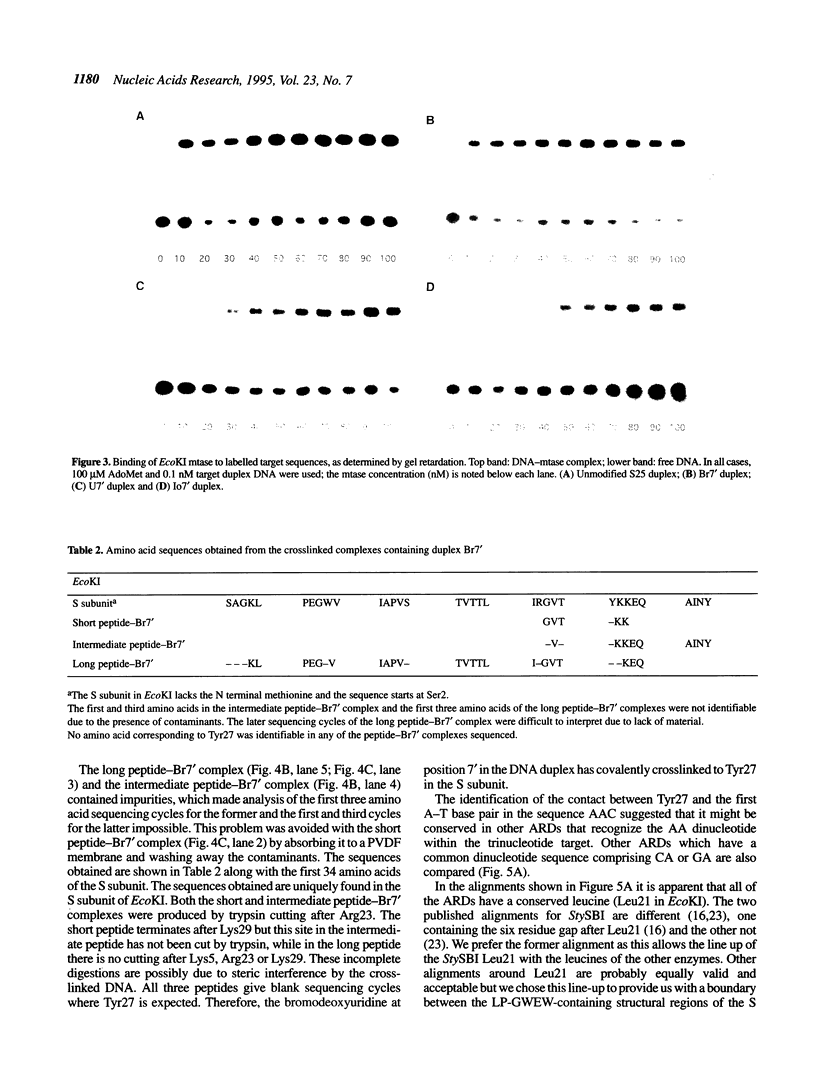
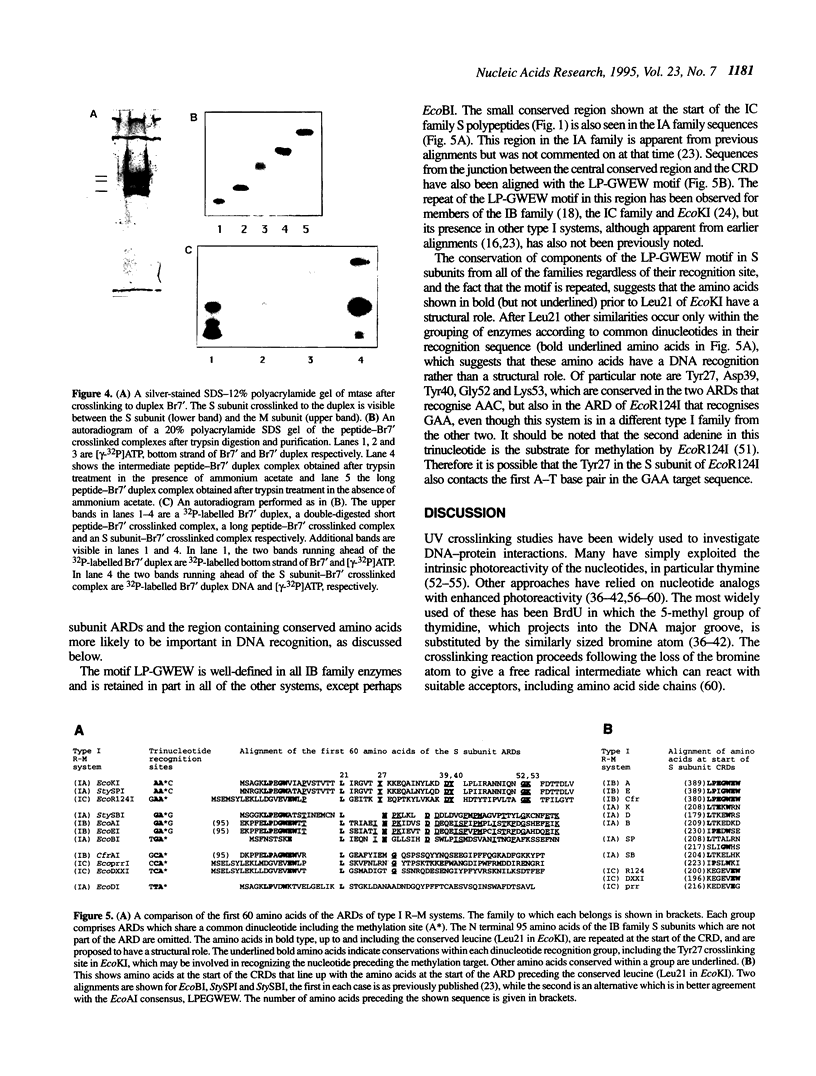
Abstract
The specificity (S) subunit of the restriction enzyme EcoKI imparts specificity for the sequence AAC(N6)GTGC. Substitution of thymine with bromodeoxyuridine in a 25 bp DNA duplex containing this sequence stimulated UV light-induced covalent crosslinking to the S subunit. Crosslinking occurred only at the residue complementary to the first adenine in the AAC sequence, demonstrating a close contact between the major groove at this sequence and the S subunit. Peptide sequencing of a proteolytically-digested, crosslinked complex identified tyrosine 27 in the S subunit as the site of crosslinking. This is consistent with the role of the N-terminal domain of the S subunit in recognizing the AAC sequence. Tyrosine 27 is conserved in the S subunits of the three type I enzymes that share the sequence AA in the trinucleotide component of their target sequence. This suggests that tyrosine 27 may make a similar DNA contact in these other enzymes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abadjieva A., Webb M., Patel J., Zinkevich V., Firman K. Deletions within the DNA recognition subunit of M.EcoR124I that identify a region involved in protein-protein interactions between HsdS and HsdM. J Mol Biol. 1994 Aug 5;241(1):35–43. doi: 10.1006/jmbi.1994.1471. [DOI] [PubMed] [Google Scholar]
- Ahmad I., Rao D. N. Interaction of EcoP15I DNA methyltransferase with oligonucleotides containing the asymmetric sequence 5'-CAGCAG-3'. J Mol Biol. 1994 Sep 30;242(4):378–388. doi: 10.1006/jmbi.1994.1588. [DOI] [PubMed] [Google Scholar]
- Allen T. D., Wick K. L., Matthews K. S. Identification of amino acids in lac repressor protein cross-linked to operator DNA specifically substituted with bromodeoxyuridine. J Biol Chem. 1991 Apr 5;266(10):6113–6119. [PubMed] [Google Scholar]
- Athanasiadis A., Vlassi M., Kotsifaki D., Tucker P. A., Wilson K. S., Kokkinidis M. Crystal structure of PvuII endonuclease reveals extensive structural homologies to EcoRV. Nat Struct Biol. 1994 Jul;1(7):469–475. doi: 10.1038/nsb0794-469. [DOI] [PubMed] [Google Scholar]
- Bickle T. A., Brack C., Yuan R. ATP-induced conformational changes in the restriction endonuclease from Escherichia coli K-12. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3099–3103. doi: 10.1073/pnas.75.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickle T. A., Krüger D. H. Biology of DNA restriction. Microbiol Rev. 1993 Jun;57(2):434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatter E. E., Ebright Y. W., Ebright R. H. Identification of an amino acid-base contact in the GCN4-DNA complex by bromouracil-mediated photocrosslinking. Nature. 1992 Oct 15;359(6396):650–652. doi: 10.1038/359650a0. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Burckhardt J., Weisemann J., Yuan R. Characterization of the DNA methylase activity of the restriction enzyme from Escherichia coli K. J Biol Chem. 1981 Apr 25;256(8):4024–4032. [PubMed] [Google Scholar]
- Cheng X., Balendiran K., Schildkraut I., Anderson J. E. Structure of PvuII endonuclease with cognate DNA. EMBO J. 1994 Sep 1;13(17):3927–3935. doi: 10.1002/j.1460-2075.1994.tb06708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleghon V., Klessig D. F. Characterization of the nucleic acid binding region of adenovirus DNA binding protein by partial proteolysis and photochemical cross-linking. J Biol Chem. 1992 Sep 5;267(25):17872–17881. [PubMed] [Google Scholar]
- Cooper L. P., Dryden D. T. The domains of a type I DNA methyltransferase. Interactions and role in recognition of DNA methylation. J Mol Biol. 1994 Mar 4;236(4):1011–1021. doi: 10.1016/0022-2836(94)90008-6. [DOI] [PubMed] [Google Scholar]
- Cowan G. M., Gann A. A., Murray N. E. Conservation of complex DNA recognition domains between families of restriction enzymes. Cell. 1989 Jan 13;56(1):103–109. doi: 10.1016/0092-8674(89)90988-4. [DOI] [PubMed] [Google Scholar]
- Dryden D. T., Cooper L. P., Murray N. E. Purification and characterization of the methyltransferase from the type 1 restriction and modification system of Escherichia coli K12. J Biol Chem. 1993 Jun 25;268(18):13228–13236. [PubMed] [Google Scholar]
- Ellenberger T. E., Brandl C. J., Struhl K., Harrison S. C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992 Dec 24;71(7):1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Erlanger B. F., Cooper A. G., Cohen W. The inactivation of chymotrypsin by diphenylcarbamyl chloride and its reactivation by nucleophilic agents. Biochemistry. 1966 Jan;5(1):190–196. doi: 10.1021/bi00865a025. [DOI] [PubMed] [Google Scholar]
- Evans R. K., Johnson J. D., Haley B. E. 5-Azido-2'-deoxyuridine 5'-triphosphate: a photoaffinity-labeling reagent and tool for the enzymatic synthesis of photoactive DNA. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5382–5386. doi: 10.1073/pnas.83.15.5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Bullas L. R., Delius H., Murray N. E. Genetic recombination can generate altered restriction specificity. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6095–6099. doi: 10.1073/pnas.81.19.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Cowan G. M., Murray N. E. EcoA and EcoE: alternatives to the EcoK family of type I restriction and modification systems of Escherichia coli. J Mol Biol. 1985 Nov 5;186(1):65–75. doi: 10.1016/0022-2836(85)90257-8. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Murray N. E. Two DNA recognition domains of the specificity polypeptides of a family of type I restriction enzymes. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9368–9372. doi: 10.1073/pnas.83.24.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gann A. A., Campbell A. J., Collins J. F., Coulson A. F., Murray N. E. Reassortment of DNA recognition domains and the evolution of new specificities. Mol Microbiol. 1987 Jul;1(1):13–22. doi: 10.1111/j.1365-2958.1987.tb00521.x. [DOI] [PubMed] [Google Scholar]
- Glover S. W., Colson C. Genetics of host-controlled restriction and modification in Escherichia coli. Genet Res. 1969 Apr;13(2):227–240. doi: 10.1017/s0016672300002901. [DOI] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Gubler M., Braguglia D., Meyer J., Piekarowicz A., Bickle T. A. Recombination of constant and variable modules alters DNA sequence recognition by type IC restriction-modification enzymes. EMBO J. 1992 Jan;11(1):233–240. doi: 10.1002/j.1460-2075.1992.tb05046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. D., Kerr L. A., Cronshaw A. D. Evidence that glutathione S-transferases B1B1 and B2B2 are the products of separate genes and that their expression in human liver is subject to inter-individual variation. Molecular relationships between the B1 and B2 subunits and other Alpha class glutathione S-transferases. Biochem J. 1989 Dec 1;264(2):437–445. doi: 10.1042/bj2640437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson F. The lesions produced by ultraviolet light in DNA containing 5-bromouracil. Q Rev Biophys. 1973 May;6(2):201–246. doi: 10.1017/s0033583500001141. [DOI] [PubMed] [Google Scholar]
- Kan N. C., Lautenberger J. A., Edgell M. H., Hutchison C. A., 3rd The nucleotide sequence recognized by the Escherichia coli K12 restriction and modification enzymes. J Mol Biol. 1979 May 15;130(2):191–209. doi: 10.1016/0022-2836(79)90426-1. [DOI] [PubMed] [Google Scholar]
- Kannan P., Cowan G. M., Daniel A. S., Gann A. A., Murray N. E. Conservation of organization in the specificity polypeptides of two families of type I restriction enzymes. J Mol Biol. 1989 Oct 5;209(3):335–344. doi: 10.1016/0022-2836(89)90001-6. [DOI] [PubMed] [Google Scholar]
- Kim Y. C., Grable J. C., Love R., Greene P. J., Rosenberg J. M. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science. 1990 Sep 14;249(4974):1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S., Kumar S., Roberts R. J., Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994 Jan 28;76(2):357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- Kneale G. G. A symmetrical model for the domain structure of type I DNA methyltransferases. J Mol Biol. 1994 Oct 14;243(1):1–5. doi: 10.1006/jmbi.1994.1624. [DOI] [PubMed] [Google Scholar]
- Labahn J., Granzin J., Schluckebier G., Robinson D. P., Jack W. E., Schildkraut I., Saenger W. Three-dimensional structure of the adenine-specific DNA methyltransferase M.Taq I in complex with the cofactor S-adenosylmethionine. Proc Natl Acad Sci U S A. 1994 Nov 8;91(23):10957–10961. doi: 10.1073/pnas.91.23.10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lin S. Y., Riggs A. D. Photochemical attachment of lac repressor to bromodeoxyuridine-substituted lac operator by ultraviolet radiation. Proc Natl Acad Sci U S A. 1974 Mar;71(3):947–951. doi: 10.1073/pnas.71.3.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffert R., Dose K. UV-induced cross-linking of proteins to plasmid pBR322 containing 8-azidoadenine 2'-deoxyribonucleotides. FEBS Lett. 1988 Nov 7;239(2):190–194. doi: 10.1016/0014-5793(88)80914-1. [DOI] [PubMed] [Google Scholar]
- Meffert R., Rathgeber G., Schäfer H. J., Dose K. UV-induced cross-linking of Tet repressor to DNA containing tet operator sequences and 8-azidoadenines. Nucleic Acids Res. 1990 Nov 25;18(22):6633–6636. doi: 10.1093/nar/18.22.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister J., MacWilliams M., Hübner P., Jütte H., Skrzypek E., Piekarowicz A., Bickle T. A. Macroevolution by transposition: drastic modification of DNA recognition by a type I restriction enzyme following Tn5 transposition. EMBO J. 1993 Dec;12(12):4585–4591. doi: 10.1002/j.1460-2075.1993.tb06147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill B. M., Williams K. R., Chase J. W., Konigsberg W. H. Photochemical cross-linking of the Escherichia coli single-stranded DNA-binding protein to oligodeoxynucleotides. Identification of phenylalanine 60 as the site of cross-linking. J Biol Chem. 1984 Sep 10;259(17):10850–10856. [PubMed] [Google Scholar]
- Meselson M., Yuan R. DNA restriction enzyme from E. coli. Nature. 1968 Mar 23;217(5134):1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- Nagaraja V., Shepherd J. C., Bickle T. A. A hybrid recognition sequence in a recombinant restriction enzyme and the evolution of DNA sequence specificity. Nature. 1985 Jul 25;316(6026):371–372. doi: 10.1038/316371a0. [DOI] [PubMed] [Google Scholar]
- Newman M., Strzelecka T., Dorner L. F., Schildkraut I., Aggarwal A. K. Structure of restriction endonuclease BamHI and its relationship to EcoRI. Nature. 1994 Apr 14;368(6472):660–664. doi: 10.1038/368660a0. [DOI] [PubMed] [Google Scholar]
- Newman M., Strzelecka T., Dorner L. F., Schildkraut I., Aggarwal A. K. Structure of restriction endonuclease bamhi phased at 1.95 A resolution by MAD analysis. Structure. 1994 May 15;2(5):439–452. doi: 10.1016/s0969-2126(00)00045-9. [DOI] [PubMed] [Google Scholar]
- Nikiforov T. T., Connolly B. A. Oligodeoxynucleotides containing 4-thiothymidine and 6-thiodeoxyguanosine as affinity labels for the Eco RV restriction endonuclease and modification methylase. Nucleic Acids Res. 1992 Mar 25;20(6):1209–1214. doi: 10.1093/nar/20.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata R., Gilbert W. Contacts between the lac repressor and the thymines in the lac operator. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4973–4976. doi: 10.1073/pnas.74.11.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradiso P. R., Nakashima Y., Konigsberg W. Photochemical cross-linking of protein . nucleic acid complexes. The attachment of the fd gene 5 protein to fd DNA. J Biol Chem. 1979 Jun 10;254(11):4739–4744. [PubMed] [Google Scholar]
- Powell L. M., Dryden D. T., Willcock D. F., Pain R. H., Murray N. E. DNA recognition by the EcoK methyltransferase. The influence of DNA methylation and the cofactor S-adenosyl-L-methionine. J Mol Biol. 1993 Nov 5;234(1):60–71. doi: 10.1006/jmbi.1993.1563. [DOI] [PubMed] [Google Scholar]
- Powell L. M., Murray N. E. S-adenosyl methionine alters the DNA contacts of the EcoKI methyltransferase. Nucleic Acids Res. 1995 Mar 25;23(6):967–974. doi: 10.1093/nar/23.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sain B., Murray N. E. The hsd (host specificity) genes of E. coli K 12. Mol Gen Genet. 1980;180(1):35–46. doi: 10.1007/BF00267350. [DOI] [PubMed] [Google Scholar]
- Suri B., Nagaraja V., Bickle T. A. Bacterial DNA modification. Curr Top Microbiol Immunol. 1984;108:1–9. doi: 10.1007/978-3-642-69370-0_1. [DOI] [PubMed] [Google Scholar]
- Taylor I., Watts D., Kneale G. Substrate recognition and selectivity in the type IC DNA modification methylase M.EcoR124I. Nucleic Acids Res. 1993 Oct 25;21(21):4929–4935. doi: 10.1093/nar/21.21.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vovis G. F., Horiuchi K., Zinder N. D. Kinetics of methylation of DNA by a restriction endonuclease from Escherichia coli B. Proc Natl Acad Sci U S A. 1974 Oct;71(10):3810–3813. doi: 10.1073/pnas.71.10.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. R., Konigsberg W. H. Identification of amino acid residues at interface of protein-nucleic acid complexes by photochemical cross-linking. Methods Enzymol. 1991;208:516–539. doi: 10.1016/0076-6879(91)08027-f. [DOI] [PubMed] [Google Scholar]
- Willis M. C., LeCuyer K. A., Meisenheimer K. M., Uhlenbeck O. C., Koch T. H. An RNA-protein contact determined by 5-bromouridine substitution, photocrosslinking and sequencing. Nucleic Acids Res. 1994 Nov 25;22(23):4947–4952. doi: 10.1093/nar/22.23.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- Winkler F. K., Banner D. W., Oefner C., Tsernoglou D., Brown R. S., Heathman S. P., Bryan R. K., Martin P. D., Petratos K., Wilson K. S. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993 May;12(5):1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfes H., Fliess A., Winkler F., Pingoud A. Cross-linking of bromodeoxyuridine-substituted oligonucleotides to the EcoRI and EcoRV restriction endonucleases. Eur J Biochem. 1986 Sep 1;159(2):267–273. doi: 10.1111/j.1432-1033.1986.tb09863.x. [DOI] [PubMed] [Google Scholar]