Abstract
Background and Aims
The combination of pegylated interferon (IFN) α and ribavirin (RBV) is standard therapy for patients with chronic HCV infection. However, it produces a sustained virologic response (SVR) in only half of treated individuals and is associated with significant side effects. Recently several single-nucleotide polymorphisms (SNPs) near the IL28B locus, also known as IFNλ3, were identified to be strong predictors of SVR in patients receiving PEG-IFN and RBV. We sought to determine whether IL28B was capable of inhibiting HCV replication and to determine the pathway by which IL28B exhibits anti-HCV activity.
Methods
Using the full-length HCV replicon OR6 and the infectious HCV clones JFH1 and Jc1, we assessed the anti-HCV effect of IL28B on HCV and characterized the key steps of the JAK-STAT pathway by real time PCR, luciferase assay, and Western blot. Finally, we evaluated the anti-HCV effect of IL28B in the presence of JAK-STAT pathway inhibitors such as blocking antibodies, a pharmacological inhibitor and siRNAs.
Results
We found that IL28B inhibits HCV replication in a dose- and time- dependent manner. Like IFNα, IL28B induces the phosphorylation of STAT1 and STAT2, ISRE-driven transcription, and expression of known ISGs. The anti-HCV effects of IL28A, IL28B and IL29 were abrogated by an IL10R2 blocking antibody, a pharmacological inhibitor of JAK1/TYK2, and by siRNA against IL28R1, STAT1, STAT2 and IRF9.
Conclusions
Our data demonstrate that IL28A, IL28B and IL29 signal through the JAK-STAT pathway to inhibit HCV. These data suggest possible applications of new approaches in HCV treatment.
Keywords: HCV, JAK, STAT, IL28B
Introduction
Hepatitis C virus (HCV) is an enveloped, single-stranded, positive sense RNA virus that belongs to the hepacivirus genus in the family Flaviviridae [1, 2]. Currently, 4% of the world’s population is chronically infected with HCV, of whom as many as 30% will develop cirrhosis within 20 years of infection and a large subset will subsequently develop liver failure and/or hepatocellular carcinoma [1, 3]. Infection with HCV has grown into a global epidemic with a death rate surpassing that of HIV/AIDS, and its complications will continue to accumulate for several decades [3].
Combination therapy with pegylated interferon (IFN) α and oral ribavirin is standard treatment for patients with chronic HCV infection. However, it eradicates HCV in only about half of the patients infected with HCV genotype 1, the most frequent genotype in the world. Moreover, severe adverse events are associated with type I IFN therapy, such as myelosuppression, influenza-like symptoms, and neuropsychiatric effects. Because these effects are dose-limiting, many patients are unable to receive higher doses of IFN that might inhibit HCV replication more effectively. These treatment-limiting adverse effects result from the very broad activity of IFN on the immune system, particularly on lymphocytes and neutrophils. It is therefore essential to identify more selective therapeutic agents for the treatment of hepatitis C.
Recently, several groups reported that several single-nucleotide polymorphisms (SNPs) (rs12979860, rs12980275, and rs8099917) near the IL28B (IFNλ3) gene locus are strongly associated with SVR to IFNα and ribavirin treatment for hepatitis C [4–8]. IL28B is a member of the type III IFN family [9], which also includes IFNλ1 (IL29) and IFNλ2 (IL28A). IFNλs bind to their cognate receptor, composed of IL28R1 and IL10R2, and then activate the receptor-associated protein kinases Jak1 and Tyk2, leading to activation of downstream STATs by phosphorylating critical serine and tyrosine residues. Activated STAT1 and STAT2 heterotrimerize with IRF9 to form the ISGF3 complex. ISGF3 then translocates to the nucleus where it binds to the IFN-stimulated response element (ISRE) within the promoter region of IFN-stimulated genes (ISGs). The human genome encodes hundreds of ISGs that are effectors of host responses to viral infection, including ISG15, MxA, and PKR. However, the specific ISGs required for inhibiting HCV replication remain unknown. In this manner, type III IFNs are thought to have considerable functional overlap with type I IFNs [10, 11], including IFNα. However, the magnitude of overlap between type I IFNA and IFNλ3 in their antiviral activity is unknown.
We sought to analyze the role of IL28B in limiting hepatitis C virus replication and its regulation of ISG-mediated antiviral pathways. Previous studies in other laboratories have shown antiviral properties for two other closely related IFNλs, IFNλ1 and IFNλ2 against HCV [12–14]. Using both an HCV full length replicon and JFH1-infected Huh7.5.1 cells, we show here that IL28B is capable of inhibiting HCV replication in a dose and time dependent manner. IL28B treatment stimulates the phosphorylation of STAT1 and STAT2. ISRE activity and several known ISGs are upregulated by IL28B. We also show that the anti-HCV effect of IL28B is impaired when key components of the JAK-STAT signaling pathway are inhibited.
Materials and methods
Cells, virus and reagents
Huh7.5.1 cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The infectious JFH1 plasmid was obtained from Dr. Takaji Wakita and inoculated as previously described [15]. The OR6 cell line [16], which harbors full-length genotype 1b HCV RNA and co-expresses Renilla luciferase, was grown in DMEM supplemented with 10% FBS and 500 µg/ml of G418 (Promega, Madison, WI). The infectious Jc1 plasmid Jc1FLAG2(p7-nsGluc2A) expressing Gaussia luciferase was obtained from Dr. Charles Rice [17]. IL28A, IL28B and IL29 were obtained from R&D systems. IL28A and IL29 are recombinant proteins generated from an NSO-derived murine myeloma cell line. IL28B is a recombinant protein generated from the CHO cell line. PEG-IFNα was obtained from Schering Corporation (Kenilworth, NJ). JAK inhibitor I was purchased from EMD Chemicals, Inc., Gibbstown, NJ, dissolved in 1% dimethyl sulfoxide (DMSO). IL10R2 blocking antibody was purchased from R&D Systems (Minneapolis, MN).
Western blotting
Cells were lysed using radioimmune precipitation assay (RIPA) buffer containing 1% NP-40, 0.1% SDS, 10 mM Tris-HCl (pH 7.4), 1 mM EDTA, 150 mM NaCl and protease inhibitor cocktail, and subsequently sonicated. Proteins were separated by SDS-PAGE with NuPAGE Novex pre-cast 4–12% Bis-Tris gradient gels (Invitrogen, Carlsbad, CA) and transferred to PVDF membranes. The primary antibodies used in this paper were mouse anti-STAT1, rabbit anti-Phospho-STAT1 (Tyr701), rabbit anti-Jak1, anti-Tyk2, anti-STAT2, anti-phospho-STAT2, (Cell Signaling Technology, Inc., Beverly, MA), mouse anti-HCV core, (Affinity BioReagents Inc., Golden, CO), anti-E2, anti-NS4A, anti-NS4B, anti-NS5A, anti-NS5B (Virogen, Watertown, MA), ISG15, MXA (Abcam), mouse anti-actin (Sigma Life Science and Biochemicals, St. Louis, MO), and IL28R1 (Novus Biologicals, Littleton, CO). Secondary antibodies were HRP-conjugated ECL donkey anti-rabbit IgG and HRP-conjugated ECL sheep anti-mouse IgG (Amersham Biosciences, Piscataway, NJ). The ECL Western Blotting Detection Kit (Amersham Biosciences, Piscataway, NJ) was used to detect chemiluminescent signals.
Luciferase Assay
HCV replication in OR6 cells or Jc1FLAG2(p7-nsGluc2A)-infected Huh 7.5.1 cells was determined by monitoring Renilla or Gaussia luciferase activity (Promega, Madison, WI).To monitor IFN signaling directed by ISRE, the plasmids pISRE-luc (500 ng/well) expressing firefly luciferase and pRL-TK (50 ng/well) expressing Renilla luciferase as an internal control were cotransfected using Fugene HD following the manufacturer’s protocol. Relative luciferase activity was assessed by the Promega dual-luciferase reporter assay system (Promega, Madison, WI).
siRNA and transfection
Indicated siRNAs were transfected into cells using Lipofectamine™ RNAiMAX Transfection Reagent (Invitrogen, Carlsbad, CA). The negative control siRNA was obtained from QIAGEN. All siRNAs used for gene knock-down were SMART pools from Dharmacon and indicated: IL28R1, M-007981-00-0005; Jak1, M-003145-02-0005; Tyk2, M-003182-02-0002; STAT1, M-003543-01-0005; STAT2, M-012064-00-0005; IRF9, M-020858-02-0005. Protein expression of each gene knock down was confirmed by Western blotting as previously described [18].
Cell Viability Assay
Cell viability was assessed using the Cell Titer-Glo Luminescent Cell Viability Assay Kit (Promega, Madison, WI) according to the manufacturer’s protocol.
Quantitative PCR
Total cellular and viral RNA was isolated post infection using RNeasy Mini columns (QIAGEN) and reverse transcribed by random priming with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems; Foster City, CA), then quantitated by real-time PCR using the DyNAmo HS SYBR Green qPCR kit (Finnzyme; Espoo, Finland). The primers are listed in Table 1.
Table 1.
Primers used for quantitative RT-PCR.
| Target gene | Primera | Nucleotide sequence |
|---|---|---|
| GAPDH | F | 5'-CAACTGGTCGTGGACAACCAT-3' |
| R | 5'-GCACGGACACTCACAATGTTC-3' | |
| IL28R1 | F | 5'-GACATGACCGGGGACTGCATG-3' |
| R | 5'-GACACACAGGTCCCCGCTGG-3' | |
| PKR | F | 5'-TCTACGCTTTGGGGCTAAT-3' |
| R | 5'-AGATGATGCCATCCCGTAG-3' | |
| STAT1 | F | 5'-GTGGAAAGACAGCCCTGCA-3' |
| R | 5'-ACTGGACCCCTGTCTTCAA-3' | |
| IRF9 | F | 5'-CCCGAAAACTCCGGAACTG -3' |
| R | 5'-CAGCACACTCCGGGAAACT-3' | |
| ISG15 | F | 5'-GGTGGACAAATGCGACGAA-3' |
| R | 5'-ATGCTGGTGGAGGCCCTTA-3' | |
| OAS1 | F | 5'-AGAAGGCAGCTCACGAAAC-3' |
| R | 5'- CCACCACCCAAGTTTCCTG-3' | |
| MxA | F | 5'-GCCGGCTGTGGATATGCTA-3' |
| R | 5'- TTTATCGAAACATCTGTGA-3' | |
| JFH1 | F | 5'- TCTGCGGAACCGGTGAGTA-3' |
| R | 5'-TCAGGCAGTACCACAAGGC-3' |
F, forward; R, reverse.
Statistics
Data analysis was performed using a 2-tailed Student’s t-test. Data are expressed as mean ± SEM of at least three sample replicates, unless stated otherwise.
Results
IL28B demonstrates antiviral activity against HCV in a full-length replicon
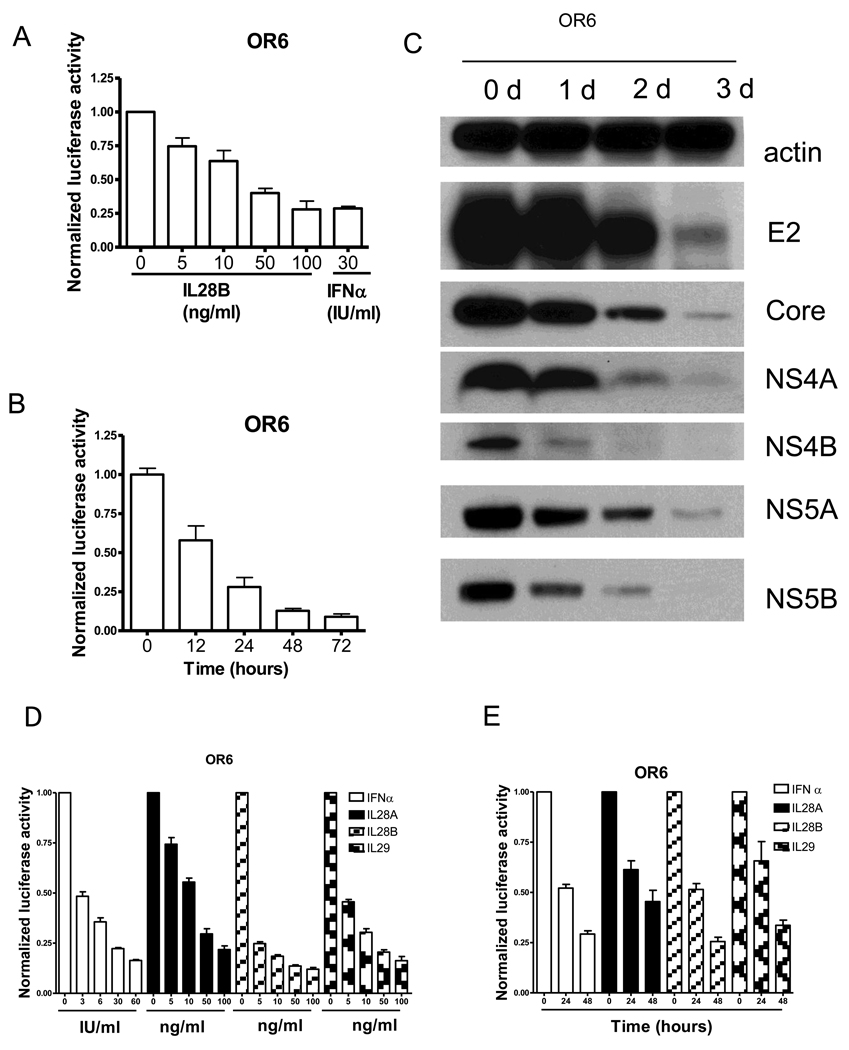
As a successful model for HCV infection, the OR6 replicon cell line harbors a full-length genotype 1b HCV RNA with Renilla luciferase as a reporter. To determine the antiviral effect of IL28B against HCV, OR6 cells were seeded in 96-well plates for 24 hours and then treated with IL28B at different doses for another 24 hours. Renilla luciferase activity reflected the amount of HCV RNA and cell viability was evaluated by assessing cellular ATP levels. As shown in Fig. 1A, IL28B suppressed HCV replication in a dose dependent manner. IL28B at 100 ng/ml inhibited HCV replication to the same extent as 30 IU/ml IFNα (72% reduction).
Figure 1. IL28B inhibits HCV replication in OR6 cells and JFH1 infection model.
(A) OR6 cells were incubated with different doses of IL28B or 30 IU/ml IFNα as indicated for 24 hours. Renilla luciferase activity and cellular ATP levels were measured. (B) OR6 cells were incubated with 100 ng/ml IL28B for different times as indicated. Renilla luciferase activity and cellular ATP levels were measured. (C) OR6 cells were incubated with 100 ng/ml IL28B for different times as indicated. Cell lysates were analyzed by immunoblotting with the indicated antibodies (D) OR6 cells were incubated with different doses of IFNα, IL28A, IL28B or IL29 as indicated for 48 hours. Renilla luciferase activity and cellular ATP levels were measured. (E) OR6 cells were incubated with 100 ng/ml IL28A, IL28B, IL29 or 30IU/ml IFNα for different times as indicated. Renilla luciferase activity and cellular ATP levels were measured. (F) JFH1-infected Huh7.5.1 cells were incubated with different doses of IL28B or IFNα as indicated for 24 hours. Total RNA was isolated and reversely transcribed and then quantitive PCR was performed. (G) JFH1-infected Huh7.5.1 cells were incubated with 100 ng/ml IL28B for different times as indicated. Total RNA was isolated and reversely transcribed and then quantitive PCR was performed. (H) JFH1-infected Huh7.5.1 cells were incubated with 100 ng/ml IL28B for different times as indicated. Cell lysates were analyzed by immunoblotting with the indicated antibodies.
We next determined the time course of IL28B’s anti-HCV effect. As Fig. 1B shows, IL28B (100 ng/ml) inhibited HCV replication in a time dependent manner, achieving 42% suppression within the first 12 hours, and 91% suppression by day 3.
To further confirm IL28B’s antiviral effect, expression levels of HCV proteins in IL28B treated OR6 cells were measured by Western blot using antibodies against HCV core, E2, NS4A, NS4B, NS5A, and NS5B. As shown in Fig. 1C, the levels of each of these HCV proteins were reduced by IL28B in the full-length OR6 replicon, confirming that IL28B antiviral for HCV.
To compare the anti-HCV effects of all three types of IFNλ, we treated OR6 cells with IFNα, IL28A, IL28B or IL29 at different doses for 48 hours. As shown in Fig. 1D and 1E, IFNα, IL28A, IL28B and IL29 all suppressed HCV replication in a dose-dependent (Fig. 1D) and time-dependent (Fig. 1E) manner. IL28B appeared to be somewhat more potent than IL28A and IL29.
IL28B inhibits infectious JFH1 replication
We then assessed IL28B’s effect on HCV replication in JFH1, an established infectious cell culture model for HCV (genotype 2a). We infected Huh7.5.1 cells with JFH1 for 72 hours and then treated the cells with various doses of IL28B or IFNα for 24 hours. As shown in Fig. 1F, normalized JFH1 RNA levels were suppressed in an IL28B dose-dependent manner, achieving 64% suppression at 10 ng/ml and 92% suppression at 100 ng/ml IL28B (Fig. 1F). IL28B at 10 ng/ml inhibited JFH1 replication in a manner comparable to 15 IU/ml IFNα, while 100 ng/ml IL28B inhibited JFH1 replication to the same extent as 150 IU/ml IFNα.
We next determined the time course of IL28B’s anti-HCV effect. As shown in Fig. 1G, IL28B (100 ng/ml) inhibited HCV replication in a time dependent manner, achieving 50% suppression at 6 hours, and 92% suppression by 24 hours.
To confirm the suppression of HCV proteins, the level of HCV core, E2, NS3, and NS5B proteins were measured by immunoblot. We found that IL28 B reduced levels of HCV proteins in a time-dependent manner (Fig. 1H).
IL28B induces phosphorylation of STAT1 and STAT2
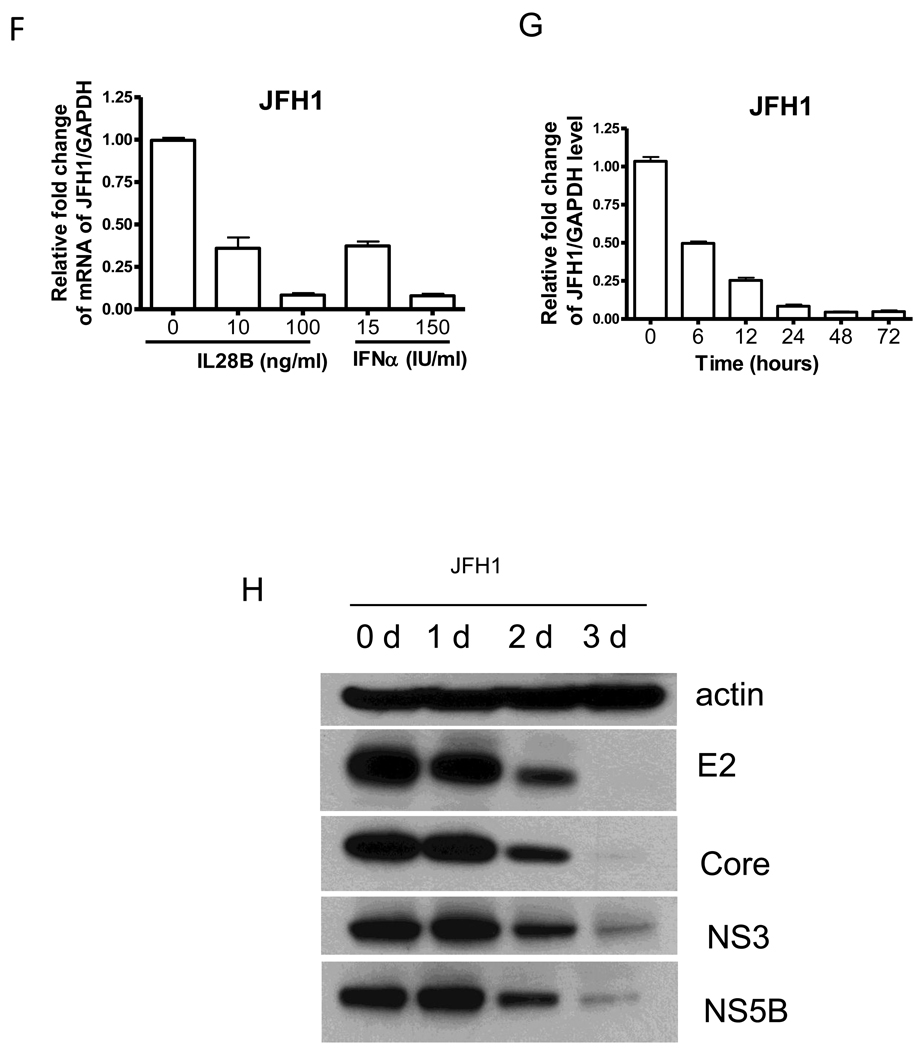
IL28R1 and IL10R2 form the cognate receptor complex for IFNλs. After IFNλs bind to their receptor, the JAK-STAT pathway is activated. We next measured phosphorylation of STAT1 and STAT2 induced by IL28B. OR6 and JFH1-infected Huh7.5.1 cells were treated with 100 ng/mL IL28B, 30 IU/ml IFNα or mock treated for 30 min, and STAT1 and STAT2 phosphorylation was assessed. As shown in Fig. 2A and B, IL28B treatment induced STAT1 and STAT2 phosphorylation comparable to IFNα, confirming that the JAK-STAT signaling pathway is activated by IL28B in these cells.
Figure 2. IL28B induces STAT1 and STAT2 phosphorylation and ISRE reporter activity.
OR6 cells (A) or JFH1-infected Huh7.5.1 cells (B) were treated with 100 ng/ml IL28B, 30 IU/ml IFNα or mock treatment for 30 min and the cell lysates were analyzed by Western blotting with the indicated antibodies. pISRE-luc encoding firefly luciferase under the control of the ISRE and pRL-TK expressing Renilla luciferase were transfected to Huh7.5.1 cells (C) or JFH1-infected Huh7.5.1 cells (D) for 48 hours and 100 ng/ml IL28B or 30 IU/ml IFNα were added to the cells for 6 hours. The firefly and Renilla luciferase activity was measured.
IL28B induces ISRE activity and expression of classical ISGs
Like type I IFNs, type III IFNs are thought to mediate signaling through the STAT1 and STAT2 components of the JAK-STAT signal transduction pathways. We used the interferon stimulated response element (ISRE) luciferase reporter assay to assess activity downstream of the STAT1/STAT2 axis. We transfected pISRE-luc and pRL-TK into uninfected Huh7.5.1 cells or JFH1-infected Huh7.5.1 cells for 48 hours and IL28B was then added to the cells for 6 hours. Firefly and Renilla luciferase activity were then measured. IL28B significantly stimulated ISRE activity in both uninfected and JFH1-infected Huh7.5.1 cells (Fig. 2C and D). In uninfected Huh7.5.1 cells, ISRE luciferase activity was about 3-fold higher with IL28B treatment than with mock (Fig. 2C). In JFH1-infected Huh7.5.1 cells, ISRE luciferase activity was about double with IL28B treatment compared to mock (Fig. 2D). The increased ISRE luciferase activity by IL28B was similar to the extent of induction by IFNα. The lesser induction of the ISRE reporter activity by IFN in the presence of HCV likely reflected HCV’s suppression of the JAK-STAT signaling pathway [19, 20].
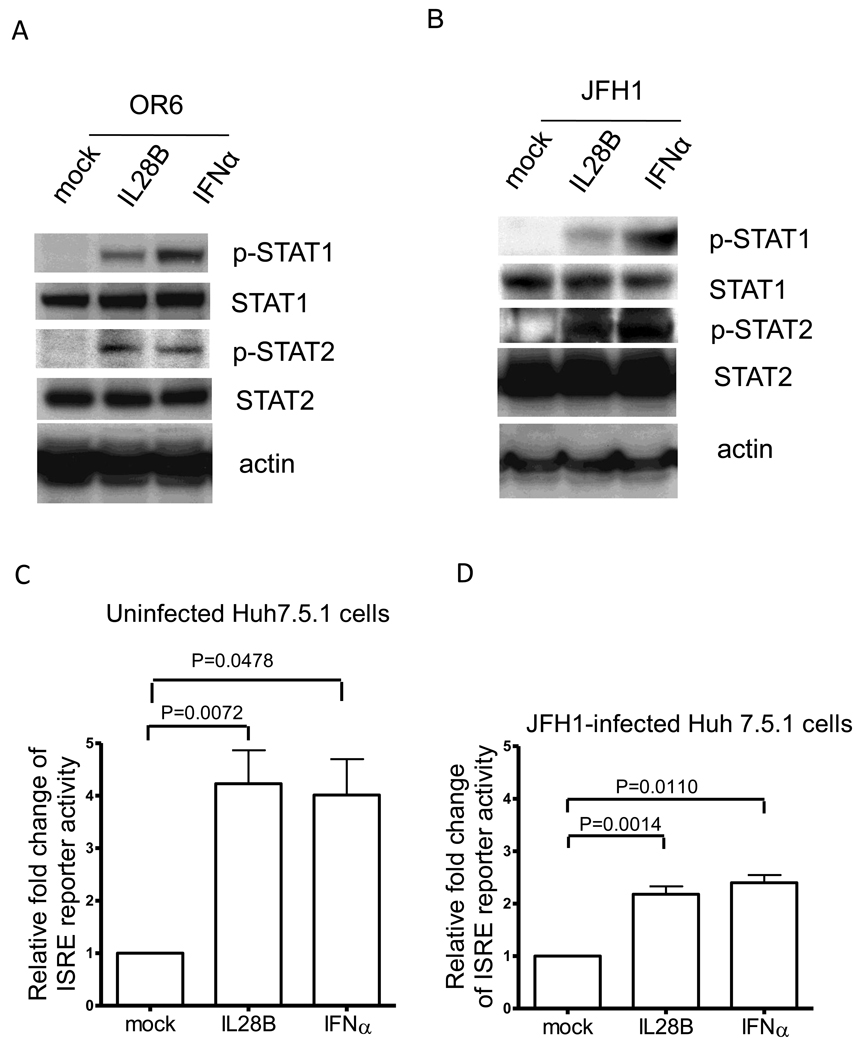
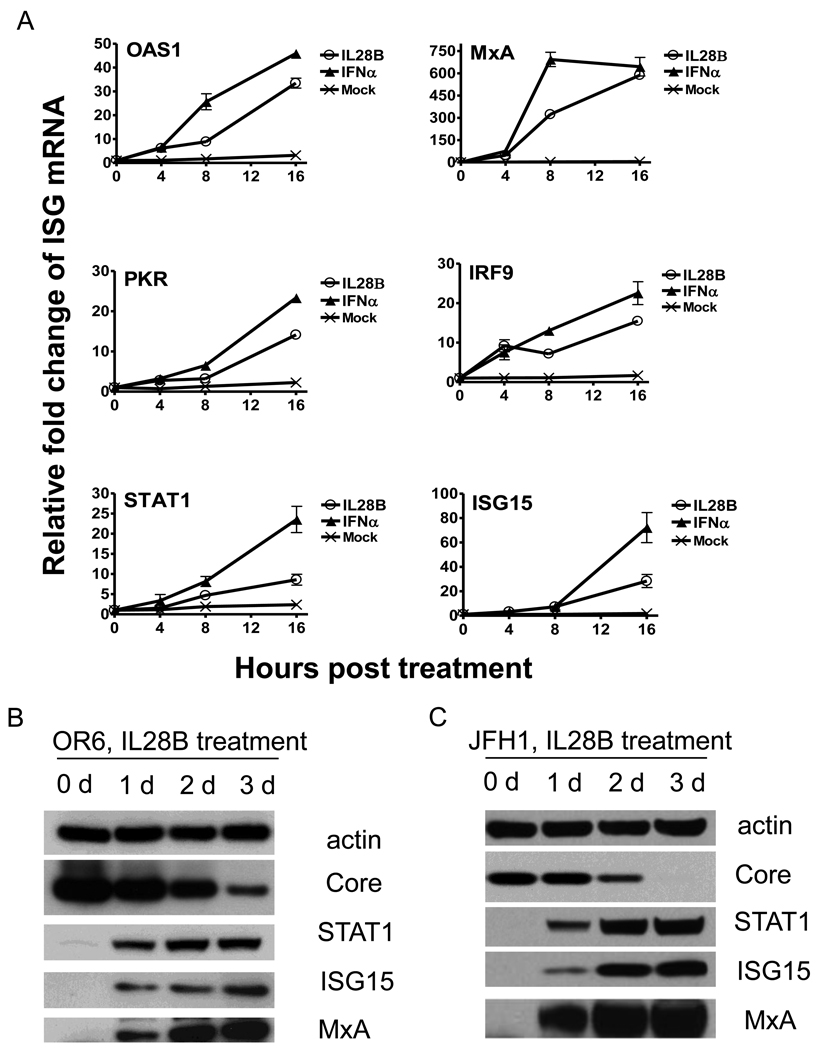
Interferons are a family of multifunctional cytokines with the ability to interfere with viral infection through induction of the expression of IFN-stimulated genes. To determine IL28B’s effect on ISGs, we analyzed expression of several classic antiviral ISGs. OR6 cells were treated with 10 ng/mL IL28B or 15 IU/ml IFNα or mock for varying lengths of time, and gene expression of several ISGs was assessed. Like IFNα, IL28B significantly increased the expression of IRF9, ISG15, MxA, OAS1, PKR, and STAT1 in a time dependent manner, while mock treatment failed to induce the expression of ISGs (Fig. 3A).
Figure 3. IL28B stimulates known ISGs.
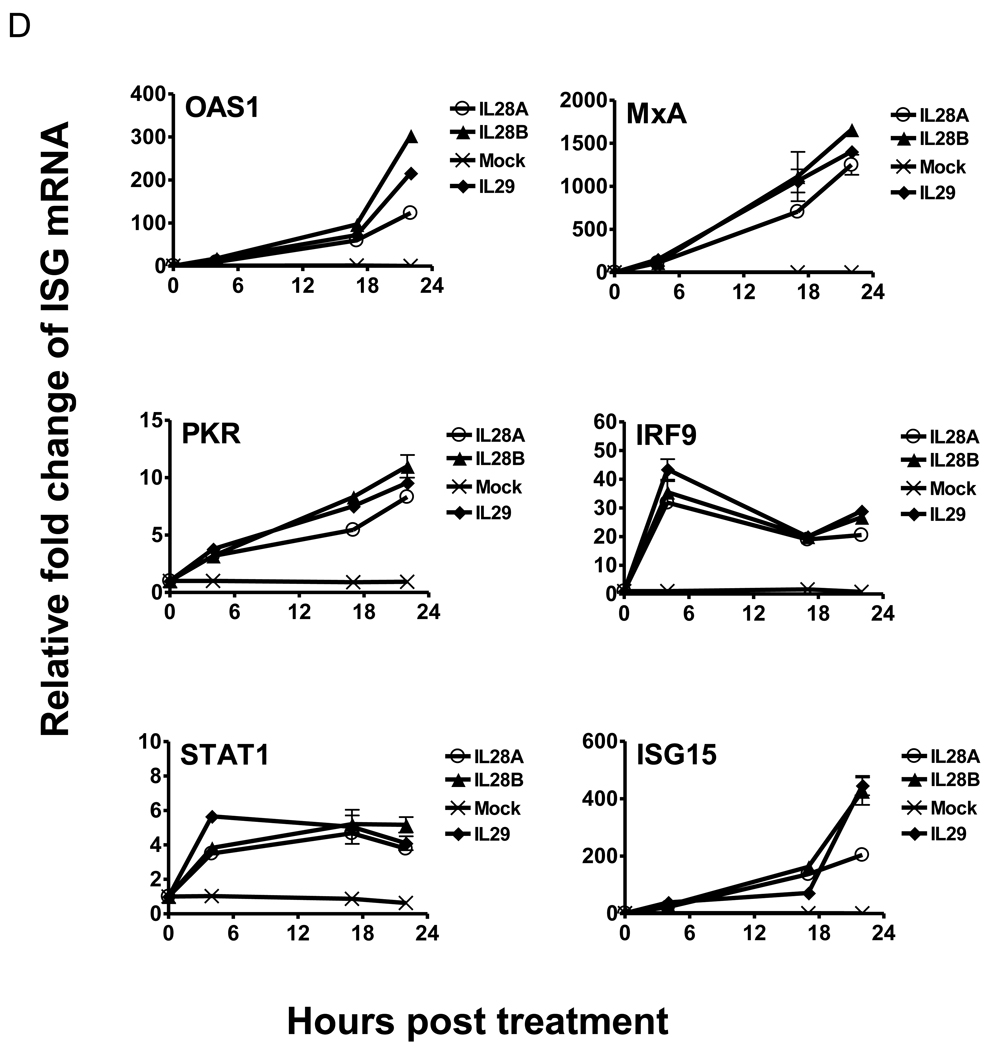
(A) OR6 cells were incubated 10 ng/ml IL28B or 15 IU/ml IFNα or mock treatment for different times as indicated. Total RNA was isolated and reverse transcribed followed by real time PCR. OR6 cells (B) and JFH1-infected Huh7.5.1 cells (C) were incubated with 100 ng/ml IL28B for different times as indicated. Cell lysates were analyzed by immunoblotting with the indicated antibodies. (D) Huh 7.5.1 cells were incubated 100 ng/ml IL28A, IL28B, IL29 or mock treatment for different times as indicated. Total RNA was isolated and reverse transcribed followed by real time PCR.
We also assessed ISG protein expression levels with IL28B stimulation. As shown in Fig. 3B and C, protein levels of STAT1, MxA, and ISG15 were significantly increased by IL28B treatment in both OR6 cells and JFH1-infected Huh7.5.1 cells.
To compare the induction of ISGs by the three types of IFNλ, we treated Huh 7.5.1 cells with 100 ng/ml IL28A, IL28B, IL29 or mock treatment for varying lengths of time, and gene expression of several ISGs was assessed. As shown in Fig. 3D, the expression pattern of IRF9, ISG15, MxA, OAS1, PKR, and STAT1 stimulated by IL28A, IL28B or IL29 are similar. These data suggested that the three types of IFNλ likely induce the same set of ISGs.
Taken together, these results imply that IL28B stimulates phosphorylation of STAT1/STAT2 and ISRE activity, thereby leading to the expression of known ISGs.
The antiviral activity of IL28B is dependent on the IFNλ receptor
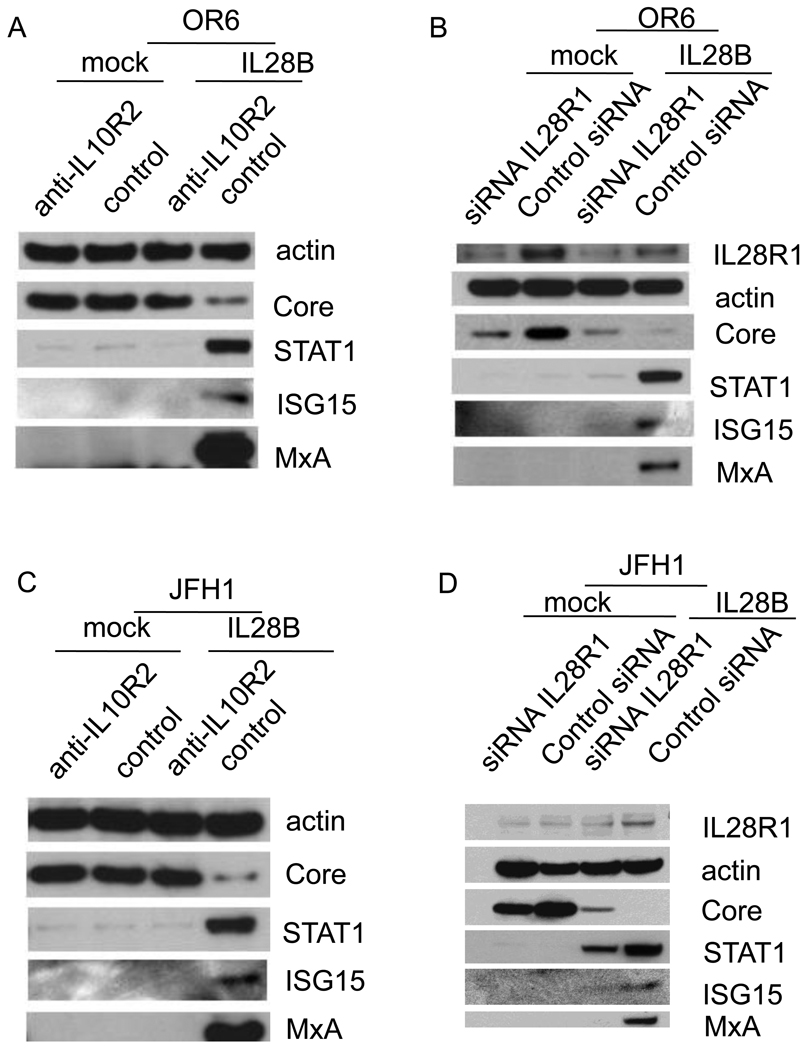
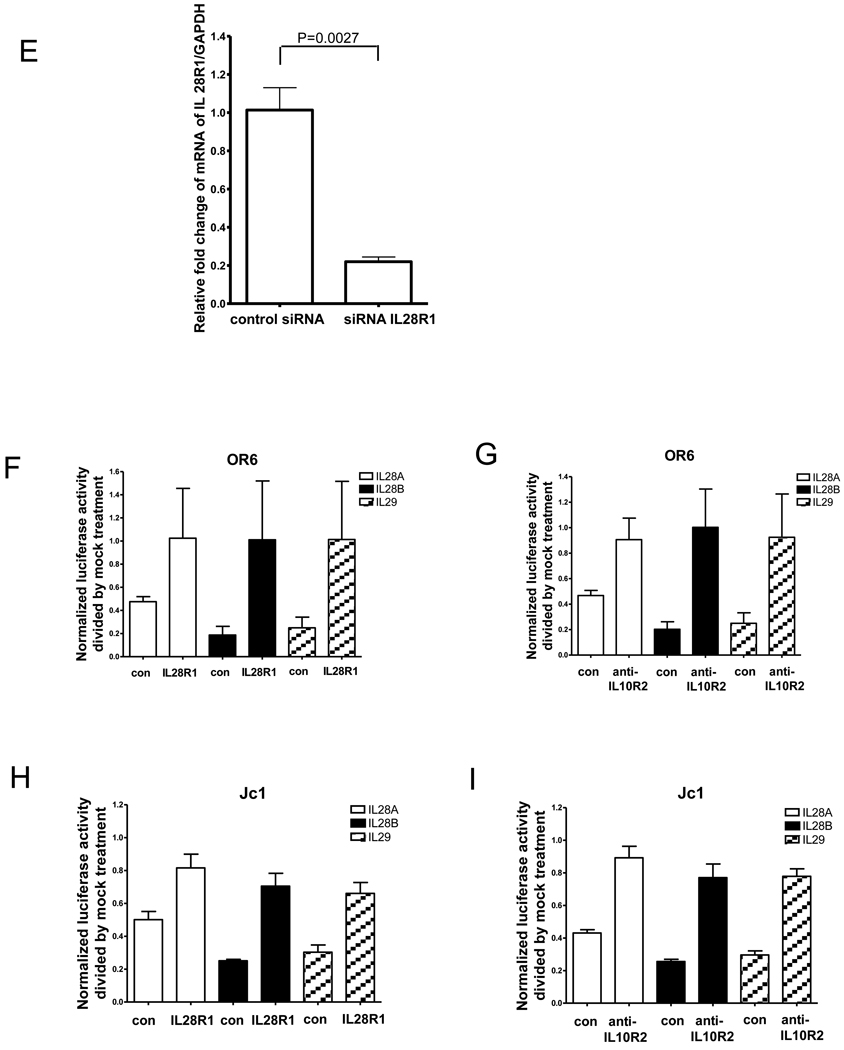
Type III IFNs bind to the cellular IFNλ receptor (IL28R1 and IL10R2), which in turn engages the tyrosine kinases Jak1 and Tyk2. We tested whether the antiviral activity of IL28B against HCV is mediated by the IFNλ receptor. We used an IL10R2 blocking antibody to inhibit IL28B signaling in OR6 and JFH1-infected cells (Fig. 4A and C). The induction of known ISGs (STAT1, MxA and ISG15) by IL28B was reduced by IL10R2 antibody. Correspondingly, the reduction of HCV core protein levels by IL28B, as assessed by Western blotting, was rescued by IL10R2 antibody (Fig. 4A and C). To inhibit IL28R1, we used an siRNA approach. IL28R1 knockdown in OR6 was validated by Western blotting as in Fig. 4B. IL28R1 knockdown in JFH1-infected Huh7.5.1 cells was validated by QPCR as in Fig. 4G. The induction of known ISGs (STAT1, MxA and ISG15) by IL28B was also reduced by silencing of IL28R1, indicating that the downstream JAK-STAT pathway was inhibited. As shown in Fig. 4B and D, protein levels of HCV core inhibited by IL28B were rescued by knocking down IL28R1. As shown in Fig. 4B, silencing IL28R1 unexpectedly caused the reduction of HCV core levels in the absence of IL28B, suggesting the possibility of siRNA-mediated off-target effects. Alternatively, IL28R1 may facilitate HCV replication, since the favorable IL28B genotype is unexpectedly associated with higher HCV viral loads [4].
Figure 4. The IFNλ receptor is required for the anti-HCV effect of IL28B.
OR6 cells (A) and JFH1-infected Huh7.5.1 cells (C) were incubated with IL10R2 blocking antibody or control goat IgG for 2 hours and were then treated with 100 ng/ml IL28B or control mock treatment for 3 days. Cell lysates were analyzed by immunoblotting with the indicated antibodies. OR6 cells (B) and JFH1-infected Huh7.5.1 cells (D) were treated with siRNA against IL28R1 for 3 days and incubated with 100 ng/ml IL28B for 3 days. Cell lysates were analyzed by immunoblotting with the indicated antibodies. (E) JFH1-infected Huh7.5.1 cells were treated with siRNA against IL28R1 for 3 days. Total RNA was isolated and reversely transcribed and then quantitive PCR was performed. OR6 cells (F) and Jc1-infected Huh7.5.1 cells (H) were incubated with either IL10R2 or control goat antibodies for 2 hours and then treated with 100 ng/ml IL28A, IL28B, IL29 or mock treatment for 3 days. Renilla (F) or Gaussia (H) luciferase activity and cellular ATP levels were measured. The normalized luciferase activity were then divided by the normalized luciferase activity from mock treatment. OR6 cells (G) and Jc1-infected Huh7.5.1 cells (I) were treated with siRNA against IL28R1 for 3 days and then treated with 100 ng/ml IL28A, IL28B, IL29 or mock treatment for 3 days. Renilla (G) or Gaussia (I) luciferase activity and cellular ATP levels were measured. The normalized luciferase activity was then divided by the normalized luciferase activity from mock treatment.
To compare the dependence of the anti-HCV effects of the three types of IFNλ on IL10R2 receptor, OR6 cells or Jc1FLAG2(p7-nsGluc2A)-infected Huh 7.5.1 cells were pre-incubated with either IL10R2 or control antibody and then treated with 100 ng/ml of IL28A, IL28B, IL29 or mock treatment for three days. As shown in Fig. 4F and G, levels of normalized luciferase activity inhibited by IL28A, IL28B, IL29 were rescued by IL10R2 antibody.
Similarly, to compare the dependence of the anti-HCV effects of the three types of IFNλ on IL28R1 receptor, OR6 cells or Jc1FLAG2(p7-nsGluc2A)-infected Huh 7.5.1 cells were treated with siRNA against IL28R1 or control siRNA for three days and then incubated with 100 ng/ml of IL28A, IL28B, IL29 or mock treatment for three days. As shown in Fig. 4H and I, levels of normalized luciferase activity inhibited by IL28A, IL28B, IL29 were rescued by siRNA against IL28R1.
Taken together, the anti-HCV effect of IL28B as well as IL28A and IL29 is dependent on its intact IFNλ receptor.
The antiviral activity of IL28B is dependent on Jak1 and Tyk2
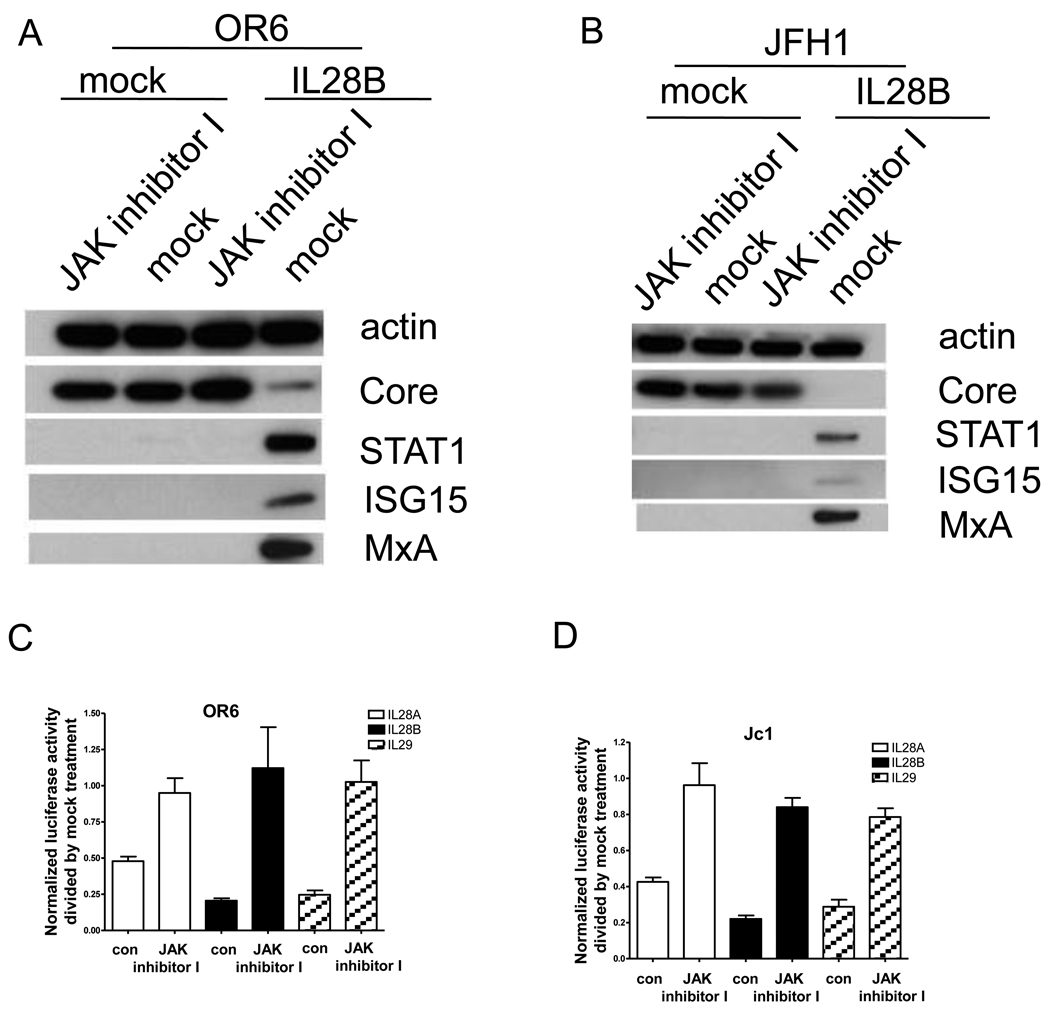
Because Jak1 and Tyk2 are required for STAT1 and STAT2 activation, we conjectured that Jak1 and Tyk2 are critical for the suppression of HCV replication by IL28B. To investigate this, OR6 cells or JFH1-infected Huh7.5.1 cells were incubated with JAK inhibitor I (inhibitor of both Jak1 and Tyk2) for 1 hour prior to treatment with IL28B and cell lysates were collected and analyzed by Western blot (Fig. 5A and B). In the presence of JAK inhibitor I, the induction of known ISGs (STAT1, MxA and ISG15) by IL28B was reduced and HCV core protein levels inhibited by IL28B were rescued by JAK inhibitor I. These data indicate that Jak1 and Tyk2 are required for IL28B’s antiviral effect.
Figure 5. Jak1 and Tyk2 are required for the anti-HCV effect of IL28B.
OR6 cells (A) and JFH1-infected Huh7.5.1 cells (B) were incubated with JAK inhibitor I or mock for 1 hour and then treated with 100 ng/ml IL28B or mock for 3 days. Cell lysates were analyzed by immunoblotting with the indicated antibodies. OR6 cells (C) and Jc1-infected Huh7.5.1 cells (D) were incubated with JAK inhibitor I or mock for 1 hour and then treated with 100 ng/ml IL28A, IL28B, IL29 or mock treatment for 3 days. Renilla (C) or Gaussia (D) luciferase activity and cellular ATP levels were measured. The normalized luciferase activity were then divided by the normalized luciferase activity from mock treatment.
To compare the dependence of the anti-HCV effects of the three types of IFNλ on Jak1 and Tyk2, OR6 cells or Jc1FLAG2(p7-nsGluc2A)-infected Huh 7.5.1 cells were pre-treated with either JAK inhibitor I or mock treatment for 1 hour and then incubated with 100 ng/ml of IL28A, IL28B, IL29 or mock treatment for three days. As shown in Fig. 5C and D, levels of normalized luciferase activity inhibited by IL28A, IL28B, IL29 were rescued by JAK inhibitor I. These data indicate that Jak1 and Tyk2 are required for the antiviral effects of all three types of IFNλ.
The antiviral activity of IL28B is dependent on STAT1, STAT2 and IRF9
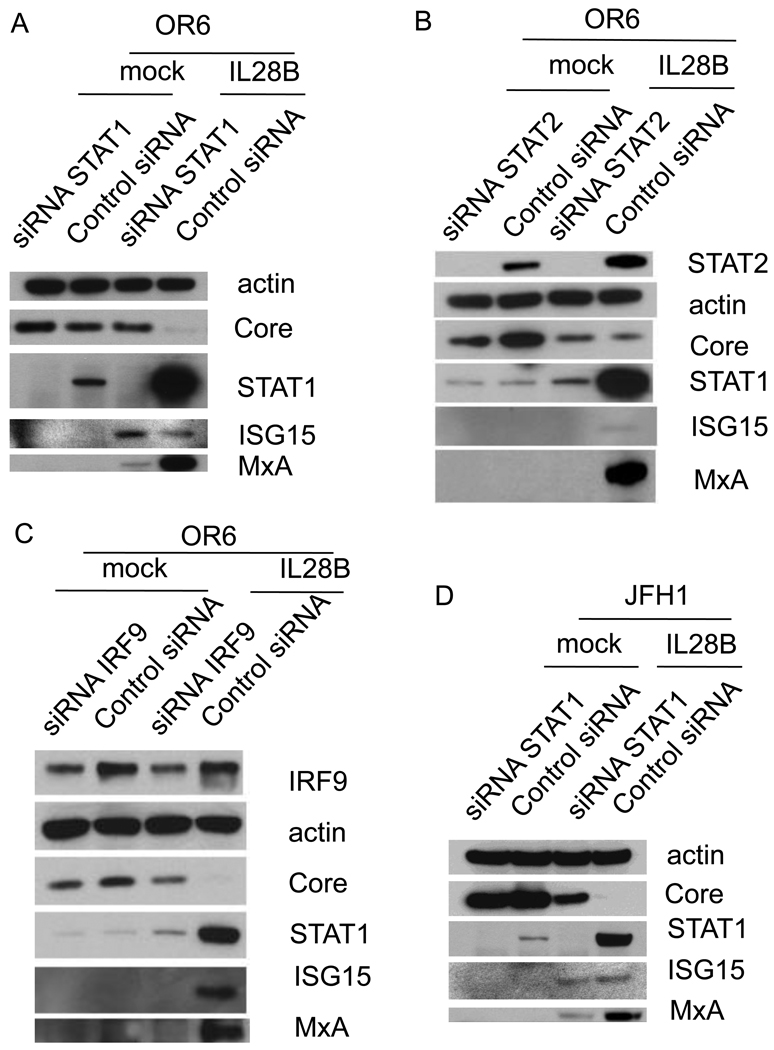
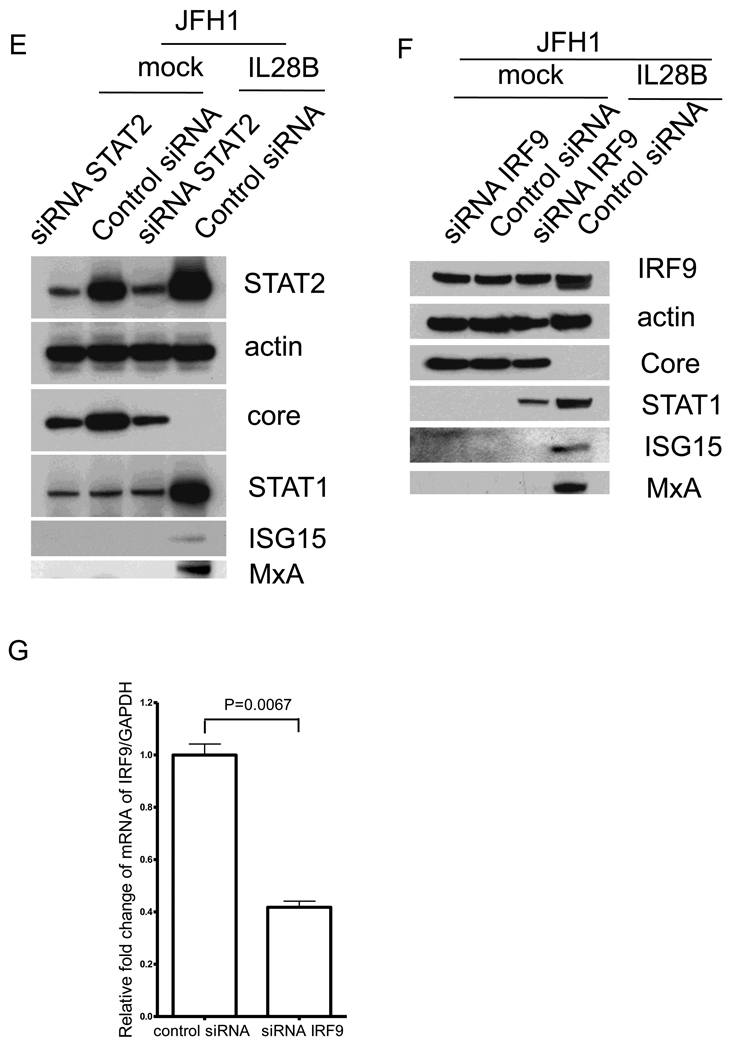
In the type I IFN signaling cascade, STAT1, STAT2 and IRF9 form the trimetric ISGF3 complex and subsequently undergo nuclear translocation. We therefore tested whether STAT1, STAT2 and IRF9 are required for the antiviral activity of IL28B. We used siRNAs to knock down STAT1, STAT2 and IRF9. In both OR6 cells and JFH1-infected Huh7.5.1 cells, the silencing of STAT1 and STAT2 was validated by Western blotting (Fig. 6A, B, D, and E). Partial knockdown of IRF9 protein was validated by Western blotting in OR6 cells (Fig. 6C). However, knockdown of IRF9 protein in JFH1-infected Huh7.5.1 cells was observed only in the presence of IL28B (Fig. 6F), despite the fact that siRNA against IRF9 was capable of silencing IRF9 mRNA in JFH1-infected Huh7.5.1 cells (Fig. 6G). This relatively weak observed silencing of IRF9 protein may be related to the abundant expression of IRF9 protein. By knocking down STAT1, the induction of STAT1 and MxA by IL28B was reduced; however, ISG15 protein levels remained similar to that of control siRNA (Fig. 6A and D). By knocking down STAT2 or IRF9, the induction of STAT1, MxA, and ISG15 by IL28B was reduced (Fig. 6B, C, E and F). HCV protein levels inhibited in the presence of IL28B were rescued by knocking down STAT1, STAT2, or IRF9. These data indicate that STAT1, STAT2 and IRF9 are required for IL28B antiviral signaling.
Figure 6. STAT1, STAT2 and IRF9 are required for the anti-HCV effect of IL28B.
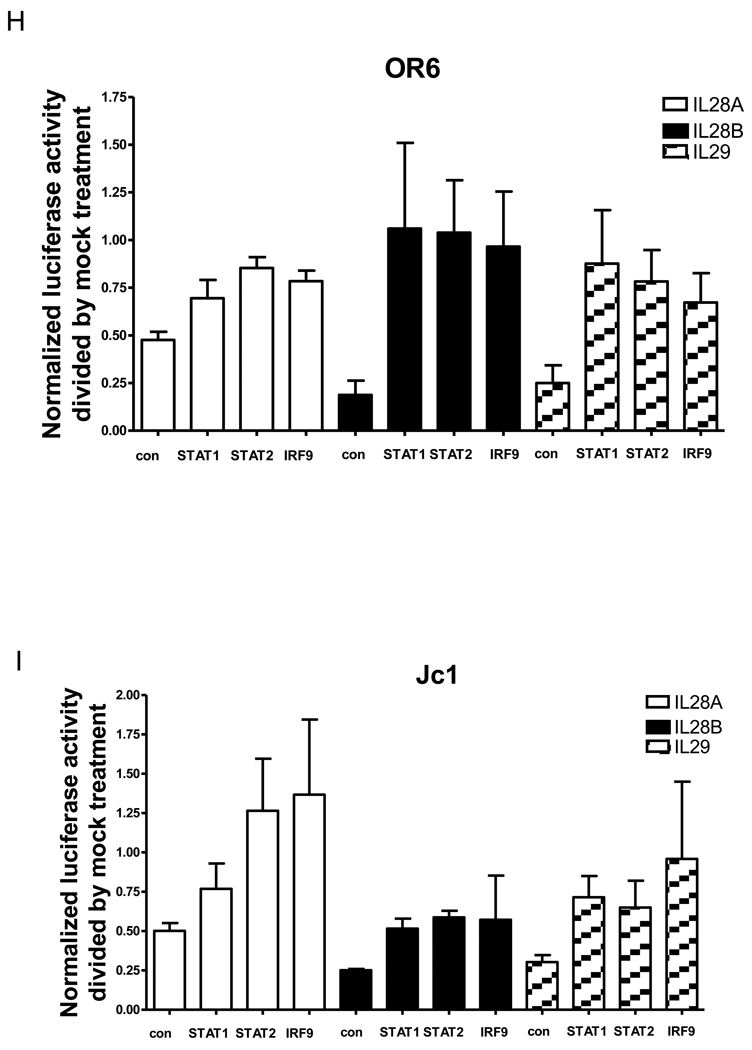
OR6 cells were treated with siRNAs against STAT1 (A), STAT2 (B) or IRF9 (C) for 3 days and incubated with 100 ng/ml IL28B or mock for 3 days. Cell lysates were analyzed by immunoblotting with the indicated antibodies. JFH1-infected Huh7.5.1 cells were treated with siRNAs against STAT1 (D), STAT2 (E) or IRF9 (F) for 3 days and incubated with 100 ng/ml IL28B or mock for 3 days. Cell lysates were analyzed by immunoblotting with the indicated antibodies. (G) JFH1-infected Huh7.5.1 cells were treated with siRNAs against IRF9 for 3 days. Total RNA was isolated and reversely transcribed and then quantitive PCR was performed. OR6 cells (H) and Jc1-infected Huh7.5.1 cells (I) were treated with siRNAs against STAT1, STAT2 or IRF9 for 3 days and then treated with 100 ng/ml IL28A, IL28B, IL29 or mock treatment for 3 days. Renilla (H) or Gaussia (I) luciferase activity and cellular ATP levels were measured. The normalized luciferase activity was then divided by the normalized luciferase activity from mock treatment.
To compare the dependence of the anti-HCV effects of the three types of IFNλ on STAT1, STAT2 and IRF9, OR6 cells or Jc1FLAG2(p7-nsGluc2A)-infected Huh 7.5.1 cells either treated with siRNAs against STAT1, STAT2, IRF9 or control siRNA for three days and then incubated with 100 ng/ml of IL28A, IL28B, IL29 or mock treatment for three days. As shown in Fig. 6H and I, levels of normalized luciferase activity inhibited by IL28A, IL28B, IL29 were rescued by siRNAs against STAT1, STAT2 or IRF9. These data indicate that STAT1, STAT2 and IRF9 are required for the antiviral effects of all three types of IFNλ.
Discussion
As the first line of defense against viral pathogens, interferons act on viral RNA translation and sense RNA synthesis directly or indirectly through activation of host interferon stimulated genes (ISGs) [21]. IFNα is the major component of current standard treatment for hepatitis C [22]. The recent discovery of the type III lambda interferon family has opened new avenues of research into novel mechanisms of antiviral activity [3, 23, 24]. Previously, IFNλ1 and λ2 have been shown to inhibit HCV replication in HCV replicon cells [14]. In another study, IFN induced genes were compared by microarrays and different clusters of genes activated by IFNλ1 were identified [13]. In this report, we have found that IL28B (IFNλ3) inhibits HCV replication for two different genotypes (1b and 2a) in a time and dose dependent manner, confirming that all three IFNλs are anti-HCV cytokines.
The mechanisms by which type III IFNs establish an antiviral state are not as well characterized as those for the type I IFNs, but are believed to be similar. We found that IL28B stimulated the phosphorylation of STAT1/STAT2 and ISRE luciferase reporter activities and subsequently induced the expression of known ISGs. Because of a more restricted distribution of the IFNλ receptor, IFNλ may be better tolerated than IFNα, which may justify the use of IFNλ as an alternative or complementary agent for hepatitis C. A recent clinical study found that weekly PEG-IFNλ1 for 4 weeks is well tolerated with minimal adverse events and hematologic effects and is associated with clear antiviral activity in patients with chronic hepatitis C [25]. Our data provide evidence that IFNλ3 could also have a role in hepatitis C treatment.
Several GWAS studies identified an association of IL28B SNPs with response to clearance of chronic HCV infection by IFNα and ribavirin [4–8, 26]. Whether these SNPs are associated with altered IL28B gene expression or receptor activation remains to be further established. Furthermore, it is not clear whether IL28B acts solely through its overlap with type I IFN or whether other signaling transduction pathways are also activated.
To elucidate mechanisms contributing to the anti-HCV effect of IL28A, IL28B, and IL29, we examined core components of the JAK-STAT pathway related to IFNλ. We systematically inhibited IL10R2, IL28R1, Jak1, Tyk2, STAT1, STAT2, and IRF9 using chemical, antibody, or siRNA inhibition. The expression of known ISGs, such as STAT1, MxA and ISG15 was measured to reflect the activation of the JAK-STAT pathway. In OR6 cells, JFH1-infected or Jc1-infected Huh7.5.1 cells, HCV suppression mediated by IL28A, IL28B, and IL29 was largely rescued when we inhibited each of these components of the JAK-STAT pathway, indicating that the JAK-STAT pathway is required for the anti-HCV effect of IL28B as well as IL28A and IL29.
In conclusion, our results demonstrate that IL28B inhibits HCV replication in three independent HCV models. Loss-of-function studies by inhibition of the JAK-STAT pathway suggest that the suppression of HCV by IL28B is predominantly mediated by this pathway. Further studies directed at understanding the specific genes induced by IFNλ and the mechanisms of their antiviral effect against HCV will provide valuable insight into HCV pathogenesis. Given that rescue of HCV by blocking JAK-STAT pathway was incomplete, these findings leave open the possibility of independent pathways induced by IL28B. However it is likely that these pathways play a less dominate role than the canonical type I IFN pathway.
Acknowledgements
This work was supported by grants AI069939, AI082630 and DK078772 (R.T.C.) from the National Institutes of Health. N.J. was supported by Grant Ji 145/1-1 from the Deutsche Forschungsgemeinschaft, Bonn, Germany. We thank Drs Nobuyuki Kato and Masanori Ikeda for the gift of OR6 cells; Dr Francis Chisari for the Huh7.5.1 cell line; Dr Takaji Wakita for the infectious HCV virus JFH1 DNA construct; Dr. Charles Rice for Jc1FLAG2(p7-nsGluc2A).
Abbreviations
- IFN
interferon
- RBV
ribavirin
- SVR
sustained virologic response
- HCV
Hepatitis C virus
- SNP
single-nucleotide polymorphism
- ISRE
interferon-stimulated response element
- ISG
IFN-stimulated genes
- DMEM
Dulbecco’s Modified Eagle’s Medium
- FBS
fetal bovine serum
- RIPA
radioimmune precipitation assay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declared that they do not have anything to disclose regarding funding from industry or conflict of interest with respect to this manuscript.
References
- 1.Bostan N, Mahmood T. An overview about hepatitis C: a devastating virus. Crit Rev Microbiol. 2010;36:91–133. doi: 10.3109/10408410903357455. [DOI] [PubMed] [Google Scholar]
- 2.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 3.National intelligence estimate: the global infectious disease threat and its implications for the United States. Environ Change Secur Proj Rep. :200033–200065. [PubMed] [Google Scholar]
- 4.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 5.Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345. doi: 10.1053/j.gastro.2009.12.056. 1345 e1331–1337. [DOI] [PubMed] [Google Scholar]
- 6.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 8.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dellgren C, Gad HH, Hamming OJ, Melchjorsen J, Hartmann R. Human interferon-lambda3 is a potent member of the type III interferon family. Genes Immun. 2009;10:125–131. doi: 10.1038/gene.2008.87. [DOI] [PubMed] [Google Scholar]
- 10.Doyle SE, Schreckhise H, Khuu-Duong K, Henderson K, Rosler R, Storey H, et al. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology. 2006;44:896–906. doi: 10.1002/hep.21312. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL, Hartmann R. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J Virol. 2007;81:7749–7758. doi: 10.1128/JVI.02438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butera M, Nelson DR, Liu C. Novel type I interferon IL-28A suppresses hepatitis C viral RNA replication. Virol J. 2005;2:80. doi: 10.1186/1743-422X-2-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 14.Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79:3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao RX, Zhang L, Peng LF, Sun E, Chung WJ, Jang JY, et al. Suppressor of cytokine signaling 3 suppresses hepatitis C virus replication in an mTOR-dependent manner. J Virol. 84:6060–6069. doi: 10.1128/JVI.02484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda M, Abe K, Dansako H, Nakamura T, Naka K, Kato N. Efficient replication of a full-length hepatitis C virus genome, strain O, in cell culture, and development of a luciferase reporter system. Biochem Biophys Res Commun. 2005;329:1350–1359. doi: 10.1016/j.bbrc.2005.02.138. [DOI] [PubMed] [Google Scholar]
- 17.Marukian S, Jones CT, Andrus L, Evans MJ, Ritola KD, Charles ED, et al. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology. 2008;48:1843–1850. doi: 10.1002/hep.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Lee SY, Beznoussenko GV, Peters PJ, Yang JS, Gilbert HY, et al. A role for the host coatomer and KDEL receptor in early vaccinia biogenesis. Proc Natl Acad Sci U S A. 2009;106:163–168. doi: 10.1073/pnas.0811631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin W, Choe WH, Hiasa Y, Kamegaya Y, Blackard JT, Schmidt EV, et al. Hepatitis C virus expression suppresses interferon signaling by degrading STAT1. Gastroenterology. 2005;128:1034–1041. doi: 10.1053/j.gastro.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Lemon SM. Induction and Evasion of Innate Antiviral Responses by Hepatitis C Virus. J Biol Chem. 2010 doi: 10.1074/jbc.R109.099556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonjardim CA. Interferons (IFNs) are key cytokines in both innate and adaptive antiviral immune responses--and viruses counteract IFN action. Microbes Infect. 2005;7:569–578. doi: 10.1016/j.micinf.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Darling JM, Fried MW. Optimizing treatment regimens in hepatitis C. Clin Liver Dis. 2006;10:835–850. doi: 10.1016/j.cld.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 23.Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ank N, Paludan SR. Type III IFNs: new layers of complexity in innate antiviral immunity. Biofactors. 2009;35:82–87. doi: 10.1002/biof.19. [DOI] [PubMed] [Google Scholar]
- 25.Muir AJ, Shiffman ML, Zaman A, Yoffe B, de la Torre A, Flamm S, et al. Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology. 2010;52:822–832. doi: 10.1002/hep.23743. [DOI] [PubMed] [Google Scholar]
- 26.McCarthy JJ, Li JH, Thompson A, Suchindran S, Lao XQ, Patel K, et al. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology. 2010;138:2307–2314. doi: 10.1053/j.gastro.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]