Introduction
M. tuberculosis Rv2704 is a member of the highly conserved YjgF/YER057c/UK114 protein superfamily. Homologues of this protein occur in eubacteria, archaea and eukaryotes. Proteins in this family are functionally diverse and are involved in variety of enzymatic and non-enzymatic functions. The high sequence and structural similarity between members of this protein family provides an intriguing dataset to rationalize the wide variety of functional roles. This protein family is an example of minimalistic changes leading to functional diversification. This feature is best exemplified by the three close homologues of YjgF proteins in mammals (human, rat and goat) with sequence identity better than 85%. These homologues perform different functions1-3. The diverse functions attributed to these proteins in this family include tumour antigen activity in the goat homologue1, translation inhibition in human and rat homologues (hp14.5 and rp14.5)2,3, endoribonuclease activity in rp14.54, calpain activation in the bovine homologue5, molecular chaperone activity in DUK1146 and involvement in the regulation of purine and removal of toxic metabolites in YjgF7,8 and isoleucine (YjgF, YER057c, Ibm1) biosynthetic pathways9-11. In addition, members of this protein family have also been shown to regulate mitochondrial maintenance (Ibm1) in yeast11. Proteins from the YjgF family in plants are involved in photosynthesis and chromoplastogenesis (CHRD)12.
The three dimensional structures of fifteen homologues of Rv2704 have been determined from bacteria, human, rat, goat and archaea7,13-18. These proteins share similar homotrimeric structures with the clefts between the monomeric subunits proposed to have some functional relevance7,15-17 (Supplementary Table I). The best characterized protein with the YjgF fold is chorismate mutase which typifies this α+β fold and trimeric quaternary association. However, this protein shares very little sequence similarity with the other members of this family that were subsequently structurally characterized17,18. Members of the Yjgf family could potentially bind various metabolites. This suggestion was based on crystal structures of some homologues bound to ligands like benzoate15, acetate17, 2-ketobutyrate7, 1,2-ethanediol7, propanoate7 and serine7. In these crystal structures, the ligands bound at the cleft between the monomeric subunits of the trimer. This prompted a comparative analysis of this intersubunit cleft and an examination of role of conserved residues lining the cleft7,15. No biological role could be assigned for these bound ligands except perhaps for 2-ketobutyrate where it was proposed that Yjgf may be involved in the removal of toxic products7. Even this functional implication, however, remains contestable with a recent report proposing that 2-ketobutyrate may not be the natural substrate for YjgF from Salmonella enterica9. These studies thus highlight the difficulties in structure-based functional assignment for proteins belonging to the YjgF family.
M. tuberculosis Rv2704 lies in the same operon as the principal sigma (σ) factor, σA (Supplementary Figure 1). σ factors are transcriptional proteins that are often regulated by their interactions with an anti-σ factor located in the same operon. Rv2704 was thus examined for its role as a putative regulator for σA. While we could not detect any interaction between Rv2704 and σA in vitro, it is still likely that it could regulate σA activity in vivo, perhaps involving metabolite interactions. Here we present the crystal structure of Rv2704 at 1.93Å resolution, solved using single isomorphous replacement with anomalous scattering (SIRAS). This protein is a trimer in solution with the overall structure similar to that of other YjgF homologues. The structure and biochemical features of Rv2704 could thus provide a template for more directed predictive methods for functional annotation in the YjgF family of proteins.
Materials and methods
Cloning, expression and purification of Rv2704
The gene encoding Rv2704 was PCR amplified from the genomic DNA of M. tuberculosis H37Rv and cloned between Nhe1 and Xho1 sites of the E. coli expression vector pET22b. After transforming the plasmid into Rosetta BL21( DE3) cells (Novagen, Inc.), the cells were grown in Luria broth in the presence of ampicillin (100 μg/ml) to an OD600nm of 0.5–0.6. The cells were then induced with 0.3 mM IPTG (final concentration). Subsequently, the growth temperature was lowered to 290 K and cells were grown for further 12–18 h before they were spun down and stored at 193 K. Cells were lysed by sonication in a buffer containing 20 mM Tris pH 7.5 and 250 mM NaCl. The cell-free lysate was incubated with Co-NTA beads and washed with 10 bed volumes of lysis buffer in the column. Recombinant Rv2704 was eluted by a gradient of elution buffer (20 mM Tris pH 7.5, 250 mM NaCl and 200 mM Imidazole). The purity of the sample was analyzed by SDS-PAGE. The molecular weight of the recombinant protein was also verified by mass spectrometry on both MALDI-TOF as well as LC-ESI mass spectrometers (Bruker Daltonics, Inc.).
Crystallization
The initial crystallization conditions were obtained using commercial screens from Hampton Research by the hanging drop method. Recombinant Rv2704 could be concentrated to ca 80 mg/ml. Initial crystallization trials with 15 mg/ml protein concentration gave precipitates in more than 80 % of conditions. Subsequently reducing the protein concentration to 3 mg/ml provided initial crystals within 3-4 days of setting up the crystallization trays. Diffraction quality crystals were obtained in a condition containing 15% PEG 8000, 0.1 M Bis-Tris Propane (pH 6.5) and any one of several additives – N-docecyl-N,N-dimethylamine-N-oxide (0.5 % w/v), 1,6 hexane diol (3% v/v), glucose monohydrate (3% w/v) or sodium fluoride (50 mM) using the microbatch method. Rv2704 crystals were cryo-protected using 15% (v/v) glycerol in mother liquor prior to flash freezing at 100 K. NaI and CsCl derivatives were made adopting the quick soak methods described earlier19,20. Transferring Rv2704 crystals from the crystallization droplet under oil to a solution on a cover slip containing cryo-protectant and derivative salts led to a substantial drop in diffraction quality. To circumvent this problem, the crystallization solution containing two-fold excess of the derivative salt was transferred to the well containing crystals (layered below mineral oil) in 1:1 ratio and gently mixed using a loop. These derivative crystals were soaked for various time points and were tested for diffraction quality and isomorphism. The optimal conditions that gave isomorphous crystals with a reasonable anomalous signal involved soaking Rv2704 crystals in 500 mM NaI for ca 3 min. No anomalous signal was observed for the CsCl soaked crystals.
Structure determination and refinement
Molecular replacement trials using the structures of different YjgF homologues were performed using PHASER21 and MOLREP22. Although, MR solutions could be obtained with different models, the Rfree did not drop below ca 35% even after repeated cycles of model building and refinement using COOT23 and REFMAC24. The electron density for the first 20 residues was poor and thus this segment could not be modeled. Phase information for the Rv2704 structure was thus obtained by Single Isomorphous Replacement with Anomalous Scattering (SIRAS) using the halide/alkali quick soak methods19,20,25-27. Although both CsCl and NaI were examined, we were successful in obtaining anomalous signal with the NaI derivative alone. Diffraction data were collected at the home source (1.5418Å) on a Microstar UltraII X-Ray generator (Bruker AXS) and a MAR345 detector (Mar Research, Inc). Data were indexed and integrated using iMOSFLM28 and were scaled using SCALA29. The online Auto-Rickshaw server21,30-34 at EMBL was used to locate the positions of the Iodine atoms. Six heavy atom sites were located with occupancies ranging from 0.4 to 1. Several cycles of model building were performed using COOT23 and the structure was refined using REFMAC24. The locations of bound water molecules were initially identified by Arp/wArp31 and were subsequently added using COOT23. The data collection, phasing and refinement statistics are summarized in Table I. The coordinates and structure factors have been deposited in the Protein Data Bank with the accession code 3I7T.
TABLE I.
Crystallographic Data and Refinement Statistics
| I. Data collection | ||
|---|---|---|
| Native | NaI derivative | |
| Wavelength | 1.5418 Å | 1.5418 Å |
| Resolution (Å) | 33.96 – 1.94 (2.05-1.94) a | 33.19 – 1.93 (2.03-1.93) |
| Unit cell parameters | a =b = c = 67.92 Å | a =b = c = 67.88 Å |
| Space group | P213 | P213 |
| Total number of reflections | 44811 (6489) | 144413 (20323) |
| Number of unique reflections | 7959 (1141) | 8092 (1149) |
| Completeness (%) | 99.7 (100) | 100 (100) |
| Multiplicity | 5.6 (5.3) | 17.8 (17.7) |
| Rmergeb (%) | 12.1 (50.3) | 10 (42.4) |
| <I>/σ(I) | 8.7 (3.1) | 21.5 (6.6) |
| II. Refinement statistics | ||
| Resolution range | 33.19 – 1.93 | |
| rmsdbonds (Å) | 0.009 | |
| rmsdangles (°) | 1.241 | |
| Rcrystc /Rfreed(%) | 20.5/24.7 | |
| No. of protein atoms | 877 | |
| No. of solvent atoms | 61 | |
| III. Ramachandran plot statistics | ||
| Residues in most favoured regions (%) | 94.1 | |
| Residues in allowed regions (%) | 5.9 | |
| Residues in generously allowed regions (%) | 0 | |
| Residues disallowed regions (%) | 0 | |
Values for outer shells are given in parentheses.
Rmerge=∑j|<I>-Ij|/∑<I> where Ij is the intensity of the jth reflection and <I> is the average intensity.
Rcryst=∑hkl|Fo – Fc|/∑hkl|Fo|
Rfree was calculated as for Rcryst but on 5 % of the data excluded from the refinement calculation.
Results and discussion
Rv2704 has a chorismate mutase-like fold
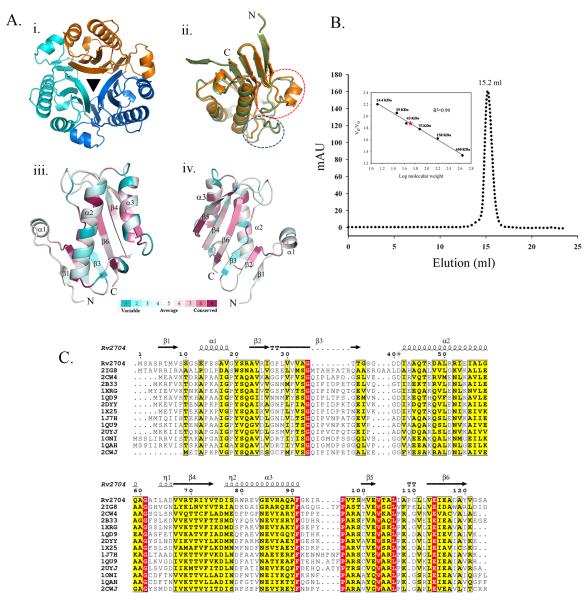
There is a monomer of Rv2704 in the asymmetric unit of the crystal. The three fold axis of the Rv 2704 trimer is perfectly aligned with the crystallographic three fold axis (Figure 1A). This observation is consistent with the data from size exclusion chromatography experiments carried out using a Superdex G75 column (GE Healthcare) which suggests that Rv2704 is a trimer in solution (Figure 1B). This 142aa long polypeptide folds into a single compact domain as is the case with chorismate mutase, the canonical member of the fold. An alignment of YjgF-like protein sequences for which structural data is available suggests poor sequence conservation in the N-terminal segment (~20aa) of Rv2704 and an eleven residue extension in the C-terminal domain. Not surprisingly, these regions show structural differences when compared to the other YjgF proteins. The N-terminal region of Rv2704 adopts a α helical conformation spanning Ser12 to Gly19. This α helix is not seen in any of the other crystal structures of YjgF proteins determined thus far (Figure 1A). An interesting observation is that this helix (α1) of Rv2704 is replaced by a loop with a high B-factor in other structures and has been proposed to play role in facilitating ligand binding15. 21 residues at the C-terminus of Rv2704 could not be modeled due to poor electron density. The introduction of an α helix thus differentiates the structure of Rv2704 from other YjgF proteins. The arrangement of the secondary structures in Rv2704 is β1α1β2β3α2β4α3β5β6. This arrangement is different from that of other YjgF proteins which have the arrangement β1β2β3α1β4α2β5β67,15,17 (Supplementary Figure 2B). The other difference between the structure of Rv2704 and YjgF proteins is in a loop that connects α2 and β3. Multiple sequence alignment and structural superimposition suggests that this connecting loop is the shortest in Rv2704 (Figures 1A and 1C).
Fig. 1.
(A) Structural features of Rv2704. i. The trimeric arrangement of Rv2704 monomers. ii. The crystal structure of Rv2704 differs from other YjgF homologs in a loop segment that adopts an α helical structure (α1) (red circle) and in a loop that connects α2 and β3 which is shorter than those in other YjgF proteins (blue circle). iii. Sequence conservation mapped on to the crystal structure of Rv2704 reveals highly conserved regions in YjgF proteins. This figure was drawn using ConSurf38. (B) Size exclusion profile of Rv2704. Rv2704 is a trimer in solution. (C) Multiple sequence alignment of YjgF proteins reveals seven invariant residues (highlighted in red) and a high overall sequence similarity (shown in yellow). The C-terminal extension of Rv2704 (Val125-Gly142 ) is not shown in this sequence alignment. This alignment and presentation were made using CLUSTALW39 and ESPript40.
Structural features of Rv2704
The comparatively large (ca 15) dataset of YjgF protein structures allows for a comparative analysis of surface features, cavities and clefts in proteins with this fold. There are two prominent clefts in Rv2704 that are also conserved in a few YjgF homologues. One cleft is located at the trimeric interface (Type-I) while the other lies at the interface of two monomers i.e. AB, BC and CA (Type-II). Type-II clefts are often characterized by the presence of bound ligands. In a comparative analysis of 226 ligand binding clefts, the first and second largest pockets (clefts) included more than 80% of the binding sites35. Indeed, it has been variously suggested that these clefts could serve as potential active sites for a catalytic role. However, no functional role for the cleft between the trimeric interface (type I) has been assigned in any of the YjgF proteins characterized till date. The type II clefts (between adjacent monomers) on the other hand, have been shown to bind various ligands– benzoate (in human hp14.5)15 or 2-ketobutyrate, acetate, ethylene glycol or serine (in E.coli TdcF)7. It is worth noting that while the type 1 cleft is less conserved (altogether absent in some cases), the type II cleft is maintained in all known YjgF homologues. Structural and sequence analyses of the residues lining the type-II cleft shows substantial variations in architecture, volume, charge distribution and sequence composition. Thus ligand specificity may not be hardwired for this binding site. For example, side-chain specific interactions are mediated by Arg105 and Glu114 in E.coli TdcF. The NH2 and NE of the conserved Arg105 forms a bi-dentate salt-bridge with the carboxylate of 2-ketobutyrate. An Arg is present at a structurally equivalent position in several YjgF proteins. In Rv2704, Arg105 is replaced by Thr98 (Supplementary Figure 2A). However, Glu114 of Rv2704 is structurally equivalent to Glu120 of E. coli TdcF. In the crystal structure of TdcF solved in complex with several ligands, a majority of the conformational changes were restricted to the loop connecting β1 and β2. This loop becomes more ordered in the ligand bound form7. This loop lining the putative active-site cleft was also noted to have a high temperature factor in the human homologue15, a finding that is consistent across all crystal structures that were examined.
Sequence-structure correlation between members of the YjgF family
The multiple sequence alignment presented in Figure 1C shows that the closest protein from the YjgF family shares 37% sequence identity over 109 aligned residues with an overall sequence identity of 28%. Structural superimposition of the Rv2704 monomer with these YjgF proteins suggests that the fold is fairly well conserved. The root mean squared deviations (rmsd) over 114 Cα atoms between these structures vary from 1.4-1.8 Å with the sequence identity ranging from 21-35% (Supplementary Table I). The seven invariant residues in this alignment differ from the conservation pattern reported earlier36. Thus the conserved Arg105 which is believed to play important role in the activity of YjgF proteins is replaced by Thr98 while the catalytic cysteine residue in E. coli YjgF18 is replaced by methionine in Rv2704 (Supplementary Fig. 2A). Another aspect, perhaps of interest from a bioinformatics perspective, is the difference in the physico-chemical properties among mammalian, archaeal and bacterial YjgF proteins. High acid stability is a characteristic feature of mammalian and archaeal YjgF proteins. Unlike homologues from human (hp14.5)15, rat (rp14.5)3 and archaea,16 Rv2704 is not stable in acidic conditions as it immediately precipitates in 5% perchloric acid solution.
The probable functions of E. coli TdcF and YabJ could be inferred from the role of the other gene products in the same operon. E. coli TdcF is a part of tdc operon which is involved in degradation of L-threonine and L-serine7, and YabJ forms part of pur operon37. Unfortunately the functional annotation of majority of the YjgF proteins is difficult as their genes lie in operons with other hypothetical genes. As Rv2704 lies in the same operon as the principal σ factor σA, the probable role of Rv2704 as an anti-σ factor was examined. σA-Rv2704 interactions were examined using a co-expression and co-purification strategy. Both genes (σA also referred to as Rv2703 and Rv2704) were cloned into the pET-Duet expression vector (Novagen, Inc.). σA was cloned in the Multiple Cloning Site 1 (MCS1) that would incorporate a poly-histidine tag in the recombinant protein and Rv2704 was cloned in MCS2. Both proteins were seen to express and a purification strategy using a Co-NTA affinity chromatography was adopted to obtain this complex. While this strategy was successful in several cases (data not shown), σA was not seen to interact with Rv2704 in this experiment. Rv2704 did not coelute with σA. In this context, it is important to note that while σA is an essential gene involved in the transcription of all house-keeping genes in M. tuberculosis, Rv2704 is not. Thus while direct interaction between Rv2704 and σA does not appear likely, the structure of Rv2704 suggests a different possibility. The finding that the members of the YjgF family bind different metabolites suggests that Rv2704 could modulate its function in the presence of different metabolites. The structural and biochemical features of Rv2704 could thus serve as a template to further examine the role of this protein in M. tuberculosis.
Supplementary Material
Acknowledgements
This work was supported in part by the Wellcome Trust, United Kingdom. The X-ray facility is supported by Grants-in-Aid from the Department of Science and Technology and the Department of Biotechnology, Government of India. BG is an International Senior Research Fellow of the Wellcome Trust, U.K.
References
- 1.Ceciliani F, Faotto L, Negri A, Colombo I, Berra B, Bartorelli A, Ronchi S. The primary structure of UK114 tumor antigen. FEBS Lett. 1996;393(2-3):147–150. doi: 10.1016/0014-5793(96)00850-2. [DOI] [PubMed] [Google Scholar]
- 2.Oka T, Tsuji H, Noda C, Sakai K, Hong YM, Suzuki I, Munoz S, Natori Y. Isolation and characterization of a novel perchloric acid-soluble protein inhibiting cell-free protein synthesis. J Biol Chem. 1995;270(50):30060–30067. doi: 10.1074/jbc.270.50.30060. [DOI] [PubMed] [Google Scholar]
- 3.Schmiedeknecht G, Kerkhoff C, Orso E, Stohr J, Aslanidis C, Nagy GM, Knuechel R, Schmitz G. Isolation and characterization of a 14.5-kDa trichloroacetic-acid-soluble translational inhibitor protein from human monocytes that is upregulated upon cellular differentiation. Eur J Biochem. 1996;242(2):339–351. doi: 10.1111/j.1432-1033.1996.0339r.x. [DOI] [PubMed] [Google Scholar]
- 4.Morishita R, Kawagoshi A, Sawasaki T, Madin K, Ogasawara T, Oka T, Endo Y. Ribonuclease activity of rat liver perchloric acid-soluble protein, a potent inhibitor of protein synthesis. J Biol Chem. 1999;274(29):20688–20692. doi: 10.1074/jbc.274.29.20688. [DOI] [PubMed] [Google Scholar]
- 5.Melloni E, Michetti M, Salamino F, Pontremoli S. Molecular and functional properties of a calpain activator protein specific for mu-isoforms. J Biol Chem. 1998;273(21):12827–12831. doi: 10.1074/jbc.273.21.12827. [DOI] [PubMed] [Google Scholar]
- 6.Farkas A, Nardai G, Csermely P, Tompa P, Friedrich P. DUK114, the Drosophila orthologue of bovine brain calpain activator protein, is a molecular chaperone. Biochem J. 2004;383(Pt 1):165–170. doi: 10.1042/BJ20040668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burman JD, Stevenson CE, Sawers RG, Lawson DM. The crystal structure of Escherichia coli TdcF, a member of the highly conserved YjgF/YER057c/UK114 family. BMC Struct Biol. 2007;7:30. doi: 10.1186/1472-6807-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitz G, Downs DM. Reduced transaminase B (IlvE) activity caused by the lack of yjgF is dependent on the status of threonine deaminase (IlvA) in Salmonella enterica serovar Typhimurium. J Bacteriol. 2004;186(3):803–810. doi: 10.1128/JB.186.3.803-810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christopherson MR, Schmitz GE, Downs DM. YjgF is required for isoleucine biosynthesis when Salmonella enterica is grown on pyruvate medium. J Bacteriol. 2008;190(8):3057–3062. doi: 10.1128/JB.01700-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enos-Berlage JL, Langendorf MJ, Downs DM. Complex metabolic phenotypes caused by a mutation in yjgF, encoding a member of the highly conserved YER057c/YjgF family of proteins. J Bacteriol. 1998;180(24):6519–6528. doi: 10.1128/jb.180.24.6519-6528.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim JM, Yoshikawa H, Shirahige K. A member of the YER057c/yjgf/Uk114 family links isoleucine biosynthesis and intact mitochondria maintenance in Saccharomyces cerevisiae. Genes Cells. 2001;6(6):507–517. doi: 10.1046/j.1365-2443.2001.00443.x. [DOI] [PubMed] [Google Scholar]
- 12.Leitner-Dagan Y, Ovadis M, Zuker A, Shklarman E, Ohad I, Tzfira T, Vainstein A. CHRD, a plant member of the evolutionarily conserved YjgF family, influences photosynthesis and chromoplastogenesis. Planta. 2006;225(1):89–102. doi: 10.1007/s00425-006-0332-y. [DOI] [PubMed] [Google Scholar]
- 13.Deaconescu AM, Roll-Mecak A, Bonanno JB, Gerchman SE, Kycia H, Studier FW, Burley SK. X-ray structure of Saccharomyces cerevisiae homologous mitochondrial matrix factor 1 (Hmf1) Proteins. 2002;48(2):431–436. doi: 10.1002/prot.10151. [DOI] [PubMed] [Google Scholar]
- 14.Deriu D, Briand C, Mistiniene E, Naktinis V, Grutter MG. Structure and oligomeric state of the mammalian tumour-associated antigen UK114. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 9):1676–1678. doi: 10.1107/s0907444903014306. [DOI] [PubMed] [Google Scholar]
- 15.Manjasetty BA, Delbruck H, Pham DT, Mueller U, Fieber-Erdmann M, Scheich C, Sievert V, Bussow K, Niesen FH, Weihofen W, Loll B, Saenger W, Heinemann U. Crystal structure of Homo sapiens protein hp14.5. Proteins. 2004;54(4):797–800. doi: 10.1002/prot.10619. [DOI] [PubMed] [Google Scholar]
- 16.Miyakawa T, Lee WC, Hatano K, Kato Y, Sawano Y, Miyazono K, Nagata K, Tanokura M. Crystal structure of the YjgF/YER057c/UK114 family protein from the hyperthermophilic archaeon Sulfolobus tokodaii strain 7. Proteins. 2006;62(2):557–561. doi: 10.1002/prot.20778. [DOI] [PubMed] [Google Scholar]
- 17.Sinha S, Rappu P, Lange SC, Mantsala P, Zalkin H, Smith JL. Crystal structure of Bacillus subtilis YabJ, a purine regulatory protein and member of the highly conserved YjgF family. Proc Natl Acad Sci U S A. 1999;96(23):13074–13079. doi: 10.1073/pnas.96.23.13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volz K. A test case for structure-based functional assignment: the 1.2 A crystal structure of the yjgF gene product from Escherichia coli. Protein Sci. 1999;8(11):2428–2437. doi: 10.1110/ps.8.11.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dauter Z, Dauter M. Entering a new phase: using solvent halide ions in protein structure determination. Structure. 2001;9(2):R21–26. doi: 10.1016/s0969-2126(01)00565-2. [DOI] [PubMed] [Google Scholar]
- 20.Dauter Z, Dauter M, Rajashankar KR. Novel approach to phasing proteins: derivatization by short cryo-soaking with halides. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 2):232–237. doi: 10.1107/s0907444999016352. [DOI] [PubMed] [Google Scholar]
- 21.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vagin A, Teplyakov A. MOLREP: an Automated Program for Molecular Replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Dauter M, Dauter Z. Phase determination using halide ions. Methods Mol Biol. 2007;364:149–158. doi: 10.1385/1-59745-266-1:149. [DOI] [PubMed] [Google Scholar]
- 26.Korolev S, Dementieva I, Sanishvili R, Minor W, Otwinowski Z, Joachimiak A. Using surface-bound rubidium ions for protein phasing. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 7):1008–1012. doi: 10.1107/s0907444901007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagem RA, Dauter Z, Polikarpov I. Protein crystal structure solution by fast incorporation of negatively and positively charged anomalous scatterers. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 7):996–1002. doi: 10.1107/s0907444901007260. [DOI] [PubMed] [Google Scholar]
- 28.Leslie A. Joint CCP4+ESF-EAMCB Newsletter on Protein Crystallography. 1992. (No6).
- 29.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Cowtan KD, Zhang KY. Density modification for macromolecular phase improvement. Prog Biophys Mol Biol. 1999;72(3):245–270. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 31.Morris RJ, Zwart PH, Cohen S, Fernandez FJ, Kakaris M, Kirillova O, Vonrhein C, Perrakis A, Lamzin VS. Breaking good resolutions with ARP/wARP. J Synchrotron Radiat. 2004;11(Pt 1):56–59. doi: 10.1107/s090904950302394x. [DOI] [PubMed] [Google Scholar]
- 32.Panjikar S, Parthasarathy V, Lamzin VS, Weiss MS, Tucker PA. Auto-Rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 4):449–457. doi: 10.1107/S0907444905001307. [DOI] [PubMed] [Google Scholar]
- 33.Sheldrick GM, Schneider TR. SHELXL: high-resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 34.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 8):965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glaser F, Morris RJ, Najmanovich RJ, Laskowski RA, Thornton JM. A method for localizing ligand binding pockets in protein structures. Proteins. 2006;62(2):479–488. doi: 10.1002/prot.20769. [DOI] [PubMed] [Google Scholar]
- 36.Parsons L, Bonander N, Eisenstein E, Gilson M, Kairys V, Orban J. Solution structure and functional ligand screening of HI0719, a highly conserved protein from bacteria to humans in the YjgF/YER057c/UK114 family. Biochemistry. 2003;42(1):80–89. doi: 10.1021/bi020541w. [DOI] [PubMed] [Google Scholar]
- 37.Weng M, Nagy PL, Zalkin H. Identification of the Bacillus subtilis pur operon repressor. Proc Natl Acad Sci U S A. 1995;92(16):7455–7459. doi: 10.1073/pnas.92.16.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33(Web Server issue):W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15(4):305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.