Abstract
In an effort to develop microalgae as a robust system for the production of valuable proteins, we analyzed some of the factors affecting recombinant protein expression in the chloroplast of the green alga Chlamydomonas reinhardtii. We monitored mRNA accumulation, protein synthesis, and protein turnover for three codon-optimized transgenes including GFP, bacterial luciferase, and a large single chain antibody. GFP and luciferase proteins were quite stable, while the antibody was less so. Measurements of protein synthesis, in contrast, clearly showed that translation of the three chimeric mRNAs was greatly reduced when compared to endogenous mRNAs under control of the same atpA promoter/UTR. Only in a few conditions this could be explained by limited mRNA availability since, in most cases, recombinant mRNAs accumulated quite well when compared to the atpA mRNA. In vitro toeprint and in vivo polysome analyses suggest that reduced ribosome association might contribute to limited translational efficiency. However, when recombinant polysome levels and protein synthesis are analyzed as a whole, it becomes clear that other steps, such as inefficient protein elongation, are likely to have a considerable impact. Taken together, our results point to translation as the main step limiting the expression of heterologous proteins in the C. reinhardtii chloroplast.
Keywords: Algal biotechnology, Chlamydomonas, Chloroplast, mRNA translatability, Protein translation, Protein turnover, Recombinant protein expression
Introduction
Microalgae have gained a great deal of attention as potential platforms for the production of valuable proteins and other biomolecules like essential oils, vitamins, and precursors for biofuel generation. It is accepted that, in many cases, genetic engineering will be mandatory for the maintenance of high biomass and compound productivities. In this scenario, the mastering of protein expression will be a central point, since many of these applications will require not only the modification of enzyme accumulation and activities but also the introduction of new enzymes and metabolic pathways.
The use of chloroplast-based expression systems for the production of recombinant proteins has received considerable attention over the past several years [1–7]. In the chloroplast of higher plants, for example, human IFN-alpha2b [8] and somatotropin [9] have been expressed to relatively high levels. Toward similar ends, we have examined recombinant protein expression in the chloroplast of the unicellular green alga Chlamydomonas reinhardtii. The ability to culture algae at very large scale in containment and to rapidly generate stable transgenic lines makes the C. reinhardtii chloroplast an attractive system for recombinant protein expression [10]. Previously, we demonstrated that optimization of codon usage in transgenes to reflect that found in the C. reinhardtii chloroplast genome had a profound effect on heterologous protein accumulation in transgenic algae [11]. Following such an approach, we have expressed several recombinant proteins, achieving accumulation levels of less than 1% of total soluble proteins in average and as high as 5% [12–15].
In higher plants, accumulation of many heterologous proteins has exceeded 20% of soluble protein [16, 17]. There are several factors that may contribute to attaining such high levels of recombinant protein accumulation, including gene copy number, the rate of transcription of the heterologous gene, message stability, translatability of the recombinant mRNA, or turnover of the recombinant protein. Impacting any one of these parameters could have a profound effect on recombinant protein accumulation.
To elucidate which of these factors are involved in determining the levels of heterologous protein accumulation in the C. reinhardtii chloroplast, we compared three transplastomic lines of C. reinhardtii, expressing a codon optimized gfp [11], luciferase [14], or a single chain antibody (hsv8-lsc) [13].
We found that recombinant protein translation was greatly reduced when compared to the endogenous atpA protein, while recombinant mRNAs accumulated quite well when compared to the endogenous atpA mRNA, with a few exceptions. In vitro toeprint and in vivo polysome analyses suggest that poor ribosome association might contribute to the observed reduction in translation but that other steps, such as translation elongation are likely to have a great impact as well. Altogether, our results point to translation as the main step limiting the expression of heterologous proteins in the C. reinhardtii chloroplast.
Materials and Methods
Strains and Culture Conditions
Chlamydomonas reinhardtii wild-type strain 137c (mt+) was obtained from the Chlamydomonas Genetic Center, Durham, NC. Wild type and transformants were grown in 50-ml Tris-Acetate-Phosphate (TAP) medium [18] on a rotary shaker at 23°C at 4500 lux. The composition of TAP medium is: 2.42 g/l Tris-Base, 0.4 g/l NH4Cl, 0.1 g/l MgSO4·7H2O, 0.05 g/l CaCl2·2H2O, 0.108 g/l K2HPO4, 0.054 g/l KH2PO4, 50 mg/l Na2EDTA, 22 mg/l ZnSO4·7H2O, 11.4 mg/l H3BO3, 5.06 mg/l MnCl2·4H2O, 5 mg/l FeSO4·7H2O, 1.61 mg/l CoCl2·6H2O, 1.57 mg/l CuSO4·5H2O, and 1.1 mg/l (NH4)6Mo7O24·4H2O. Final pH was adjusted to 7.0 with glacial acetic acid. All transformants were grown in 50-ml TAP on a rotary shaker at 4500 lux in either continuous light or a 12 h light/12 h dark regime. Cultures were maintained at a cell density of either 1 × 106 cells/ml or 1 × 107 cells/ml for at least 48 h prior to harvest. For this purpose, cell densities were determined three times a day by cell counting in a haemocytometer. The volumes of culture at the determined cell densities that would yield 50 ml of culture at the desired cell densities (106 or 107) were used as inocula of the diluted cultures. To avoid a possible effect on protein/mRNA accumulation of nutrients coming from different volumes of fresh TAP media between samples, cells from all conditions to be used as inocula were centrifuged and resuspended in 50 ml of fresh TAP. Since we had previously determined that C. reinhardtii cells growing exponentially in TAP media duplicate as often as every 6 h, this method allowed for little fluctuation of cell density around the desired value.
Construction of Transgenes and Transformation into C. reinhardtii Chloroplasts
Classical molecular techniques were followed for all DNA and RNA manipulations as described elsewhere [19, 20]. The atpA 5′-UTR was amplified via PCR, using C. reinhardtii genomic DNA as template. The PCR primer complementary to the 5′ end of the atpA 5′-UTR beginning at position −540 relative to the translational start site has the sequence 5′-ggatcccatttttataactgg-3′ and contains a 5′-BamHI restriction site. The PCR primer complementary to the 3′ end of the atpA 5′-UTR begins at position +3 relative to the translation initiation site and has the sequence 5′-catatgaaaaaagaaaaaataaataaaag-3′ and contains a 3′-NdeI restriction site. The resulting 544-bp PCR product was cloned into pCR2.1 TOPO (Invitrogen, Carlsbad, CA, USA) to generate p5atpA. The plasmid, p5atpA, was digested with BamHI and NdeI. The resulting fragment was ligated into BamHI/NdeI partially digested p322 containing codon optimized gfp, hsv8-lsc, or lux coding regions each followed by the rbcL 3′-UTR which were described previously [11, 13, 14]. The three resulting plasmids, termed pAGR, pAHR, and pALR contain the atpA 5′-UTR driving gfp, hsv8-lsc, or lux, respectively, followed by the rbcL 3′-UTR (Fig. 1a). Plasmids pAGR, pAHR, and pALR were co-transformed with plasmid p228 (conferring resistance to spectinomycin) into C. reinhardtii strain 137c(+) chloroplasts by particle bombardment [21].
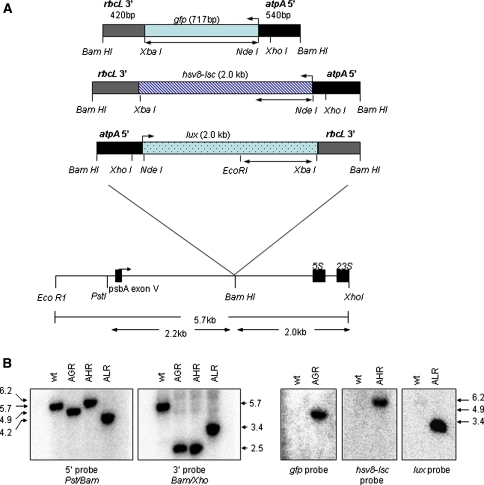
Fig. 1.
Maps of constructs encoding pAGR, pAHR and pALR, and generation of homoplasmic C. reinhardtii lines. a Relevant restriction sites delimiting the atpA 5′-UTR (BamHI/NdeI) from either gfp, hsv8-lsc, or lux coding regions (NdeI/XbaI) and the rbcL 3′-UTR (XbaI/BamHI). The size of each gene is indicated. The site of integration into the C. reinhardtii chloroplast genome of the chimeric genes spans the 5.7-kb EcoRI to XhoI fragment of the inverted repeat region. Double-headed arrows indicate regions corresponding to probes used in Southern analysis. b Wild-type and transgenic DNAs were digested simultaneously with EcoRI and XhoI and subjected to Southern blot analysis. Filters were hybridized with the radioactive probes indicated
Southern and Northern Blots
Southern blots, Northern blots, and 32P labeling of DNA for use as probes were performed using methods described [19]. Radioactive probes used for southern blots included the 2.2-kb BamHI/PstI fragment of p322 (probe 5′ p322) and the 2.0-kb BamHI/XhoI fragment of p322 (probe 3′ p322). The gene-specific probes included the NdeI/XbaI fragment of the gfp coding, an Nde/Xba fragment of a derivative of hsv8-lsc, hsv8scfv, which contains complete homology to the light chain and heavy chain variable regions of hsv8-lsc and an EcoRI/Xba fragment of lux (Fig. 1b). Additional radioactive probes used in Northern blot analysis included a 916-bp EcoRI fragment from p228 containing a portion of the C. reinhardtii 16S ribosomal RNA and a 540-bp atpA 5′-UTR BamHI/NdeI fragment. Northern and Southern blots were visualized utilizing a Packard Cyclone Storage Phosphor System equipped with Optiquant software.
Normalization and Analysis of mRNA Visualized on Northern Blots
Signals were visualized following hybridization with the gene-specific probes. Using Optiquant software, total digital light units (DLU) were measured for regions of the same area in each lane of the blot as well as an arbitrary space for background subtraction. Each blot was then stripped and re-hybridized with the 16S probe. Total DLU were measured and background subtracted. The 16S message thus served as an internal control to which chimeric signals were normalized. Utilization of the atpA 5′-UTR probe allowed a direct comparison between gfp mRNA levels and the endogenous atpA message as the sizes of these two mRNAs were sufficiently different to allow their resolution on a Northern blot while those of hsv8-lsc and lux were not.
Isolation of Polysome-Associated mRNA
Cells were grown in 250 ml of TAP in 1-l flasks to either 1 × 106 or 1 × 107 cells/ml on a rotary shaker at 23°C at 4500 lux. For each time point, polysomal RNA was isolated from cells as described previously [20], with the addition of an equal volume phenol:chloroform extraction prior to the ethanol and sodium acetate precipitation.
Protein Expression and Western Blotting
Purified C. reinhardtii and E. coli expressed proteins were prepared and quantified as described [11, 20]. Western blots were performed as described. For westerns, rabbit anti-rbcL, atpA, GFP (Clonetech, Mountain View, CA, USA), or LUX (gift of Susan Golden) primary antibodies were used prior to an alkaline phosphatase-labeled goat anti-rabbit secondary antibody (Sigma, St Louis, MO, USA). HSV8-lsc proteins were detected using a murine anti-Flag primary antibody (Sigma, St Louis, MO, USA) and an alkaline phosphatase conjugated goat anti-mouse secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Pulse Labeling and Immunoprecipitation of 14C-Labeled Proteins
C. reinhardtii cells were grown to the designated final density in TAP medium. Four hours prior to harvest, cells were re-suspended in 50 ml of TAP-HCl (acetate-deficient medium). After shaking for 2 h and 50 min, cycloheximide was added to a final concentration of 10 μg/μl and the cells incubated for a further 10 min. 14C acetate was then added to a final concentration of 1 μCi/ml. The reaction was quenched after 1 h by the addition of sodium acetate to a final concentration of 50 mM. Cultures were harvested immediately and pellets frozen in liquid N2. Proteins were isolated from whole cells by sonication using a microtip at 30% power. Cells were pulsed three times for 20 s in Buffer A (50 mM Tris pH 7.5, 150 mM NaCl, 0.1% Tween 20, 1 mM PMSF, Complete protease inhibitor-Roche). Soluble proteins were separated from membrane fractions by centrifugation at 48,000×g in a TLA-100 Beckman rotor for 45 min at 4°C. The membrane fractions were resuspended in Buffer B (0.5 mM Tris pH 7.5, 10% SDS, 10% 2-mercaptoethanol). Proteins were quantified as described previously [11, 20]. For analysis of atpA synthesis, 20 μg of membrane or soluble proteins were electrophoresed through 4–12% Bis–Tris NuPAGE gels (Invitrogen). After staining with coomassie, the gels were treated with Enhance (NEN life Science, Boston, MA, USA) for 1 h, washed in water for 45 min then dried overnight on cellulose acetate sheets. For analysis of HSV8-lsc synthesis, recombinant protein was immunoprecipitated from 450 μg of 14C-labeled total soluble proteins from strain AHR, using Anti-Flag M2-Agarose Affinity Gel (Sigma) with >90% efficiency, as determined by western analysis. For analysis of GFP synthesis, recombinant protein was immunoprecipitated from 1000 μg of 14C-labeled total soluble protein from strain AGR, using the Immunoprecipitation Kit-Protein A (Roche, Palo Alto, CA, USA) with approximately 50% efficiency. Proteins were again separated and visualized as described above. Dried gels were exposed to phosphor imaging screens for 1 week. Synthesized products were visualized and quantified using the Pakard Cyclone Storage Phosphor System equipped with Optiquant software. Total DLU were measured and background subtracted. Counts measured for atpA from the soluble and membrane fractions were combined. Synthesis of recombinant proteins relative to the endogenous atpA protein was calculated (Table 1) by multiplying the counts obtained by [(1/efficiency of immunoprecipitation) × (MW atpA/MW recombinant protein)] ÷ μg total soluble protein.
Table 1.
Effect of light regime on recombinant protein synthesis relative to atpA (%)
| Cell density | Light regime | GFP/atpA (%) | Hsv8-lsc/atpA (%) |
|---|---|---|---|
| 106 | ED | 12.5 | 2.4 |
| 1 h | 8.1 | 4.1 | |
| EL | 5.5 | 0.6 | |
| L | 1.1 | 0.4 | |
| 107 | ED | 12.5 | 0.4 |
| 1 h | 30.8 | 1.2 | |
| EL | 34.1 | 1.1 | |
| L | 6.6 | 0.4 |
In Vivo Protein Turnover
Turnover of heterologous and endogenous proteins was assayed by growing C. reinhardtii strains expressing GFP, HSV8-lsc, or LUX to 1 × 106 cells/ml at 4500 lux. After maintenance of cultures at this density for 48 h, chloramphenicol was added to a final concentration of 100 μg/ml to inhibit chloroplast translation [22]. Samples were then pulled at times indicated and proteins were isolated and visualized as described for Western blotting. Membrane proteins for the visualization of atpA were prepared as described elsewhere [11]. The effectiveness of chloramphenicol at blocking chloroplast protein synthesis was determined on the AGR strain (−CAM and +CAM 15-min treatments), from which polysome isolation was done as described.
Extension Inhibition (Toeprint) Assays
Linear fragments containing the T7 polymerase promoter cloned upstream of the atpA 5′-UTR plus 120 nt of either atpA or GFP coding regions were used as templates in in-vitro transcription reactions containing 40 mM Tris-HCl pH 8, 2 mM spermidine, 20 mM MgCl2, 4 mM rNTPs, 20 U RNAse inhibitor (SUPERase-In, Ambion, Austin, TX), 10 mM DTT, and 20 U T7 RNA polymerase (Roche). The reactions were carried out at 37°C for 90 min. Transcribed RNAs were phenol:chloroform-extracted and precipitated with 0.1 volume of 3 M sodium acetate pH 7.5 and 5 volumes of 100% ethanol. Twenty picomoles of the following oligos: 5′-gcaataccgtcacctacttgg-3′ (for atpA) and 5′-ccataagttgcgtcacc-3′ (for gfp) were labeled with T4 polynucleotide kinase (20 U, New England Biolabs, Ipswich, MA) according to the manufacturer’s instructions. Labeled oligos were purified from unincorporated radioactive isotope using Quick Spin Sephadex G-25 columns (Roche). Toeprint experiments were done using 20 nM in vitro transcribed RNAs and 0.5 picomol of labeled oligos. The oligos were annealed to the RNAs at 60 °C for 3 min and cooled down on ice. The binding reactions were done in 10 mM Tris-HCl pH 7.5, 60 mM ammonium acetate, 10 mM MgCl2 and 6 mM β-mercaptoethanol in the presence or in the absence of commercial formyl-Methionine tRNA (10 pmol) and C. reinhardtii chloroplast 30S subunits (100 or 200 nM) isolated according to previous reports [23].The S30 fractions minus ribosomes corresponded to the top fraction of sucrose density gradients, which contains lighter cellular components (e.g., translation factors) but lacks heavier material, such as ribosomal subunits, ribosomes, or polysomes. This S30 minus ribosome fraction contains all chloroplast soluble components but is depleted of ribosomes, 30 and 50S ribosomal subunits and pre-ribosomal particles, which sediment at higher density fractions [23, 24]. Incubations were carried out at 37°C for 30 min. Primer extensions were done with 20 U Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) at 37°C for 10 min. The reactions were stopped with 6 μl of 95% formamide, 0.5 mM EDTA 0.1% xylene cyanol and 0.1% bromophenol blue and loaded in 8% acrylamide-TBE-urea gels. For sequencing reactions, the strands of 2.5 μg of supercoiled DNA from both constructs were separated using 0.5 mM EDTA and 200 mM NaOH for 5 min at room temperature. DNAs were precipitated in 1.5 volumes of ammonium acetate and 7.5 volumes of 100% ethanol, incubated at −80°C for 15 min and spun down at 12,000×g for 30 min. After washing the pellets with 70% ethanol, they were resuspended in 6 μl H2O and incubated with 1 picomol labeled oligo in Sequenase reaction buffer (USB, Cleveland, OH). The samples were annealed at 60°C for 10 min and cooled down at room temperature over 30 min. Primer extension reactions were done using Sequenase™ Version 2.0 DNA Sequencing Kit according to the manufacturer’s instructions at room temperature for 5 min, stopped with dideoxynucleotides and further incubated at 37°C for 5 min. Four microliters of formamide-containing dye were used as stop solution. Half of the reactions were separated side by side with the toeprint reactions in 8% acrylamide-TBE-urea gels.
RNAse Protection Assays
RNA probes antisense to the atpA 5′-UTR and the 5′ end of atpA coding region were made as follows: DNAs were amplified by PCR using forward oligos with a BamHI site and reverse oligos with an EcoRI site proceeded by the T7 RNA polymerase promoter as follows: for probe 1, the forward oligo was 5′-ccgaattccctgtggacgtcccccccttcccc-3′ and, for probe 2, 5′-gggaattcgatcttaccactcactatttttgttg-3′. For both the probes, the reverse oligo was 5′-ggggatcctaatacgactcactatagggcaataccgtcacctacttgg-3′. PCR products were digested with EcoRI and BamHI and cloned into pUC18. All clones were linearized with BamHI. DNA templates were in vitro transcribed using T7 RNAse polymerase (Roche) according to the manufacturer’s instructions using α32P-UTP as the radioactive ribonucleotide. Full-length RNA probes were gel-purified from 8% acrylamide-TBE-urea gels in RNA elution buffer: 0.5 M sodium acetate, 0.5% SDS and 1 mM EDTA and precipitated with 0.4 M ammonium acetate and 2 volumes of 100% ethanol.
Twenty-five micrograms of C. reinhardtii total RNAs was resuspended in 20 μl RNAse protection assay hybridization buffer (40 mM PIPES pH 6.4, 1 mM EDTA pH 8.0, 0.4 M NaCl and 80% formamide) containing 1 × 105 cpm of the corresponding RNA probe. RNAse protection assays were done as described by Beligni et al. [25]. The intensity of the bands obtained for the gfp mRNA using both probes was determined by phosphoimaging and corrected by the amount of uridines present in its corresponding protected fragments compared to the amount of uridines in the protected fragments of the atpA mRNA. Thus, for probe 1, the signal of the gfp mRNA was multiplied by 1.48 and, for probe 2, by 2.1 to reflect the difference in the uridine content.
Results
Southern Blot Analysis of Transgenic C. reinhardtii
For this study, we examined the expression of three different codon-optimized genes to ensure that pleiotropic effects of any one coding region did not inadvertently influence our results. The three plasmids, pAGR, AHR, and ALR are diagrammed in Fig. 1. Expected integration occurs between the psbA and 5S rRNA genes, within a region that exists as two inverted copies in opposite locations on the C. reinhardtii chloroplast genome [26]. For comparison, all three plasmids contain the same 5′ (A, atpA) and 3′ (R, rbcL) untranslated regions (UTRs). The reason for choosing the atpA 5′ was based on the ability of this promoter/UTR to successfully drive expression of a wide range of transgenes in C. reinhardtii chloroplast at moderate levels. In a previous report, we demonstrated that the 5′ promoter/UTR was responsible for defining the levels of accumulation of transgenic proteins in the C. reinhardtii chloroplast, while the identity of the 3′-UTR had little or no effect, as long as a 3′-UTR was present [27]. For a large number of transgenes tested, the combination atpA 5′/rbcL 3′ resulted in transgenic protein accumulation to levels similar to those of the atpA 5′/atpA 3′ combination ([27], also unpublished data). The coding regions were composed of a codon optimized gfp (G), hsv8-lsc (H), or lux (L), all described previously [11, 13, 14]. For transformation, C. reinhardtii cells were co-transformed with plasmid p228 conferring ribosomal spectinomycin resistance, and one of the reporter plasmids pAGR, pAHR, or pALR. Primary transformants capable of growth on spectinomycin were screened by Southern blot analysis for integration of the chimeric gene, and positive transformants were taken through additional rounds of selection to isolate homoplasmic lines. After confirmation of recombinant protein expression by Western blot analysis, one homoplasmic line expressing the highest level of recombinant protein from each transformation was selected for further analysis. Accumulation of recombinant protein typically differs by no more than 3- to 4-fold among independent transformants (S.F. unpublished observations). Construct orientations with relevant restriction sites at the site of integration into the C. reinhardtii chloroplast genome are shown in Fig. 1a. Analysis of many different transgenes inserted at the BamHI site of vector p322 has shown no detectable difference in expression as a result of orientation of the transgene with respect to the psbA gene ([27], and unpublished results).
Genomic DNA isolated from wild type and each transformant was simultaneously digested with EcoRI and XhoI, resolved on agarose gels and subjected to Southern blot analysis (Fig. 1b). Hybridization with a 32P-labeled probe specific to the 5′ end of p322 identified the predicted 5.7-kb band in the wt lane, as well as the predicted 4.9-, 6.2-, and 4.2-kb band in strains AGR, AHR, and ALR, respectively. Hybridization with a 32P-labeled probe specific to the 3′ end of p322, again identified the 5.7-kb band in the wt lane, and the predicted 2.5-kb band for both AGR and AHR and a 3.4-kb band for ALR. Failure of either the 5′- or 3′-specific probes to hybridize to a wt band of 5.7 kb in AGR, AHR, or ALR demonstrates that each strain is homoplasmic. Hybridization with gene-specific probes shown in Fig. 1a, identified the predicted bands of 4.9, 6.2, and 3.4 kb in strains AGR, AHR, and ALR, respectively. As expected, there was no signal with any of the gene-specific probes in the wild-type lanes.
The Effect of Cell Density and Light on Recombinant Protein Accumulation
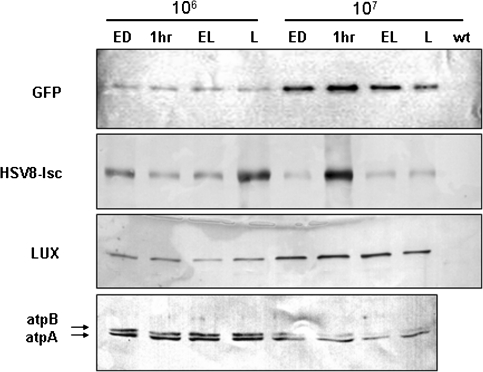
Western blot analysis was used to examine the effect of light cycling on the accumulation of the three recombinant proteins in C. reinhardtii. Proteins were extracted at the times indicated, including wild-type cells as a control. We have previously demonstrated that cell culture density affects recombinant protein accumulation. This effect is mediated also by light rather than density per se. This was shown in a previous report, in which Chlamydomonas reinhardtii GFP transgenic strains maintained at 106 cells/ml and 450 lux produced practically identical amount of protein than the same strains maintained at 107 cells/ml under 4500 lux. In contrast, the same transgenic strains kept at 106 and 4500 lux, accumulated roughly 10% as much GFP as cells kept in the two conditions mentioned above. This fact was repeatedly seen with other strains (unpublished work), and as a whole indicates that the high cell density acts to reduce light intensity within the growing culture, in essence creating a low light environment [11]. Consequently, though we will refer to cultures as grown at 106 and 107 cells/ml, we wish to point out that light has a profound effect on the variations we observe. Cells from each strain were grown at both cell densities in either continuous light or cycled through 12 h of light followed by 12 h of dark. Proteins were isolated from cycled cultures at the end of the dark (ED), 1 h into the light (1 h) and at the end of the light (EL), as well as from cultures grown in continuous light (L). One microgram of total soluble protein (tsp) from strain AGR, 20 μg from strain AHR, and 5 μg from strain ALR and wt were separated by electrophoresis through 12% SDS/PAGE gels and transferred to nitrocellulose. Westerns were carried out with anti-GFP, anti-Flag (HSV8 detection), anti-LUX, or anti-atpA antibodies. Wild-type cells showed no signal with any of the recombinant protein-specific antibodies, as expected (Fig. 2). Wild-type cells show accumulation of endogenous atpA protein to be uniform throughout the light cycle at a given cell density. However, accumulation of atpA was approximately 3-fold higher in cultures grown at 106 when compared to 107 cells/ml (Fig. 2, bottom panel), as determined by titration on Western blots. In addition, the accumulation of atpA protein was not significantly altered by the presence of the chimeric proteins in any of the three recombinant strains (not shown).
Fig. 2.
Effect of light regime on recombinant protein accumulation. Equal loading of protein was determined by Bradford assay. Protein accumulation was assayed by Western blot from the three transgenic strains grown at 1 × 106 and 1 × 107 cells/ml for samples taken at the end of the dark (ED), 1 h into the light (1 h) and at the end of the light (EL), as well as continuous light (L). One microgram of total soluble protein was assayed for GFP from the AGR strain, 20 µg for HSV8 from the AHR strain and 5 µg for both LUX (from the ALR strain) and atpA (from wild type). The blots are representative for three independent experiments
The accumulation of GFP and LUX proteins also appeared to be constant throughout the light/dark cycle, showing less than a 2-fold difference between time points. For these recombinant proteins, accumulation was higher in cultures grown at 107 cells/ml, where cells accumulated 5-fold more GFP and 3-fold more LUX than cultures grown at 106 cells/ml (Fig. 2). Conversely, the overall accumulation of HSV8-lsc protein was slightly higher at low cell density growth, with a significant fluctuation during the light–dark cycle, particularly at high cell density. For example, a 5-fold increase in accumulation of HSV8-lsc was reproducibly observed at 1 h into the light at high density (Fig. 2).
In Vivo Turnover of Recombinant Proteins
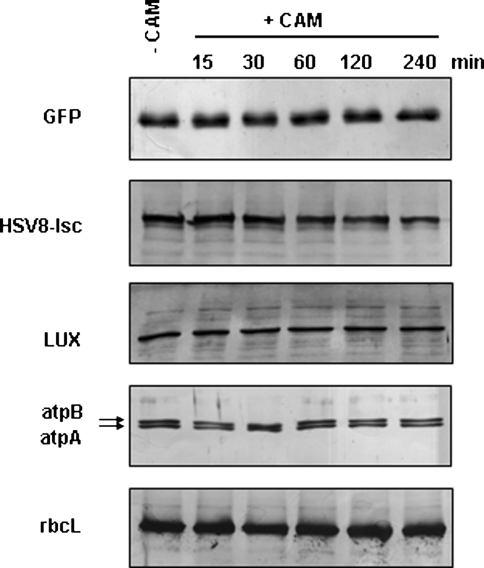
We were interested in determining the turnover of each of our recombinant proteins relative to endogenous, chloroplast encoded, C. reinhardtii proteins. The transgenic strains AGR, AHR, and ALR were grown to 1 × 106 cells/ml at 4500 lux. With the purpose of inhibiting chloroplast protein synthesis, chloramphenicol, a broad-spectrum antibiotic widely used as a blocker of prokaryotic translation elongation, was added to the strains under study. Aliquots were removed at times ranging from 15 min to 4 h. Proteins were prepared for Western blot analysis, and, in addition to examining GFP, HSV8-lsc, and LUX proteins, strain AGR was analyzed for turnover of atpA and rbcL proteins as well. As can be seen in Fig. 3, GFP and LUX exhibit a slight decrease over the 4-h time course, while the two endogenous proteins, atpA and rbcL show no discernable decrease in protein accumulation over the same time period. The level of HSV8-lsc clearly decreased over 4 h, and based on Western blot titration of HSV8-lsc, we estimate this reduction to be roughly 3-fold. The effectiveness of chloramphenicol to block translation in this particular experiment was confirmed by polysome isolation (not shown).
Fig. 3.
Stability of recombinant versus endogenous proteins. Transgenic strains AGR, AHR, and ALR were grown to 1 × 106 cells/ml at 4500 lux. Chloramphenicol was added to block chloroplast translation and aliquots were removed at times indicated (in minutes). Proteins were prepared as described in “Methods” and separated on 12% SDS PAGE. Western blots were carried out using the primary antibodies indicated. Five micrograms of total soluble protein was assayed for GFP, LUX, atpA, and rbcL; 20 µg was assayed for HSV8. The blots are representative for three independent experiments
Chimeric Transcripts Accumulate Maximally Under Conditions of Low Light
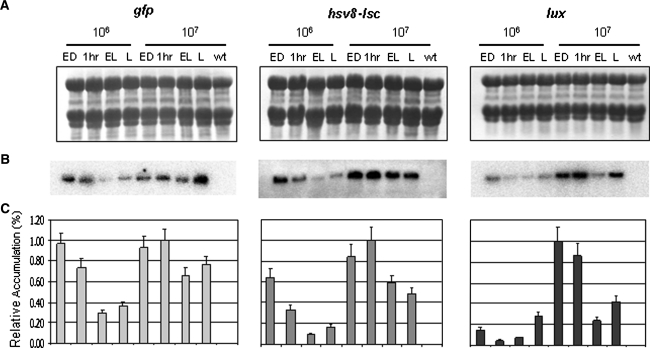
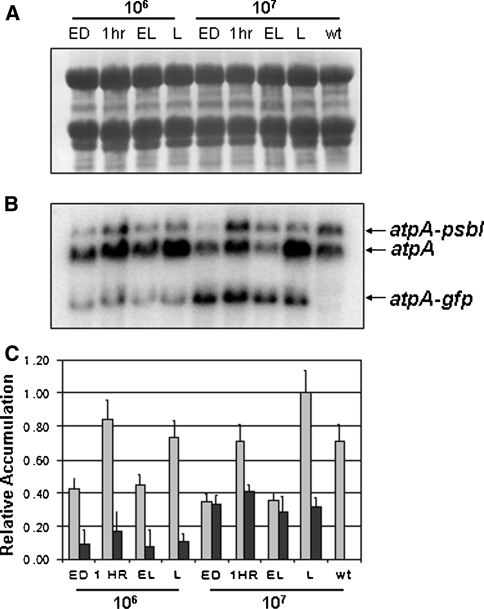
Northern blot analysis was used to determine the effects of light cycling and intensity on the accumulation of recombinant mRNAs. Ten micrograms of RNA was resolved on denaturing agarose gels and subjected to Northern blot analysis. As a control, 10 μg of total RNA isolated from wild-type cells maintained at 1 × 107 cells/ml under continuous light was also resolved on each Northern blot. Uniform transfer of each RNA sample was confirmed by methylene blue staining of the membrane (Fig. 4a). Blots were hybridized with a 32P-labeled probe specific to each chimeric coding region (Fig. 4b). Unique bands of 1.5 kb for strain AGR and 2.5 kb for both AHR and ALR were visualized, while no signal was observed in any of the wild-type samples. For normalization purposes, each blot was stripped and re-probed with a 16S rRNA probe (not shown). Signals were quantified and normalized and the counts obtained were expressed as percentages of accumulation relative to the highest signal obtained for the chimeric transcript on each blot (Fig. 4c). All three strains accumulate higher levels of recombinant transcripts when cultures are grown at higher cell densities whether in continuous light or in a light/dark cycle. In general, for each of the transgenic strains tested, heterologous mRNA accumulation at lower cell densities (higher light) was maximal at the end of the dark phase, followed by a gradual decrease in accumulation throughout the light phase. At 107 cells/ml (lower light), however, strains AGR and AHR showed a slight increase in chimeric mRNA accumulation 1 h into the light (Fig. 4c).
Fig. 4.
Effect of light regime on recombinant transcript accumulation. Ten micrograms of total RNA was assayed for each time point from strains AGR (gfp), AHR (hsv8-lsc), ALR (lux), and wild type at the indicated cell densities. Wild-type RNA was isolated from cells grown at 1 × 107 cells/ml in continuous light. a Uniform transfer of RNA to the membrane was determined by methylene blue staining. b Blots were hybridized with a 32P-labeled probe specific to each chimeric coding region. c Relative accumulation was quantified and normalized as described in “Methods” and reported as percentages of accumulation relative to the highest signal obtained on each blot. The blots are representative for three independent experiments
GFP Transcript Accumulation Relative to atpA
In order to directly compare the accumulation of recombinant mRNAs relative to the endogenous atpA mRNA, we focused on strain AGR encoding GFP. The endogenous atpA message is part of a polycistronic mRNA whose primary transcripts are 2.5 and 2.1 kb [28]. Because the hsv8-lsc and lux transcripts are also approximately 2.5 kb, a single radioactive probe corresponding to the atpA 5′-UTR cannot resolve these recombinant mRNAs from the endogenous atpA mRNA by Northern blot analysis. The gfp mRNA is approximately 1.7 kb in size and can easily be separated from the atpA message on Northern blots.
Ten micrograms of RNA from strain AGR was resolved on denaturing gels and transferred to nylon membrane. Uniform transfer of each RNA sample was confirmed by methylene blue staining of the membrane (Fig. 5a). Hybridization with a BamHI/NdeI 32P-labeled probe specific to the atpA 5′-UTR identified bands corresponding to the 2.5 and 2.1 kb endogenous atpA transcripts, as well as an additional band of 1.7 kb corresponding to the atpA–gfp transcript (Fig. 5b). As expected, RNA isolated from wild-type cells showed only the endogenous atpA transcripts. In the gfp transgenic line, the total amounts and ratios of the atpA and atpA–psbI transcripts varied considerably between different conditions, being these variations reproducible between independent experiments (not shown). In the same transgenic line, normalized counts, expressed as a percentage of the highest signal quantified on the blot (Fig. 5c), showed that gfp mRNA accumulates to anywhere from 35% (L) to 95% (ED) of the level of the endogenous atpA mRNA at high density growth. At low cell density, however, gfp mRNA levels reach only 5–18% the level of atpA mRNA (Fig. 5c).
Fig. 5.
Effect of light regime on endogenous and recombinant transcript accumulation. Ten micrograms of total RNA was isolated from strain AGR grown under the conditions indicated. a Uniform transfer of RNA to the membrane was determined by methylene blue staining. b Blots were hybridized with a 32P-labeled probe specific to the atpA 5′-UTR to visualize the 2.5-kb dicistronic atpA–psbI mRNA, the 2.1-kb monocistronic atpA mRNA and the 1.7-kb chimeric gfp mRNA. c Relative accumulation was determined as described in “Methods” and is reported as percentages of accumulation relative to the highest signal obtained from the blot. Light bars represent atpA mRNA and dark bars represent gfp mRNA accumulation. The blots are representative for three independent experiments
Synthesis of Recombinant Proteins Compared to Endogenous Proteins
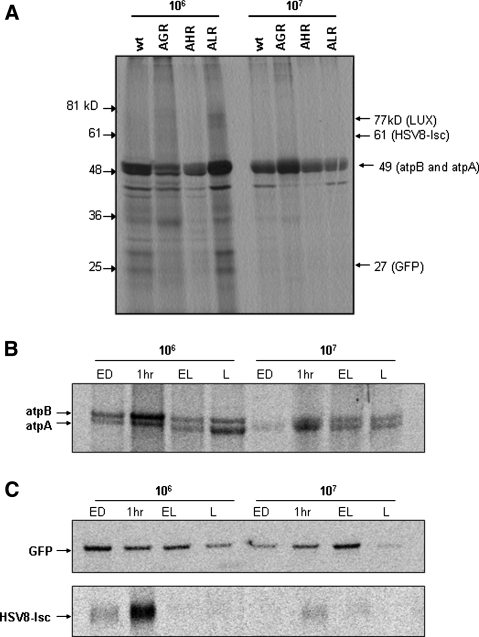
Analysis of protein synthesis by [14C] pulse-labeling failed to detect synthesis of any of the three recombinant proteins, even when the labeling time was as long as 1 h (Fig. 6a), whereas the 56-kDa endogenous atpA protein was easily visualized in membrane fractions when cells were labeled for as little as 15 min. In order to visualize recombinant protein synthesis, we labeled cells for 1 h and immunoprecipitated [14C]-labeled GFP and HSV8-lsc from 1000 (GFP) or 450 μg (HSV8-lsc) of tsp. AtpA synthesis was also measured by [14C] labeling of strain AGR for 1 h, except 20 μg tsp, from both membrane and soluble fractions, was resolved directly on SDS PAGE. Figure 6b shows the membrane proteins with newly synthesized atpA and atpB indicated. Thus, we were able to directly compare the levels of atpA, GFP, and HSV8-lsc synthesis by normalizing their synthesis relative to atpA. Synthesis of atpA was quantified using Optiquant software directly from SDS PAGE and expressed as digital light units (DLU)/μg tsp (Table 1). Immunoprecipitated recombinant proteins were quantified subsequent to electrophoresis and the resulting digital light units (DLU) were normalized to account for differences in the efficiency of immunoprecipitation and in the size of the recombinant protein and, hence, for the amount of [14C] label incorporated. The anti-LUX antibody was a gift and, therefore, we could not obtain sufficient antibody to carryout immunoprecipitations of the LUX protein.
Fig. 6.
Effect of light regime on endogenous and recombinant protein synthesis. Synthesis of all newly synthesized proteins were quantified using optiquant software and equal loading of protein was determined by Bradford assay. a Each strain was grown in continuous light and labeled with [14C] acetate for 1 h prior to harvest. 50 μg of total soluble protein from each strain was separated via 12% SDS PAGE, and exposed to phosphor imaging screens for visualization of newly synthesized proteins. No bands were detected in the 27-, 61-, or 73-kD regions corresponding to the GFP, HSV8-lsc, and LUX proteins, respectively. b Strain AGR was labeled for 1 h with [14C] acetate at each time point indicated. 20 μg of protein from the membrane fractions was separated through 4–12% bis–tris NuPAGE gels and exposed to phosphor imaging screens to visualize atpA synthesis. c GFP and HSV8-lsc were immunoprecipitated from 1 to 0.5 mg, of total soluble protein prepared from strains AGR and AHR, respectively, labeled with [14C] acetate for 1 h. Proteins were separated by electrophoresis through 4–12% bis–tris NuPAGE gels and exposed to phosphor imaging screens. The blots are representative for three independent experiments
After normalization to atpA synthesis (Fig. 6b), GFP and HSV8-lsc syntheses were expressed as a percentage of atpA synthesis (Table 1). Synthesis of atpA was approximately three to eight times higher when cells were grown at lower when compared to higher cell density conditions and showed a marked induction at the onset of light, regardless of the culture density. Recombinant protein synthesis was considerably lower than that of atpA. Synthesis of GFP was, at best, approximately one-third that of atpA, while HSV8-lsc synthesis was lower still, rising only to about 4% of the level of synthesis of endogenous atpA. Similar to atpA synthesis, however, synthesis of HSV8-lsc was induced at the beginning of the light period in both cell density conditions, and was higher overall when cells were grown at low cell density (higher light). Alternatively, GFP synthesis was highest at high cell density (lower light) and failed to exhibit an induction in synthesis at lower cell densities, remaining fairly constant throughout the light phase (Table 1).
Association of gfp mRNA with Ribosomes
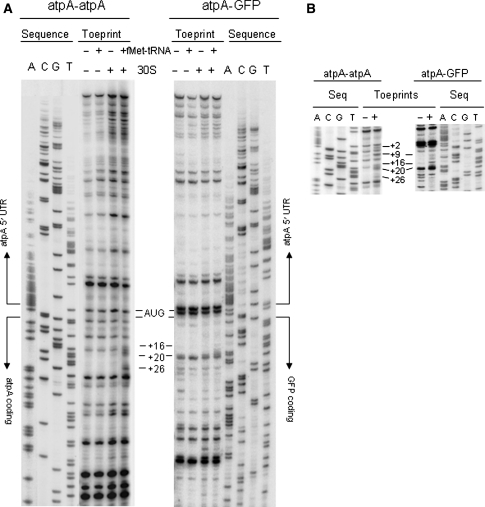
Based on our findings that gfp message can accumulate to levels comparable to the endogenous atpA mRNA, yet its synthesis remains markedly lower than that of atpA, we were interested in determining the level of gfp mRNA associated with ribosomes relative to the atpA mRNA. In order to test the ability of the gfp coding region driven by atpA 5′-UTR to form translation initiation complexes relative to the endogenous atpA, we performed in vitro primer extension inhibition (toeprint) assays. The promoterless atpA 5′-UTRs cloned upstream of the first 120 nt of either the gfp or atpA coding regions were in vitro transcribed and primer extended in the presence or absence of C. reinhardtii chloroplast 30S subunits, fMet-tRNAs or both. Both constructs produced toeprints at positions +16 to +20 (Fig. 7a, +30S +tRNA), which correspond to ternary complexes (mRNA-30S-fMet-tRNA) in which the initiator AUG (+1) is base-pairing with the fMet-tRNA and positioned in the P site of the 30S subunit. However, the intensity of this toeprint was more than 5-fold lower for the atpA–gfp mRNA, as quantified by densitometry and normalized to the full-length product. This is especially remarkable for the +16 and +17 bands, which are the most commonly reported toeprints obtained with bacterial 30S subunits [29, 30]. In addition, one extra toeprint was observed at +26 for the atpA–atpA construct that was not present in the atpA–gfp mRNA. The addition of the fractions that contain chloroplast soluble components (S30 fractions minus ribosomes) to the toeprint reactions, produced no visible modifications to the toeprints obtained with the 30S subunit and fMet-tRNA alone (data not shown).
Fig. 7.
Toeprint analyses of translation initiation complexes on atpA and atpA–gfp RNAs. In vitro transcribed atpA–atpA (left panels) and atpA–gfp (right panels) RNAs (20 nM) were extended from [γ-32P] ATP-labeled oligos that anneal 120-nt downstream of the initiator AUGs, either alone or in the presence of tRNAMet, 30S subunit or both. Sequencing reactions were run alongside. The treatments are indicated at the top of each panel. The positions of the toeprints respective to the initiator AUG (+1) are indicated with lines on the side. a Toeprint reactions using 100-nM 30S subunit, b Inset showing the toeprint area of experiments similar to (a) with 200-nM 30S subunit. The figure shows representative autoradiographs obtained from three independent experiments
When the amount of C. reinhardtii 30S subunit was doubled to 200 nM, the same +16–20 and +26 toeprints were obtained for the atpA–atpA construct, as well as two extra toeprints at positions +2 and +9, the last of which has been usually attributed to the formation of binary complexes (mRNA-30S) [31]. The +16–20 toeprints improved by 2-fold for the atpA–gfp construct when compared to 100 nM 30S subunit for the same mRNA, but were still 3-fold lower than those of the atpA–atpA mRNA (Fig. 7b). It is interesting to note that the presence of the first 120 nt of the gfp coding region downstream of the atpA 5′-UTR affected the pattern in which the reverse transcriptase paused on the 5′-UTR compared to the endogenous atpA (Fig. 7a, −30S −tRNA). As an example, the strongest pause in the AG construct was a doublet on positions −2 and +1, right on the initiator AUG.
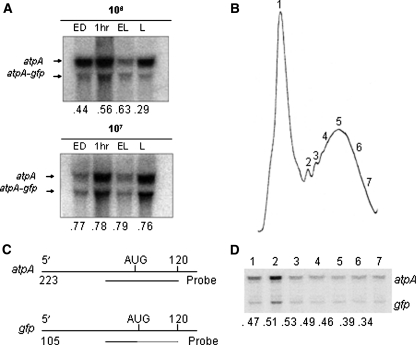
The ability of the transgenic gfp mRNA to associate to ribosomes in vivo was also determined by polysome analysis. Again, the size difference between the atpA and gfp transcripts allowed us to quantify and directly compare polysome-associated gfp mRNAs versus atpA mRNAs by utilizing the atpA 5′-UTR as probe. Polysomes were isolated via sucrose density centrifugation from strain AGR and kept as a single pool. Polysomal RNA was isolated for each time point and 20 µg was resolved on denaturing agarose gels and subjected to Northern blot analysis. Hybridization with a 32P-labeled probe specific to the atpA 5′-UTR identified bands corresponding to the endogenous atpA polysome-associated transcripts, as well as a band corresponding to the atpA–gfp polysome-associated message (Fig. 8a). Signals obtained from Northern blots were quantified and normalized. Counts obtained from gfp polysome-associated mRNAs were directly compared with counts from endogenous atpA polysome-associated mRNAs from the same sample. Polysome-associated gfp mRNAs accumulate to approximately 80% of polysome-associated atpA mRNAs when cells are maintained at higher densities whether in a light–dark cycle or in continuous light (Table 2). This ratio drops to between 30 and 60% when cells are maintained at lower cell densities (high light).
Fig. 8.
Association of atpA and gfp mRNAs to polysomes. a Twenty micrograms of total polysomal RNA isolated from strain AGR at each time point indicated was subjected to Northern blot analysis. Blots were hybridized with a 32 P-labeled probe specific to the atpA 5′-UTR, which detects the endogenous atpA transcripts as well as the chimeric atpA–gfp mRNA. b Polysomes isolated from cells grown to 1 × 106 cells/ml were separated by size by sucrose gradient centrifugation. RNAs were isolated from the fractions indicated [1–7]. c A probe antisense to the atpA 5′-UTR was designed in such a way that the atpA and gfp mRNAs would yield bands of different sizes upon digestion of the probe-RNA hybrids with a ribonuclease A + T1 mixture by RNAse protection assay (end products are shown as full lines, digested fragments as dotted lines, number of nucleotides are indicated on the left). d Results of RNAse protection assays using the probe depicted in (c) and RNAs coming from fractions 1–7 from the polysome profile shown in (b). The numbers below each panel indicate the percentage of probe protected by the gfp mRNA compared to the atpA mRNA. These values were determined by quantifying the signal intensity of each band and correcting the gfp signal to the atpA signal by the number of uridines in each protected band (see “Methods”)
Table 2.
mRNA accumulation, polysome association, and the ability to support protein synthesis differ significantly for recombinant and endogenous proteins
| Cell density | Light regime | mRNA accumulation GFP/atpA (%) |
Polysome-associated mRNA GFP/atpA (%) |
Protein synthesis GFP/atpA (%) |
|---|---|---|---|---|
| 106 | ED | 21.6 | 43.8 | 12.5 |
| 1 h | 19.9 | 56.0 | 8.1 | |
| EL | 17.4 | 63.4 | 5.5 | |
| L | 14.7 | 28.5 | 1.1 | |
| 107 | ED | 96.3 | 77.5 | 12.5 |
| 1 h | 57.3 | 77.7 | 30.8 | |
| EL | 80.8 | 79.3 | 34.1 | |
| L | 31.9 | 75.5 | 6.6 |
Signals obtained for GFP total mRNA accumulation (Fig. 4) are expressed as percentages of total atpA mRNA accumulation for each corresponding time point. Signals obtained from GFP mRNA associated with polysomes (Fig. 8) are expressed as percentages of atpA mRNA associated with polysomes for each corresponding time point. The final column shows GFP synthesis expressed as a percentage of atpA synthesis for each corresponding time point (Fig. 5). Values are the mean of three independent experiments. Standard deviations (SDs) were <15% of each mean value
In an effort to determine how polysome pools differed between the two mRNA types, polysome pools from AGR cells were separated by size on sucrose density gradients (Fig. 8b). Polysomal RNA was isolated from all fractions and 25 µg of samples 1–7, indicated on the polysome profile, was used in RNAse protection assays using the probe depicted in Fig. 8c. The fragments protected from ribonuclease degradation due to the formation of double-stranded RNA hybrids between the probe and the polysome-associated mRNAs are smaller for the gfp mRNA, allowing for quantification of transgenic RNAs associated to polysomes relative to endogenous atpA mRNAs associated to polysomes. As shown in Fig. 8d, the amount of transgenic gfp mRNAs in polysomes was always lower than the amount of endogenous atpA mRNAs. The percentage of the gfp to atpA signal varied moderately (34–54), with heavier polysomes being slightly at disadvantage for the atpA–gfp chimeric mRNA, as indicated at the bottom of each panel on Fig. 8d.
Discussion
The purpose of this study is to identify some of the factors that impact recombinant protein accumulation in the C. reinhardtii chloroplast, with the purpose of designing improved strategies for heterologous protein expression. We monitored three C. reinhardtii transplastomic strains expressing chloroplast codon-optimized green fluorescent protein (GFP) [11], a large single chain antibody (HSV8-lsc) [13], and bacterial luciferase (LUX) [14], whose accumulation ranged from 0.1 to 1% of total soluble protein. The three transgenes were driven by the endogenous C. reinhardtii atpA promoter and 5′ untranslated region (UTR), and the rbcL 3′-UTR.
Eberhard et al. [32] demonstrated that plastid copy number in C. reinhardtii appeared to have little effect on endogenous protein synthesis. In addition, the gene copy number of our transgenes is actually 2-fold higher than most endogenous C. reinhardtii genes, as we insert transgenes into the inverted repeat region. Thus, the difference between recombinant and endogenous protein accumulation cannot be simply due to a gene copy number effect.
Protein steady state levels depend on the balance between synthesis and degradation. As a starting point, we examined the turnover of the three recombinant proteins relative to the endogenous atpA and rbcL proteins. RbcL and atpA showed no reduction in accumulation over 4 h. Among the recombinant proteins, both GFP and LUX were also relatively stable, showing a mild decrease in protein level over 4 h. HSV8-lsc, however, showed a 3-fold reduction in protein level over the 4-h period. Some plastid proteins have been reported to exhibit accelerated turnover. In those cases, the half-life of these proteins ranged from 5 to 60 min depending on the condition [33]. In our work, recombinant protein turnover did not become evident until 120 min after the initiation of the experiment. Thus, while we cannot rule out protein turnover as a contributing factor to GFP and LUX accumulation, and while it certainly appears to be a factor for HSV8-lsc accumulation, it does not explain the bulk of the differences in accumulation between endogenous and recombinant proteins.
We also analyzed the synthesis of the three recombinant proteins by pulse-labeling, in comparison with the endogenous atpA protein. The synthesis of atpA shows an increase throughout the light phase, being substantially higher at low cell density (high light) than at high cell density (lower light) (Fig. 6b). Recombinant protein synthesis differed from that of atpA in two important ways. First, the overall rate of mRNA translation was considerably lower for all three recombinant proteins than that of atpA (Fig. 6a; Table 1). This suggests that poor translation is one of the factors limiting recombinant protein accumulation and that the steady state levels of recombinant protein are defined to a higher extent by translation than protein degradation. Second, the pattern of GFP and HSV8-lsc protein synthesis did not follow that of the endogenous atpA (Fig. 6a; Table 1). This is especially remarkable for GFP: at lower cell density/high light conditions, GFP synthesis was 12% of the synthesis of atpA at best, peaking up at the end of the dark period and decreasing gradually during the light phase (Table 1). In contrast, at high cell density/low light GFP synthesis reached a 34% of atpA synthesis during the light phase and again remained around 12% at end of the dark phase (Table 1). Hsv8 synthesis was always lower than 5% for all time points, but peaked up at the beginning of the light phase for both density/light conditions (Table 1).
To establish if there is a correlation between recombinant message and recombinant protein levels, we analyzed recombinant mRNA accumulation. First, we found that the chimeric mRNAs did not follow the accumulation patterns of the endogenous atpA mRNA (compare Fig. 4c with 5c). This result supports previous studies [34, 35] done on the reporter gene uidA in C. reinhardtii chloroplasts. We also found that, overall, heterologous transcripts accumulate to noticeably lower levels when cultures were grown at low cell density/high light conditions (Figs. 4b, c). All three strains accumulate mRNA to maximal levels near the end of the dark period, then decline throughout the light phase, an effect quite similar, although not as extreme, as that observed by Salvador et al. [36] for chimeric rbcL–gus transcripts.
In order to directly assess the level of recombinant mRNAs relative to endogenous mRNAs that contain the same 5′-UTR, we assayed mRNAs isolated from strain AGR expressing the chimeric atpA–gfp mRNA. Although recombinant mRNAs tended to accumulate to very low levels at low cell density, gfp mRNA levels reached those of the endogenous atpA mRNA at high cell density, at least at the ED and EL points (Fig. 5c). Since GFP protein synthesis composed only 12.5 and 34.1% of atpA protein synthesis at those same time points, this suggests that mRNA levels are not the major, general reason for the low levels of accumulation of this recombinant protein in the chloroplast. This is also evidenced by the fact that, in this study, protein levels remain constant for both GFP and LUX for the different time points within a cell density/light intensity condition, while mRNA levels fluctuate by as much as 5-fold throughout the light phase. The lack of general correlation between message and protein levels in the chloroplast has been demonstrated previously by others [32, 37, 38]. Again, these results point to poor translation as the main reason for limited recombinant protein accumulation, although mRNA levels can influence translation under specific conditions. As an example, GFP message availability might be a problem at low cell densities (high light), when mRNA content, mRNA translation, and protein accumulation are all far below the levels of the endogenous atpA.
It is interesting to note that the pattern of GFP synthesis at high cell density is similar to that for atpA, in that it increases throughout the light phase. This suggests that this chimeric mRNA maintains one feature of endogenous mRNAs: light-activated translation. This could also provide an explanation on why GFP synthesis is considerably lower, when expressed as a percentage of atpA protein synthesis, at low cell density/high light than high cell density/low light. We can reinforce the idea, proposed previously, that recombinant mRNAs compete with the atpA mRNAs for various specific translational activators that bind the atpA 5′-UTR. It has been proposed that, for chloroplast endogenous mRNAs, the coding regions might interact with the 5′-UTRs, helping define their secondary structure and, therefore, influencing on the recruitment of translational factors and ribosomes [27]. It is expected that the introduction of a different coding region might affect this interaction, therefore perturbing the ability of the 5′-UTR of the recombinant mRNA to compete as efficiently for these factors. At higher light, as the synthesis of atpA and other chloroplast-encoded proteins increases, these factors might become more limiting, thus negatively impacting on the ability of recombinant mRNAs to support any increase in synthesis. One could go further and speculate that some recombinant mRNAs have sequences that impart a general disability to recruit translation factors, independently of the light conditions. This could be the case for the hsv8-lsc mRNA. It is interesting to note that light-activated translation continued to operate for this chimeric mRNA, despite its very low level of protein synthesis, suggesting that translational efficiency and light-regulated translation are separate events, as suggested previously [27].
Translation initiation, the rate-limiting step of the universal process of protein synthesis, proceeds through sequential, tightly regulated steps. In bacteria, the correct mRNA start site and the reading frame are selected when the initiation codon is decoded in the peptidyl site of the 30S ribosomal subunit by the fMet-tRNAfMet anticodon [39]. The formation of this 30S initiation complex is supposed to be a key event in the recognition of an mRNA as a “translatable” one. With the purpose of investigating deeper into the reason for poor recombinant protein translation, we assayed the ability of the chimeric atpA–gfp mRNA to form initiation complexes with C. reinhardtii chloroplast 30S subunits in in-vitro toeprint assays. The much lower intensity of the atpA–gfp mRNA toeprints, compared to the atpA–atpA mRNA, suggests that the chimeric RNA is somehow impaired in the formation of 30S initiation complexes. Our data also suggest that the coding region is an important factor determining how well a 5′-UTR binds to ribosomes. In our toeprint experiments, the presence of the gfp coding region downstream of the atpA 5′-UTR affected the pattern in which the reverse transcriptase paused on the 5′-UTR compared to the endogenous atpA, possibly due to differences in the topography of the atpA 5′-UTR between both constructs. As mentioned earlier, the strongest pauses for the atpA–gfp mRNA are bands at positions −2 and +1, near and on the initiator AUG. One could speculate that these pauses are related to a particular RNA structure and that its presence on the initiation codon affects translation initiation.
The redistribution of mRNAs between ribosome-free and polysome-bound fractions has been widely used to ascertain translational control of a given mRNA by Northern hybridization and in protein expression profiling analysis, since ribosome loading of a transcript is a robust indicator for translation efficiency [40, 41]. We thus analyzed the ability of the AGR and atpA mRNAs to associate with chloroplast ribosomes in vivo. Northern blots and RNAse protection assays on polysome fractions of the AGR strain demonstrate that the recombinant gfp mRNA loads onto ribosomes to a lesser extent than its endogenous counterpart. In addition, the lower percentages of gfp to atpA signal found for heavier polysomes compared to lighter polysomes (compare 34% and 53%) suggest that the chimeric RNA might have a slight disadvantage to recruit an increasing number of ribosomes on the same RNA molecule. It has been pointed out that, when translational arrest happens at the point of initiation, transcripts are largely restricted to non-polysomal fractions. In contrast, when translation of mRNAs is selectively inhibited at an early point in chain elongation, transcripts should be largely restricted to association with small polysomes, mainly because the blockage is supposed to interfere with subsequent ribosome recruitment [33, 42]. Based upon the low level of GFP synthesis we observed relative to atpA (Fig. 6; Table 1), gfp mRNA-polysome association was not as low as expected for translation initiation or early elongation to be the major steps limiting heterologous protein synthesis. In contrast, here we show a case of mRNAs that can actually associate to polysomes to a certain degree but have very low translational capabilities even when in the bound form, as has been reported previously for membrane protein and secretory protein synthesis [43] and for the translation of some cellular mRNAs of Drosophila tissue culture [44]. In chloroplasts, the same pattern has been observed for the synthesis of the PsaA and PsaB proteins in dark-grown barley seedlings [33]. In all those situations, synthesis has been proposed to be obstructed, not during early elongation, but rather through delays at discrete points throughout the whole chain elongation, even at points when most of the protein has been completed [43].
In summary, we propose that the accumulation of heterologous proteins in the C. reinhardtii chloroplast is largely defined by protein synthesis. We also show evidence that suggests that ribosome association might be a contributing factor but that delayed elongation at several points on recombinant mRNA molecules is likely to play an even more important role. However, we cannot rule out an effect of other steps of translation, such as termination or ribosome recycling, in defining translational efficiency. Future study will require a careful analysis of all translation steps, a study of the nature of potential 5′-UTR-coding region interactions and the identification of trans-acting factors that impact translation of chloroplast mRNAs at its different steps.
Acknowledgments
The authors wish to thank Amy Beth Cohen for technical help and Sue Golden for her kind gift of LUX antisera. This work was supported by funds from the U.S. Department of Energy (93ER70116) and the National Institutes of Health (GM54659) to S.P.M.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Bock R, Khan MS. Taming plastids for a green future. Trends in Biotechnology. 2004;22:311–318. doi: 10.1016/j.tibtech.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Kumar S, Daniell H. Engineering the chloroplast genome for hyperexpression of human therapeutic proteins and vaccine antigens. Methods in Molecular Biology. 2004;267:365–383. doi: 10.1385/1-59259-774-2:365. [DOI] [PubMed] [Google Scholar]
- 3.Maliga P. Progress towards commercialization of plastid transformation technology. Trends in Biotechnology. 2003;21:20–28. doi: 10.1016/S0167-7799(02)00007-0. [DOI] [PubMed] [Google Scholar]
- 4.Boehm R. Bioproduction of therapeutic proteins in the 21st century and the role of plants and plant cells as production platforms. Annals of the New York Academy of Sciences. 2007;1102:121–134. doi: 10.1196/annals.1408.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fletcher SP, Muto M, Mayfield SP. Optimization of recombinant protein expression in the chloroplasts of green algae. Advances in Experimental Medicine and Biology. 2007;616:90–98. doi: 10.1007/978-0-387-75532-8_8. [DOI] [PubMed] [Google Scholar]
- 6.Mayfield SP, Manuell AL, Chen S, Wu J, Tran M, Siefker D, Muto M, Marin-Navarro J. Chlamydomonas reinhardtii chloroplasts as protein factories. Current Opinion in Biotechnology. 2007;18:126–133. doi: 10.1016/j.copbio.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Brandsma M, Tremblay R, Maxwell D, Jevnikar AM, Huner N, Ma S. A novel expression platform for the production of diabetes-associated autoantigen human glutamic acid decarboxylase (Hgad65) BMC Biotechnology. 2008;8:87. doi: 10.1186/1472-6750-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arlen PA, Falconer R, Cherukumilli S, Cole A, Cole AM, Oishi KK, Daniell H. Field production and functional evaluation of chloroplast-derived interferon-alpha2ß. Plant Biotechnology Journal. 2007;5:511–525. doi: 10.1111/j.1467-7652.2007.00258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Staub JM, Garcia B, Graves J, Hajdukiewicz PT, Hunter P, Nehra N, Paradkar V, Schlittler M, Carroll JA, Spatola L, Ward D, Ye G, Russell DA. High-yield production of a human therapeutic protein in tobacco chloroplasts. Nature Biotechnology. 2000;18:333–338. doi: 10.1038/73796. [DOI] [PubMed] [Google Scholar]
- 10.Franklin SE, Mayfield SP. Prospects for molecular farming in the green alga Chlamydomonas. Current Opinion in Plant Biology. 2004;7:159–165. doi: 10.1016/j.pbi.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Franklin S, Ngo B, Efuet E, Mayfield SP. Development of a gfp reporter gene for Chlamydomonas reinhardtii chloroplast. The Plant Journal. 2002;30:733–744. doi: 10.1046/j.1365-313X.2002.01319.x. [DOI] [PubMed] [Google Scholar]
- 12.Rasala BA, Muto M, Lee PA, Jager M, Cardoso RM, Behnke CA, Kirk P, Hokanson CA, Crea R, Mendez M, Mayfield SP. Production of therapeutic proteins in algae, analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnology Journal. 2010;8:719–733. doi: 10.1111/j.1467-7652.2010.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayfield SP, Franklin SE, Lerner RA. Expression and assembly of a fully active antibody in algae. Proc Natl Acad Sci USA. 2003;100:438–442. doi: 10.1073/pnas.0237108100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayfield SP, Schultz J. Development of a luciferase reporter gene, luxct, for Chlamydomonas reinhardtii chloroplast. The Plant Journal. 2004;37:449–458. doi: 10.1046/j.1365-313X.2003.01965.x. [DOI] [PubMed] [Google Scholar]
- 15.Mayfield SP, Franklin SE. Expression of human antibodies in eukaryotic micro-algae. Vaccine. 2005;23:1828–1832. doi: 10.1016/j.vaccine.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 16.De Cosa B, Moar W, Lee SB, Miller M, Daniell H. Overexpression of the Bt cry2aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nature Biotechnology. 2001;19:71–74. doi: 10.1038/83559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molina A, Hervas-Stubbs S, Daniell H, Mingo-Castel AM, Veramendi J. High-yield expression of a viral peptide animal vaccine in transgenic tobacco chloroplasts. Plant Biotechnology Journal. 2004;2:141–153. doi: 10.1046/j.1467-7652.2004.00057.x. [DOI] [PubMed] [Google Scholar]
- 18.Gorman DS, Levine RP. Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. The Proceedings of the National Academy of Sciences USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: A laboratory manual. New York: Cold Spring Harbor; 1989. [Google Scholar]
- 20.Cohen A, Yohn C, Bruick R, Mayfield S. Translational regulation of chloroplast gene expression in Chlamydomonas reinhardtii. Methods in Enzymology. 1998;297:192–208. doi: 10.1016/S0076-6879(98)97015-X. [DOI] [Google Scholar]
- 21.Boynton JE, Gillham NW, Harris EH, Hosler JP, Johnson AM, Jones AR, Randolph-Anderson BL, Robertson D, Klein TM, Shark KB, et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988;240:1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- 22.Kuras R, Wollman FA. The assembly of cytochrome b6/f complexes: An approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO Journal. 1994;13:1019–1027. doi: 10.1002/j.1460-2075.1994.tb06350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaguchi K, Prieto S, Beligni MV, Haynes PA, McDonald WH, Yates JR, III, Mayfield SP. Proteomic characterization of the small subunit of Chlamydomonas reinhardtii chloroplast ribosome: Identification of a novel S1 domain-containing protein and unusually large orthologs of bacterial S2, S3, and S5. Plant Cell. 2002;14:2957–2974. doi: 10.1105/tpc.004341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi K, Beligni MV, Prieto S, Haynes PA, McDonald WH, Yates JR, III, Mayfield SP. Proteomic characterization of the Chlamydomonas reinhardtii chloroplast ribosome. Identification of proteins unique to the 70 S ribosome. The Journal of Biological Chemistry. 2003;278:33774–33785. doi: 10.1074/jbc.M301934200. [DOI] [PubMed] [Google Scholar]
- 25.Beligni MV, Yamaguchi K, Mayfield SP. Chloroplast elongation factor Ts pro-protein is an evolutionarily conserved fusion with the S1 domain-containing Plastid-Specific Ribosomal Protein-7. Plant Cell. 2004;16:3357–3369. doi: 10.1105/tpc.104.026708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider M, Darlix JL, Erickson J, Rochaix JD. Sequence organization of repetitive elements in the flanking regions of the chloroplast ribosomal unit of Chlamydomonas reinhardii. Nucleic Acids Research. 1985;13:8531–8541. doi: 10.1093/nar/13.23.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barnes D, Franklin S, Schultz J, Henry R, Brown E, Coragliotti A, Mayfield SP. Contribution of 5′- and 3′-untranslated regions of plastid mRNAs to the expression of Chlamydomonas reinhardtii chloroplast genes. Molecular Genetics and Genomics. 2005;274:625–636. doi: 10.1007/s00438-005-0055-y. [DOI] [PubMed] [Google Scholar]
- 28.Drapier D, Suzuki H, Levy H, Rimbault B, Kindle KL, Stern DB, Wollman FA. The chloroplast atpA gene cluster in Chlamydomonas reinhardtii. Functional analysis of a polycistronic transcription unit. Plant Physiology. 1998;117:629–641. doi: 10.1104/pp.117.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang JT, Green CB, Wolf RE., Jr Inhibition of translation initiation on Escherichia coli gnd mRNA by formation of a long-range secondary structure involving the ribosome binding site and the internal complementary sequence. Journal of Bacteriology. 1995;177:6560–6567. doi: 10.1128/jb.177.22.6560-6567.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Day JM, Janssen GR. Isolation and characterization of ribosomes and translation initiation factors from the gram-positive soil bacterium Streptomyces lividans. Journal of Bacteriology. 2004;186:6864–6875. doi: 10.1128/JB.186.20.6864-6875.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Philippe C, Eyermann F, Benard L, Portier C, Ehresmann B, Ehresmann C. Ribosomal protein S15 from Escherichia coli modulates its own translation by trapping the ribosome on the mRNA initiation loading site. The Proceedings of the National Academy of Sciences USA. 1993;90:4394–4398. doi: 10.1073/pnas.90.10.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eberhard S, Drapier D, Wollman FA. Searching limiting steps in the expression of chloroplast-encoded proteins: Relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. The Plant Journal. 2002;31:149–160. doi: 10.1046/j.1365-313X.2002.01340.x. [DOI] [PubMed] [Google Scholar]
- 33.Klein RR, Mason HS, Mullet JE. Light-regulated translation of chloroplast proteins. I. Transcripts of psaA-psaB, psbA, and rbcL are associated with polysomes in dark-grown and illuminated barley seedlings. Journal of Cell Biology. 1988;106:289–301. doi: 10.1083/jcb.106.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishikura K, Takaoka Y, Kato K, Sekine M, Yoshida K, Shinmyo A. Expression of a foreign gene in Chlamydomonas reinhardtii chloroplast. Journal of Bioscience and Bioengineering. 1999;87:307–314. doi: 10.1016/S1389-1723(99)80037-1. [DOI] [PubMed] [Google Scholar]
- 35.Kasai S, Yoshimura S, Ishikura K, Takaoka Y, Kobayashi K, Kato K, Shinmyo A. Effect of coding regions on chloroplast gene expression in Chlamydomonas reinhardtii. Journal of Bioscience and Bioengineering. 2003;95:276–282. doi: 10.1016/s1389-1723(03)80029-4. [DOI] [PubMed] [Google Scholar]
- 36.Salvador ML, Klein U, Bogorad L. 5’ sequences are important positive and negative determinants of the longevity of Chlamydomonas chloroplast gene transcripts. The Proceedings of the National Academy of Sciences USA. 1993;90:1556–1560. doi: 10.1073/pnas.90.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choquet Y, Wollman FA. Translational regulations as specific traits of chloroplast gene expression. FEBS Letters. 2002;529:39–42. doi: 10.1016/S0014-5793(02)03260-X. [DOI] [PubMed] [Google Scholar]
- 38.Zerges W. Translation in chloroplasts. Biochimie. 2000;82:583–601. doi: 10.1016/S0300-9084(00)00603-9. [DOI] [PubMed] [Google Scholar]
- 39.Simonetti A, Marzi S, Myasnikov AG, Fabbretti A, Yusupov M, Gualerzi CO, Klaholz BP. Structure of the 30S translation initiation complex. Nature. 2008;455:416–420. doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- 40.Leibold EA, Munro HN. Cytoplasmic protein binds in vitro to a highly conserved sequence in the 5’ untranslated region of ferritin heavy- and light-subunit mRNAs. The Proceedings of the National Academy of Sciences USA. 1988;85:2171–2175. doi: 10.1073/pnas.85.7.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikulits W, Pradet-Balade B, Habermann B, Beug H, Garcia-Sanz JA, Mullner EW. Isolation of translationally controlled mRNAs by differential screening. The FASEB Journal. 2000;14:1641–1652. doi: 10.1096/fj.14.11.1641. [DOI] [PubMed] [Google Scholar]
- 42.Chua NH, Blobel G, Siekevitz P, Palade GE. Periodic variations in the ratio of free to thylakoid-bound chloroplast ribosomes during the cell cycle of Chlamydomonas reinhardtii. Journal of Cell Biology. 1976;71:497–514. doi: 10.1083/jcb.71.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lipp J, Dobberstein B, Haeuptle MT. Signal recognition particle arrests elongation of nascent secretory and membrane proteins at multiple sites in a transient manner. The Journal of Biological Chemistry. 1987;262:1680–1684. [PubMed] [Google Scholar]
- 44.Ballinger DG, Pardue ML. The control of protein synthesis during heat shock in Drosophila cells involves altered polypeptide elongation rates. Cell. 1983;33:103–113. doi: 10.1016/0092-8674(83)90339-2. [DOI] [PubMed] [Google Scholar]