome-wide Binding Affecting Global Chromatin and Nuclear Structure
As noted above, many of the MyoD binding events are not directly associated with regional gene transcription, but rather with regional histone modifications associated with active or accessible chromatin [34]. Genome-wide changes in chromatin also occur in response to Myc binding [45]. Therefore, a major biological role of these factors, and perhaps other genome-wide binding factors, might not be to directly regulate transcription, but rather to re-organize the chromatin to make regions generally more accessible for factors expressed later in development. Such a role is supported by several studies of genome-wide influence on chromatin structure of general regulatory factors in yeast [46–48].
Although it might seem unusual to suggest that some transcription factors have a role in regional chromatin organization at some sites and function as typical transcription factors at others, these represent two related functions of many transcription factors and it is reasonable to imagine that they can be deployed independently. For example, at genes transcriptionally regulated by MyoD, MyoD recruits histone acetyltransferases and chromatin remodeling complexes prior to mediating transcriptional initiation, which often occurs following the binding of an additional transcription factor [33,49,50]. Therefore, the initial steps of transcription factor-mediated chromatin modifications can be distinguished from subsequent steps of transcriptional activation.
The suggestion that some transcription factors might have a role in regional chromatin organization that is independent of regional transcription is reminiscent of CTCF, which was originally identified as a transcription factor and is now recognized to have a broad role in chromatin organization. CTCF has also been found to have tens of thousands of binding sites in human and mouse cells [38,51]. The greatest portion of CTCF sites was located in intergenic regions and many were at the border of distinct chromatin regions, consistent with a role in demarcating different chromatin domains [51,52]. Furthermore, CTCF binding sites were flanked by arrays of well-positioned nucleosomes enriched in specific histone types (H2A.Z) and specific histone modifications, suggesting additional roles in broad changes in chromatin composition and structure [53].
Related to the model that some transcription factors might influence chromatin on a global scale is the idea that some of these factors might contribute to other aspects of regional nuclear organization. Apart from its role in affecting chromatin structure, CTCF may also mediate long-range chromatin interactions [54,55]. Also, as previously noted, both MyoD and Myc mediate broad epigenetic reprogramming within the nucleus, and it is reasonable to speculate that this activity might alter nuclear architecture and be important for their biological function. The ability to study changes in nuclear organization has recently become more accessible through the development of techniques such as Hi-C [56], and it will be interesting to determine whether the major role of some transcription factors is to re-organize the architecture of the nucleus.
Selective advantage model to explain widespread binding
The relationship between the feed-forward network motif and the evolution of new transcriptional regulatory networks is another theoretical model for understanding a potential biological role for genome-wide binding. Feed-forward regulation is the dominant motif for regulating complex biological pathways, with the ability to temporally regulate the expression of its targets while retaining the ability to rapidly cease target expression [10,57,58]. Feed-forward circuits have been found to occur repeatedly in S. cerevisiae, and have arisen via convergent evolution, suggesting their widespread utility [59].
Genome-wide transcription factor binding and feed-forward mechanisms might have led to the evolution of distinct regulatory networks from a common network, a theory that can be understood using MyoD as an example. MyoD directly binds and regulates genes expressed throughout the program of skeletal myogenesis. At many targets, binding alone is not sufficient for transcriptional activation, but instead requires cooperation with factors that MyoD also regulates, thereby achieving temporal patterning through the feed-forward circuit. The evolution of a feed-forward circuit can be easily understood as the refinement of an initial single-input motif (Figure 2). For example, a primitive MyoD-like factor might have initially activated all the genes necessary for a primitive muscle cell phenotype, providing some selective advantage for this initial event. Subsequently, feed-forward regulation could be superimposed on the single-input motif to gradually improve and regulate the final output.
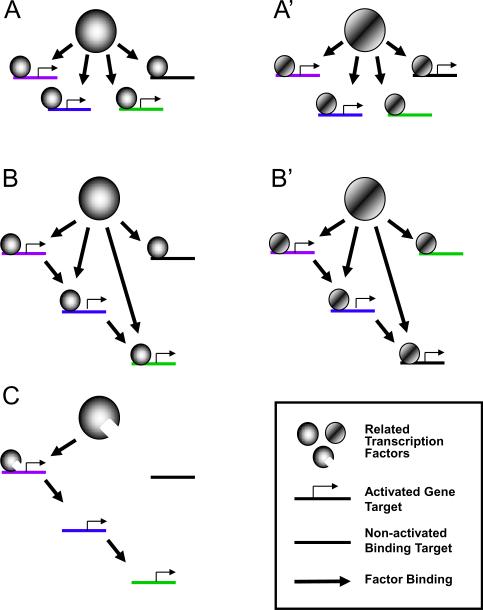
Figure 2. Genome-wide binding and the evolution of transcriptional networks.
The ability of certain transcription factors to bind widely throughout the genome could permit the evolution of new transcriptional regulatory networks in a relatively limited number of events. This could mean that genome-wide binding might actually serve an evolutionary advantage in cells, permitting them to more easily acquire new networks and phenotypes, as a result of the different genes involved in those networks. (A) Schematic representation of a transcription factor that binds to many sites throughout the genome and regulates transcription at a subset of these sites in a single input motif, in which it alone regulates the expression of the targets at which it binds. (A') Duplication and sequence divergence of this factor can give rise to a family member with similar DNA binding characteristics but transcriptional regulation of an overlapping yet distinct set of genes. The more promiscuous the binding of factor A and A', the greater the subset of genes they have the potential to influence and the greater potential for target diversity between A and A'. Therefore, changing from A to A' could lead to the generation of a new complex program by a single factor modification. (B and B') If the cellular phenotype conferred by the set of genes regulated in A and A' have some selective advantage, then the single input motif can be refined by the gradual super-imposition of a feed-forward motif to achieve temporal regulation and more robust kinetics. (C) It is also possible for feed-forward motifs to degenerate into simple cascades of regulated genes over time if subsequent mutations in the original factor limit the set of genes that can be directly bound, further separating the two networks that originally came from a common progenitor.
One prediction of this model is that factors with the potential to regulate complex transcriptional programs would bind throughout the genome because mutations in factors that sample a large portion of the genome would have the highest probability of generating a new network by changing the expression of large numbers of genes. Again using MyoD as an example, MyoD binds within a regulatory distance of more than one-half of all genes [34]. Altering the activation potential of MyoD through a translocation or mutation could drastically alter genome-wide transcription and potentially generate a novel complex phenotype from a single genetic event. In this model, genome-wide binding of a subset of transcription factors might reflect an evolutionary advantage rather than a cell-type specific function.
Concluding Remarks
Comparing the findings from genome-wide transcription factor binding studies supports two general types of transcription factor binding. In some studies, the transcription factors tend to bind in the neighborhood of genes that they regulate, whereas in others the factors bind throughout the genome and relatively equivalently at both regulated and apparently non-regulated genes. A major caveat in suggesting that these might represent different biological strategies is the problem inherent to comparing results from different studies. Differences in sample preparation, data acquisition, and data processing can result in dramatically different conclusions that do not directly reflect the biology of the factors studied. Having acknowledged this important caveat, some factors appear to have binding profiles that reflect their regulatory network. For these factors it should be possible to infer their function based on knowledge of their binding sites, and, ultimately, it might be possible to compute their regulatory networks directly from knowledge of the organism's DNA sequence. The binding profiles of other factors appear much too dispersed across the genome to accurately correlate binding with regional transcription. For these factors, it might be impossible to infer their regulatory networks from DNA sequence, or even from knowledge of where they are physically bound. It remains to be determined whether these genome-wide binding events have one or more biological functions that are distinct from regulating regional transcription. Although speculative, this raises the intriguing possibility that the majority of binding events of some transcription factors might not be the direct regulation of transcription, but rather a currently unrecognized role in genome-wide biology.
Box 1. Transcriptional Regulatory Networks.
Transcription factors interact in a sequence-specific fashion with DNA to either increase or decrease transcription of gene targets. Transcription factors often bind and regulate multiple targets simultaneously, and targets, in turn, are frequently regulated by multiple factors. Regulatory networks can be constructed to describe these interactions, and represent the interactions that occur at multiple factor-target levels. Networks can be comprised of various motifs, which represent the regulatory approaches taken by one or more factors at specific targets. Multiple types of motifs have been described, but two common ones include the feed-forward loop and multi-input motif (Figure 1). Using these and other commonly found motifs (eg. auto-regulatory loops in which a gene product downregulates its own production), transcription factors are able to establish complex and dynamic mechanisms of gene regulation.
Acknowledgements
S.J.T. was supported by NIH NIAMS R01AR045113. K.L.M. was supported by a Developmental Biology Predoctoral Training Grant T32HD007183 from the National Institute of Child Health and Human Development. A.P.F was supported by a grant from the University of Washington Child Health Research Center, NIH U5K12HD043376-08.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weintraub H. Summary: genetic tinkering--local problems, local solutions. Cold Spring Harb Symp Quant Biol. 1993;58:819–836. doi: 10.1101/sqb.1993.058.01.089. [DOI] [PubMed] [Google Scholar]
- 2.Ptashne M, Gann A. Genes & signals. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2002. p. xvi.p. 192. [Google Scholar]
- 3.Fry CJ, Peterson CL. Chromatin remodeling enzymes: who's on first? Curr Biol. 2001;11:R185–197. doi: 10.1016/s0960-9822(01)00090-2. [DOI] [PubMed] [Google Scholar]
- 4.Cosma MP. Ordered recruitment: gene-specific mechanism of transcription activation. Molecular Cell. 2002;10:227–236. doi: 10.1016/s1097-2765(02)00604-4. [DOI] [PubMed] [Google Scholar]
- 5.Guarente L, et al. Mutant lambda phage repressor with a specific defect in its positive control function. Proc Natl Acad Sci U S A. 1982;79:2236–2239. doi: 10.1073/pnas.79.7.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis RL, et al. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- 7.Turcotte B, Guarente L. HAP1 positive control mutants specific for one of two binding sites. Genes Dev. 1992;6:2001–2009. doi: 10.1101/gad.6.10.2001. [DOI] [PubMed] [Google Scholar]
- 8.Leung TH, et al. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 9.Meijsing SH, et al. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee TI, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 11.Lieb JD, et al. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 12.Zeitlinger J, et al. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell. 2003;113:395–404. doi: 10.1016/s0092-8674(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 13.Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- 14.Gaudet J, et al. Whole-genome analysis of temporal gene expression during foregut development. PLoS Biol. 2004;2:e352. doi: 10.1371/journal.pbio.0020352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong M, et al. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 2010;6:e1000848. doi: 10.1371/journal.pgen.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandmann T, et al. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev Cell. 2006;10:797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Cunha PM, et al. Combinatorial binding leads to diverse regulatory responses: Lmd is a tissue-specific modulator of Mef2 activity. PLoS Genet. 2010;6:e1001014. doi: 10.1371/journal.pgen.1001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson DS, et al. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 19.Cui JY, et al. ChIPing the cistrome of PXR in mouse liver. Nucleic Acids Research. 2010 doi: 10.1093/nar/gkq654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfenning AR, et al. Genome-wide identification of calcium-response factor (CaRF) binding sites predicts a role in regulation of neuronal signaling pathways. PLoS ONE. 2010;5:e10870. doi: 10.1371/journal.pone.0010870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeitlinger J, et al. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li XY, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandmann T, et al. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 2007;21:436–449. doi: 10.1101/gad.1509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang Z, Biggin MD. Eve and ftz regulate a wide array of genes in blastoderm embryos: the selector homeoproteins directly or indirectly regulate most genes in Drosophila. Development. 1998;125:4471–4482. doi: 10.1242/dev.125.22.4471. [DOI] [PubMed] [Google Scholar]
- 25.Orian A, et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes & Development. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez PC, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 28.Cheng Y, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19:2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palii C, et al. Differential genomic targeting of the transcription factor TAL1 in alternate hematopoietic lineages. EMBO J. doi: 10.1038/emboj.2010.342. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frankel N, et al. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kassouf MT, et al. Genome-wide identification of TAL1's functional targets: insights into its mechanisms of action in primary erythroid cells. Genome Res. 2010;20:1064–1083. doi: 10.1101/gr.104935.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergstrom DA, et al. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- 33.Penn BH, et al. A MyoD-generated feed-forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes & Development. 2004;18:2348–2353. doi: 10.1101/gad.1234304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao Y, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006;16:962–972. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin S, Riggs AD. The general affinity of lac repressor for E. coli DNA: implications for gene regulation in procaryotes and eucaryotes. Cell. 1975;4:107–111. doi: 10.1016/0092-8674(75)90116-6. [DOI] [PubMed] [Google Scholar]
- 37.MacArthur S, et al. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 2009;10:R80. doi: 10.1186/gb-2009-10-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 39.Bergstrom DA, Tapscott SJ. Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family. Molecular and Cellular Biology. 2001;21:2404–2412. doi: 10.1128/MCB.21.7.2404-2412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao Y, et al. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 2006;25:502–511. doi: 10.1038/sj.emboj.7600958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vakoc CR, et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 42.Ragoczy T, et al. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes & Development. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schoenfelder S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yochum GS, et al. A beta-catenin/TCF-coordinated chromatin loop at MYC integrates 5' and 3' Wnt responsive enhancers. Proc Natl Acad Sci USA. 2010;107:145–150. doi: 10.1073/pnas.0912294107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knoepfler PS, et al. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Badis G, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ganapathi M, et al. Extensive role of the general regulatory factors, Abf1 and Rap1, in determining genome-wide chromatin structure in budding yeast. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 50.Aziz A, et al. Regulating a master regulator: Establishing tissue-specific gene expression in skeletal muscle. Epigenetics. 2010;5:691–695. doi: 10.4161/epi.5.8.13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 53.Fu Y, et al. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hadjur S, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mishiro T, et al. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cordero OX, Hogeweg P. Feed-forward loop circuits as a side effect of genome evolution. Mol Biol Evol. 2006;23:1931–1936. doi: 10.1093/molbev/msl060. [DOI] [PubMed] [Google Scholar]
- 58.Shen-Orr SS, et al. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 59.Conant GC, Wagner A. Convergent evolution of gene circuits. Nat Genet. 2003;34:264–266. doi: 10.1038/ng1181. [DOI] [PubMed] [Google Scholar]