Abstract
LR11 (SorLA) is a recently identified neuronal protein that interacts with amyloid precursor protein (APP), a central player in the pathology of the Alzheimer’s disease (AD). AD is a neurodegenerative disease and the most common cause of dementia in the elderly. Current estimates suggest that as many as 5.3 million Americans are living with AD. Recent investigations have uncovered the pathophysiological relevance of APP intracellular trafficking in AD. LR11 is of particular importance due to its role in regulating APP transport and processing. LR11 is a type I transmembrane protein and belongs to a novel family of Vps10p receptors. Using a new expression vector, pMTTH, (MBP- MCS1(multiple cloning site)-Thrombin protease cleavage site-MCS2-TEV protease cleavage site-MCS3-His6), we successfully expressed, purified and reconstituted the LR11 transmembrane (TM) and cytoplasmic (CT) domains into bicelles and detergent micelles for NMR structural studies. This new construct allowed us to overcome several obstacles during sample preparation. MBP fused LR11 TM and LR11TMCT proteins are preferably expressed at high levels in E. coli membrane, making a refolding of the protein unnecessary. The C-terminal His-tag allows for easy separation of the target protein from the truncated products from the C-terminus, and provides a convenient route for screening detergents to produce high quality 2D 1H-15N TROSY spectra. Thrombin protease cleavage is compatible with most of the commonly used detergents, including a direct cleavage at the E. coli membrane surface. This new MBP construct may provide an effective route for the preparation of small proteins with TM domains.
Keywords: Membrane protein overexpression, LR11, SorLA, Amyloid precursor protein binding protein, Alzheimer’s disease
Introduction
LR11 (SorLA) is a recently identified neuronal protein that interacts with amyloid precursor protein (APP) [1-3]. The sequential hydrolysis of APP by β- and γ-Secretases produces 40/42-residue peptides called amyloid-β (Aβ) [4-7]. The accumulation of Aβ in the brain is closely associated with the development of Alzheimer’s disease (AD), a progressive neurodegenerative disease characterized by a global cognitive decline involving memory, orientation, judgment and reasoning [8-13]. Due to increasing longevity, AD is becoming the most common form of dementia in the elderly. Current estimates suggest that as many as 5.3 million Americans are living with AD and projections are that more than $20 trillion will be spent on treatment costs over the next 40 years (http://www.alz.org). The relevance of LR11 in AD was first implicated in a study by Dodson et al. which indicated that the expression of LR11 is consistently low in the brains of patients suffering from sporadic AD [14, 15]. This was further substantiated by the association of variants in LR11 genes with AD [16]. In vitro, cell culture, knockout mouse models, and clinical investigations all support the concept that LR11 plays a crucial role in APP trafficking and is a key regulator of APP processing [14, 17, 18].
LR11/SorLA is a 250 kDa, highly conserved type-1 transmembrane protein that is predominately expressed in the neurons of the cortex and hippocampus, regions of the brain that are associated with memory. It contains a vacuolar protein sorting 10 protein (Vps10p) homology domain, β-propeller and epidermal growth factor (EGF) domains, a cluster of 11 complement-type repeat domains, six fibronectin type III repeats, a single transmembrane domain (TM), and a cytoplasmic domain (CT) [19, 20]. LR11 regulates APP trafficking between the trans-Golgi network (TGN) and early endosomes by sequestering APP in the TGN, and consequently reduces the amount of APP that can be processed to Aβ and other products in post-Golgi compartments and at the cell surface. LR11 also shuttles APP from early endosomes back to the TGN by interacting with cargo molecules such as GGAs and PACS-1, and further reduces the amount of APP in late endosomes where most Aβ peptides are produced [21-24]. These regulatory roles of LR11 require its proper location to the TGN, which is critically dependent on multiple motifs in its CT and the interactions of these motifs with adaptor proteins [22, 24]. Furthermore, the LR11 CT may directly interact with the C-termini of APP and β-secretase [18, 25], and regulate transcription after cleavage by γ-secretase [26, 27].
Little is known about the structures of the LR11 TM and CT domains alone or in complex with their biological partners. The preparation of proteins with TM domains in sufficient quantity for structural analysis is difficult [28]. One method to address this challenge is to express these proteins as fusion constructs with more soluble proteins such as maltose binding protein (MBP), glutathione S-transferase (GST), thioredoxin, or staph-nuclease [29]. MBP fused proteins, in particular for low molecular weight membrane proteins, express well and frequently appear in the membrane fraction [30]. However, when our laboratory expressed the LR11 TM and CT domains fused to MBP, we observed several products that correspond to truncated forms of the full-length construct degraded from the C-terminus. These “premature” products hampered protein purification. To resolve this problem, we prepared a new expression vector, pMTTH, MBP- MCS1(multiple cloning site)-Thrombin protease cleavage site-MCS2-TEV protease cleavage site-MCS3-His6. This vector retained the high level expression of MBP, allowed for easy separation of the full-length proteins from “premature” products, and offered a convenient route for detergent optimization, a necessary step in sample preparation for NMR structural studies. High quality 2D 1H-15N TROSY spectra have been obtained on the resulting recombinant proteins reconstituted in bicelles and detergent micelles and preliminary NMR chemical shift analysis supports the predicted secondary structure of the TM helix.
Material and Methods
Plasmid construction
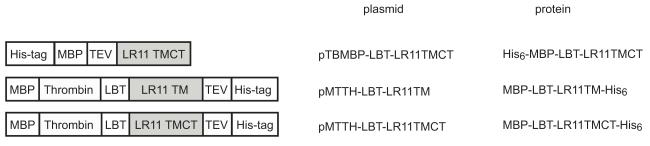
The codon usage of an LR11 fragment including TM and CT domains was optimized and synthesized for E. coli expression. Plasmid pMTTH (supplemental Figure S1) was derived from plasmid pTBMBP (His6-MCS1-MBP-TEV cleavage site-MCS2), provided by Dr. Cross’ lab [29]. Three clones were constructed by PCR (Figure 1): LR11 TM and CT domains (residues 2132 to 2214) insert into the SspI sites of the vector pTBMBP, LR11 TM domain (residues 2132 to 2160) insert into the BamHI/HindIII sites of the vector pMTTH, and LR11 TM and CT domains insert into the BamHI/HindIII sites of the vector pMTTH. All selected clones were verified by DNA sequencing.
Figure 1.
Schematic representations of the expression constructs used in this study. MBP: maltose binding protein; LBT: lanthanide binding tag (YIDTNNDGWYEGDELLA) [36]; TEV: TEV protease cleavage site; Thrombin: thrombin protease cleavage site; TM: transmembrane domain; CT: cytoplasmic domain. In this study, the LR11 TM construct includes residues 2132 to 2160 and the LR11-TMCT construct includes residues 2132 to 2214.
Expression of His6-MBP-LBT-LR11TMCT, MBP-LBT-LR11TM-His6 and MBP-LBT-LR11TMCT-His6
Each recombinant plasmid, pTBMBP-LBT-LR11TMCT, pMTTH-LBT-LR11TM, or pMTTH-LBT-LR11TMCT, was separately introduced into E. coli BL21 CodonPlus (DE3) RIPL competent cells (Stratagene) for protein expression. Cells were grown in 1 to 2 mL LB medium overnight and then inoculated in 250 to 320 mL of LB medium for production of unlabeled proteins or M9 medium (3 g/L KH2PO4, 6 g/L Na2HPO4, 0.5 g/L NaCl, 0.2 mM MgSO4, 7 mg/L (NH4)2Fe(SO4)2·6H2O, and 0.01 mg/L thiamine hydrochloride) supplemented with D-glucose (or D-glucose-13C6) (4 g/L) and 15NH4Cl (1 g/L) for 15N (or 15N/13C) labeled samples. Cells were induced at A600nm 0.6-0.9 with 2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 16 °C for ~27 hours. Cells were harvested by centrifugation and stored at −80 °C until use.
Purification of MBP-LBT-LR11TMCT-His6, MBP-LBT-LR11TM-His6 and His6-MBP-LBT-LR11TMCT
For purification of MBP-LBT-LR11TMCT-His6 and MBP-LBT-LR11TM-His6, cells were incubated on ice for 20 min and then resuspended in a lysis buffer consisting of 20 mM Tris·HCl, 500 mM NaCl, 20 mM imidazole, pH 8.0, 0.5 mM PMSF (phenylmethanesulfonyl fluoride), protease inhibitor cocktail (Sigma), lysozyme (30 mg/L, Fisher) and benzonase nuclease (250 units/L, Novagen). The sample was incubated at ~10 °C for another 20 min and subsequently sonicated on ice for a total of 6 min with 3s on and 7s off. The cell lysate was centrifuged at ~30,000g for 30 min. The insolubles were discarded and the supernatant either was mixed with a stock solution of DPC to a final concentration of 1% DPC (referred to below as the low speed supernatant fraction, which includes the soluble fraction and membrane fraction), or further centrifuged at ~160,000g for 1 hour to obtain the membrane pellet. Proteins in the pellet were extracted with a wash buffer of 20 mM Tris·HCl, 500 mM NaCl, 20 mM imidazole, pH 8.0 containing 2% DPC for 1 hour (referred to below as the membrane fraction). DPC extraction of the low speed supernatant fraction (or membrane fraction) was loaded onto a column containing 3 or 5 mL Ni-NTA resin, respectively. After washing with 100 mL wash buffer containing 0.15% DPC, the LR11 fusion protein was eluted with a buffer consisting of 20 mM Tris-HCl, 300 mM NaCl, 250 mM imidazole, pH 8.0 and 0.2% DPC. Unless specified, proteins prepared from the low speed supernatant fraction were used in this study.
For the purification of His6-MBP-LBT-LR11TMCT, the experimental steps were similar to the procedures described above except a buffer containing 1% Triton X-100 instead of 1% DPC was used to extract proteins, and the wash and elution buffers contained 0.1% Triton X-100 instead of DPC. In addition, the elute from a Ni-NTA column was further purified with an amylose column and eluted with a buffer of 20 mM Tris·HCl, 200 mM NaCl, 10 mM maltose, pH 7.4 and 0.1% Triton X-100.
Enzymatic cleavage and detergent optimization of MBP-LBT-LR11TM-His6, MBP-LBT-LR11TMCT-His6 and His6-MBP-LBT-LR11TMCT
The eluted protein of MBP-LBT-LR11TMCT-His6 or MBP-LBT-LR11TM-His6 was first dialyzed against a buffer of 140 mM NaCl, 10 mM phosphate, pH 7.3, 3 mM β-mercaptoethanol and 0.05% DPC for ~24 hours at 4 °C with 3,500 Da Spectra/Por dialysis tubing. The MBP fusion protein was then digested with ~15 units of thrombin (GE Healthcare, 27-0846-01) per mg of sample for 1 to 2 days at room temperature with slow rotation. After the enzymatic cleavage of MBP, the sample was loaded onto a Ni-NTA column. The column was washed with ~20 bed volumes of lysis buffer containing either 0.1% DPC, 0.1% LDAO, 0.1% DDM, 0.15% LMPG, or 1% bicelles ([DMPC]/[DHPC]=0.25). The target protein (LBT-LR11TMCT-His6 or LBT-LR11TM-His6) was eluted with the elution buffer containing 0.2% DPC, 0.3% LDAO, 0.2% DDM, 0.2% LMPG, or 2% bicelles. These samples were exchanged into an NMR buffer of 20 mM phosphate, 100 mM NaCl, pH 7.0 and one of the above detergents with 3,500 Da Spectra/Por dialysis tubing and concentrated to 500 μL with an Amicon Ultra-15 (MWCO = 3000 Da). Additional detergent was added to each sample to adjust its final concentration to 3.2% LADO, 1.3% LMPG, 4.5% DPC or 10% bicelles, respectively. The purity of these samples was assessed by SDS-PAGE.
The eluted protein of His6-MBP-LBT-LR11TMCT from the amylose column was digested with ~0.3 mg of His-tagged TEV protease per mg of fusion protein at room temperature for ~6 hours in 50 mM Tris·HCl, pH 8.0, 1 mM DTT, 100 mM NaCl, and 0.1% Triton X-100. After enzymatic cleavage, the solution was re-passed over a Ni-NTA column, and the target protein, LBT-LR11TMCT, was collected in the flow-through.
NMR spectroscopy
All NMR spectra were recorded at 37 °C. Experiments for backbone resonance assignment of LBT-LR11TM-His6 were collected on a sample of ~1 mM 15N,13C labeled protein in 4.5% DPC solution on a Bruker 600 MHz instrument unless otherwise specified. TROSY-HNCA, TROSY-HNCA-intra [31], TROSY-HNCACB and CBCA(CO)NH data were acquired with t1,max of 5.7 to 6.6 ms (13C), t2,max of 14 to 22 ms (15N), t3,max of 107 ms (1H), 24 to 48 scans per increment and a 1.5 s recycle delay. 3D 15N-edited NOESY data was collected on a Bruker 850 MHz spectrometer with t1,max of 8.6 ms (1H), t2,max of 8.7 ms (15N), t3,max of 75 ms (1H), 80 ms mixing time, 16 scans per increment and a 1.5 s recycle delay. NMR data were processed with NMRPipe and analyzed using Sparky software. Processing scripts were optimized for each dataset, but typically each FID was apodized with a shifted sine-bell function, and zero-filled in all dimensions. Linear prediction was implemented to double the data size in the t2 dimension to improve the spectral resolution. Sequential connectivities were established by TROSY-HNCA and TROSY-HNCACB experiments, which provided intraresidue and sequential cross-peaks of Cα and Cα/Cβ, respectively (refer to Figure S3 for examples of strip plots). The symmetric HN-HN NOEs were used to resolve and validate the connectivity when available. The chemical shifts of Cα, Cβ, N, and C’ were used to predict backbone torsion angles by TALOS [32]. The assignment data have been deposited in the BioMagResBank (BMRB accession code: 17444).
Results and Discussion
Sequence conservation of the LR11 TM and CT domains
The primary sequences of LR11 TM and CT domains in mammals share > 95% identity [33]; thus we included homologs from more distant organisms for the alignment analysis (Figure 2). These proteins are highly conserved, pointing to their functional significance. The LR11 CT domain harbors multiple conserved motifs essential for its functions. The first region, F(A/V)(N/S)SHY, is similar to the internalization signal of coated pit receptors [20]. The second region, F(S/A)DD(V/E)P(L/M)(V/I)(I/V)A, is a GGA binding motif [23] [34]. In addition, an acid cluster (DDLGEDDED) in the CT domain has been shown to interact with the PACS-1 protein [24]. The proper location and activity of LR11 are dependent on functional interactions with GGA, PACS-1 and AP-1, adaptor proteins that mediate Golgi to endosome transports.
Figure 2.
Sequence alignments of LR11 TM and CT domains from various species including Homo sapiens (BAG63842), Canis familiaris (XP_536545), Gallus gallus (XP_001232946), Taeniopygia guttata (XP_002191173) and Nasonia vitripennis (NP_001123523) by ClustalW [45] and decorated using ESPript [46]. Strictly conserved residues have a red background, well conserved residues are in red, and residues conserved between species are in blue boxes.
Production of His6-MBP-LBT-LR11TMCT
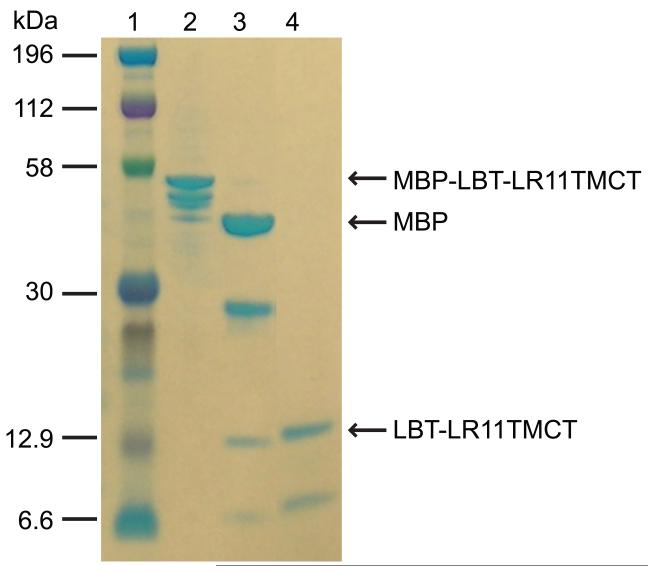
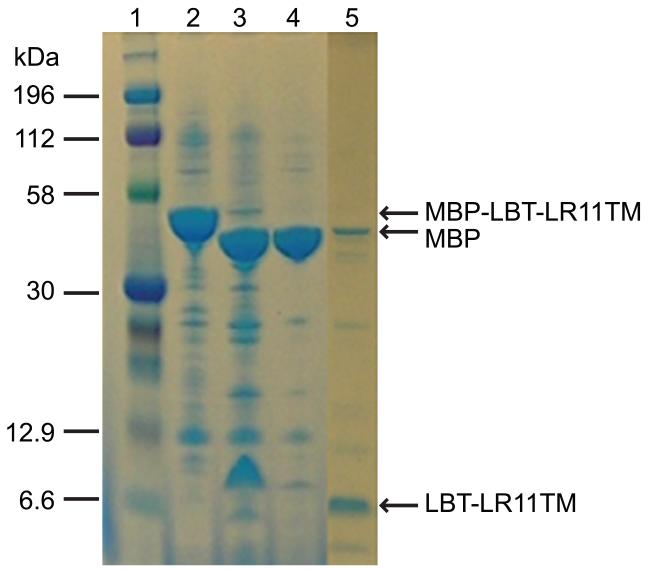
The expression of proteins with transmembrane domains is not straightforward. Since direct expression of a His-tagged LR11 TMCT construct was not successful, we used an MBP-fusion expression system. A His6-MBP-LBT-LR11TMCT (Figure 1) construct was prepared with the pTBMBP plasmid. This construct includes a small lanthanide-binding peptide tag (LBT, YIDTNNDGWYEGDELLA) fused to the N-terminus of the LR11 TM domain. The LBT tag is known to express well and its binding of a paramagnetic ion with anisotropic magnetic susceptibility provides a means to align a protein in a magnetic field in ordr to obtain orientational information for NMR structural studies [35-37]. This fusion construct showed good expression. However, several proteins which have slightly smaller sizes than the targeted His6-MBP-LBT-LR11TMCT were found in the final elute of the amylose column. In an SDS-PAGE gel (lane 2 of Figure 3), at least three additional bands right below the top band, which correspond to the targeted protein, were clearly visible. These impurities amount to >30 % of the total protein and are truncated forms of the full-length construct from the C-terminus since they are cleavable by the TEV protease (lane 3 of Figure 3). Extensive explorations of the expression conditions, such as reduction of the IPTG induction concentration from 2 to 0.05 mM, varying growth temperatures from 8 to 37 °C, elongating induction times from several hours to several days, and expression of the proteins in different E. coli strains, did not eliminate these “premature” products. These by-products complicated the purification process. The final purified sample showed two major bands on the SDS-PAGE gel (lane 4 of Figure 3) and produced an inhomogeneous NMR spectrum (data not shown). In addition, the yield of full-length protein was rather low (~1.5 mg product per liter culture).
Figure 3.
SDS-PAGE results for the purification of His6-MBP-LBT-LR11TMCT. Lanes: 1, protein marker; 2, elute of His6-MBP-LBT-LR11TMCT from amylose column; 3, TEV cleavage of the protein sample in lane 2; 4, flow-through of a Ni-NTA column for the sample in lane 3.
Production and detergent screens of MBP-LBT-LR11TMCT-His6 and MBP-LBT-LR11TM-His6
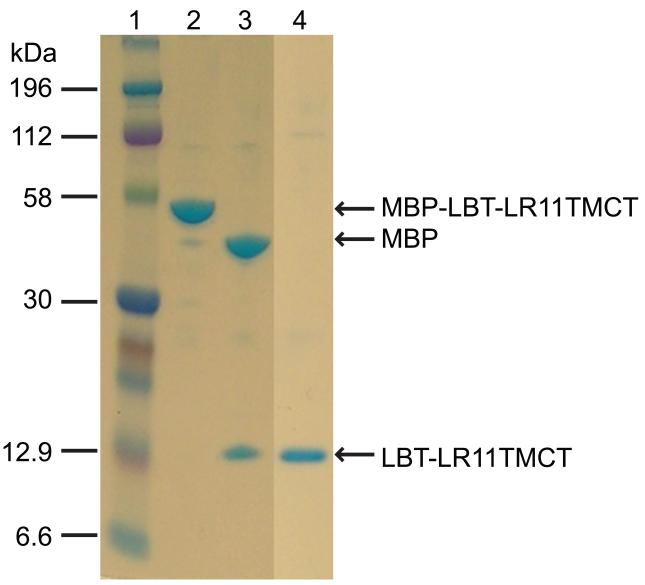
To overcome the difficulties of protein purification associated with these “premature” products, we prepared a new construct, the pMTTH vector, from the vector PTBMBP, moving the His6-tag from the N- to the C-terminus (Figure 1S). The MBP-LBT-LR11TMCT-His6 construct showed an increased level of expression and more importantly, the C-terminal His6 tag allowed separation of the targeted protein from the “premature” by-products. A single step purification with the Ni-NTA column generated the pure protein (lane 2, Figure 4). Subsequent thrombin cleavage and passage back over the Ni-NTA column produced the LBT-LR11TMCT-His6 with a yield of ~5 mg per liter culture.
Figure 4.
SDS-PAGE results for the purification of MBP-LBT-LR11TMCT-His6. Lanes: 1, protein marker; 2, elute of MBP-LBT-LR11TMCT-His6 from a Ni-NTA column; 3, thrombin cleavage of the protein sample in lane 2; 4, elute from a Ni-NTA column for the sample in lane 3 with an elution buffer containing 0.2% DPC.
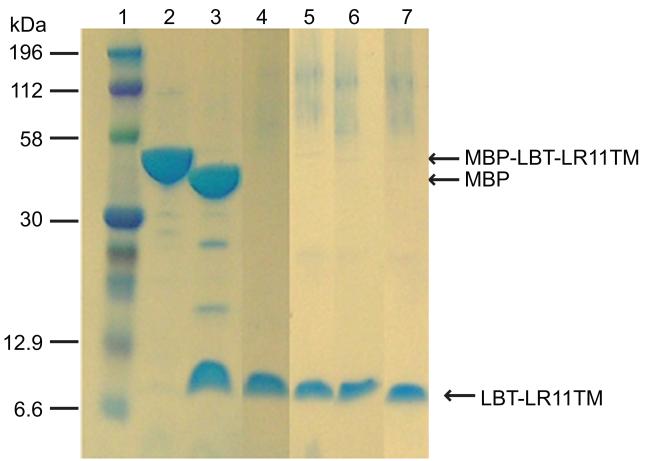
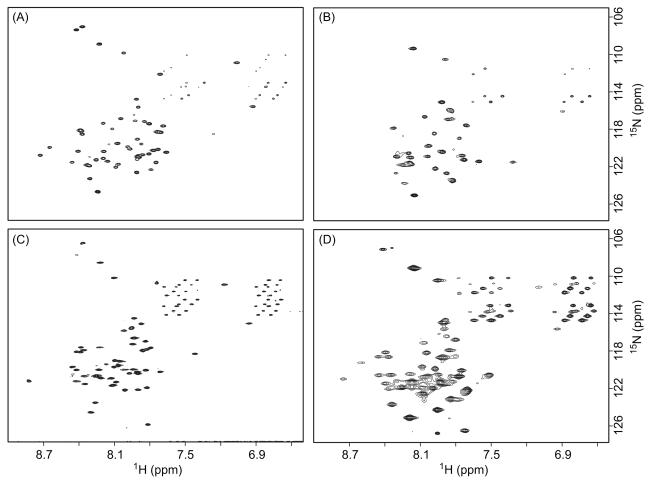
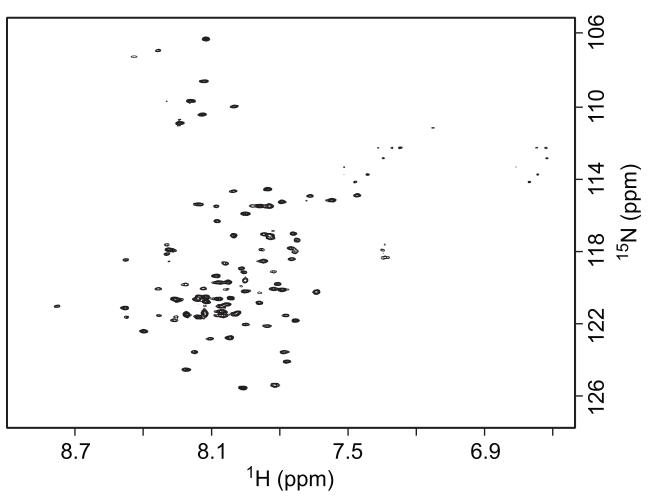
Membrane proteins are often reconstituted into detergent micelles for solution NMR studies, and screening for proper detergents is an essential step. Recent investigations highlight the importance of using detergent micelles to conserve the native structure of certain proteins [38-41]. It is widely accepted that a well-resolved NMR spectrum does not always represent the native conformation of the protein in its biological environment. Therefore, the selection of detergents should be guided by functional assays whenever possible. In the absence of such assays, it is becoming common practice to use a 2D 1H-15N TROSY spectrum from bilayer model systems, such as bicelles and lipid-protein nanodiscs, as a reference in the selection of detergent mimics for structural studies [42, 43]. The extraction and purification of the LBT-LR11TM-His6 protein (Figure 1) was used to explore several commonly used procedures and detergents for the following reasons. First, this protein is expressed at a high level (~20 mg per liter culture). Second, in comparison with LBT-LR11TMCT-His6, the shorter construct of LBT-LR11TM-His6 has fewer expected resonances and allows for easier identification of resonances from the LR11TM domain. The C-terminal His6- tag also provided a convenient route for detergent optimization with a Ni-NTA column after removal of MBP. Figure 5 shows the SDS-PAGE gel results of LBT-LR11TM-His6 eluted with 0.2% DPC, 0.3% LDAO, 0.2% LMPG, and 0.2% DDM (lanes 4 to 7, respectively). 2D 1H-15N TROSY spectra from the purified protein reconstituted in bicelles and detergent micelles are shown in Figure 6. The TROSY spectrum of the protein in DPC micelles most resembles the spectrum from bicelles and displays typical chemical shift dispersion for a helical protein. The backbone torsion angles obtained by analyzing assigned chemical shifts of Cα, Cβ, C’, and N using the TALOS program (listed in supplemental Table S1) are consistent with the expected helical structure of the TM domain. Subsequently, LBT-LR11TMCT was purified and reconstituted into DPC micelles and its TROSY spectrum is shown in Figure 7. The spectrum is well resolved and ~100 out of 113 expected resonances are observed, including 8 out of 9 Glys in the sequence. The amide proton of the N-terminal Gly, a remaining residue after thrombin cleavage, is likely not detected due to its fast exchange with water.
Figure 5.
SDS-PAGE results for the purification of MBP-LBT-LR11TM-His6. Lanes: 1, protein marker; 2, elute of MBP-LBT-LR11TM-His6 from a Ni-NTA column; 3, thrombin cleavage of the protein sample in lane 2; 4 to 7, elutes of LBT-LR11TM-His6 from Ni-NTA columns with an elution buffer containing 0.2% DPC, 0.3% LDAO, 0.2% LMPG, and 0.2% DDM, respectively.
Figure 6.
2D 1H-15N TROSY spectra of 15N-LBT-LR11TM-His6 protein in the NMR buffer containing (A) 3.2% LDAO, (B) 1.3% LMPG, (C) 4.5% DPC and (D) 10% bicelles ([DMPC]/[DHPC]=0.25). Spectra (A) and (B) were recorded on a Bruker 600 MHz spectrometer with t1,max of 82 ms (15N), t2,max of 107 ms (1H) and 24 scans per increment. Spectrum (C) was recorded on a Bruker 850 MHz spectrometer with t1,max of 97 ms (15N), t2,max of 75 ms (1H) and 4 scans per increment. Spectrum (D) was recorded on a Bruker 600 MHz spectrometer with t1,max of 69 ms (15N), t2,max of 95 ms (1H) and 16 scans per increment. The protein concentrations for spectra (A) and (B) were ~0.5 mM, and for spectra (C) and (D) were ~1 mM.
Figure 7.
A 2D 1H-15N TROSY spectrum of the 15N-LBT-LR11TMCT-His6 (~0.5 mM) in the NMR buffer containing 4.5% DPC recorded on a Varian 900 MHz spectrometer with t1,max of 85 ms (15N), t2,max of 96 ms (1H) and 4 scans per increment.
“In situ” thrombin cleavage of MBP fusion proteins at E. coli membranes
In the current study, thrombin cleavage of the MBP fusion proteins was successful in the presence of several commonly used detergents including 0.2% DPC, 0.3% LDAO, 0.2% LMPG and 0.2% DDM, although some difficulties were reported in previous publications [30]. The thrombin cleavage site in our constructs locates 22 residues preceding the first residue from LR11 (15 residues from the LBT tag); thus it may be easily accessible by the protease. Since most of the recombinant proteins were found in the membrane fraction in this study (Figure S3), we also tested the efficiency of thrombin cleavage at E. coli membranes. Membrane pellets prepared from high-speed centrifugation (~160,000g) of the low spin supernatant fraction were re-suspended in a phosphate buffer (lane 2 of Figure 8) and incubated with the thrombin protease for ~21 hours. As shown in an SDS-PAGE gel (lane 3 of Figure 8), >95% of the fusion protein was cleaved. Subsequently, DPC detergent was added to the solution to extract the target protein from the E. coli membrane and the solution was loaded onto a Ni-NTA column. MBP and other impurities were found in the flow-through (lane 4 of Figure 8). The target protein was eluted from the column after several washes (lane 5 of Figure 8), and its spectrum (Figure S5) is nearly identical to the one shown in Figure 6c. The successful “in situ” removal of MBP from the target protein at E. coli membrane surfaces provides an alternate solution for the separation of fusion protein in cases where the protease is not compatible with the detergent micelles or the cleavage site becomes inaccessible after membrane extraction.
Figure 8.
“In situ” thrombin cleavage of MBP-LBT-LR11TM-His6 at E. coli membrane surface. Lanes: 1, protein marker; 2, resuspension of MBP-LBT-LR11TM-His6 membrane fraction; 3, thrombin cleavage of the protein sample in lane 2; 4: flow-through of the sample in lane 3 from a Ni-NTA column; 5: elute of the sample in lane 3 from a Ni-NTA column.
In summary, we have successfully prepared the LR11 TM and CT domains for NMR studies. Using an MBP fused construct with a His tag at the C-terminus, we overcame several obstacles related to sample preparation. The MBP fusion system dramatically increased protein yields and, moreover, the protein was expressed in the E. coli membrane. This is favorable since refolding of the protein is not necessary. The C-terminal His tag allowed easy separation of the target protein from the “premature” products and provided a convenient route for screening detergents. In addition, the thrombin protease cleavage is compatible with most of the commonly used detergents, including a direct cleavage at the E. coli membrane surface. The MBP fusion expression system appears to be an effective approach for the preparation of small proteins with TM domains [30, 44] and its unique advantage in expressing the target proteins in the membrane is particularly appealing, as demonstrated here.
Supplementary Material
Acknowledgments
We are grateful for financial support from the National Institutes of Health (5R01GM081793-03) and the Penn State University College of Medicine. We thank Dr. J. M. Flanagan at the Penn State University College of Medicine for providing the plasmid of His-tagged TEV protease and helpful discussions, Dr. A. Benesi at the NMR facility of the Penn State University, University Park for the assistance of the use of 850 MHz instrument, Dr. J. Glushka at the Complex Carbohydrate Research Center of the University of Georgia for the assistance of the use of 900 MHz instrument, Drs. J. J. Lah and A. I. Levey at the Emory University for providing the LR11 gene and helpful discussions, and Dr. T. A. Cross at the Florida State University for providing the pTBMBP plasmid.
Abbreviations used:
- TM
transmembrane domain
- CT
cytoplasmic domain
- APP
amyloid precursor protein
- AD
Alzheimer’s disease
- MBP
Maltose binding protein
- LBT
Lanthanide binding tag
- DPC
dodecylphosphocholine
- LDAO
lauryldimethylamine oxide
- DDM
n-dodecyl-beta-D-maltoside
- LMPG
lysomyristoyl-phosphotidyl-glycerol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Small SA, Gandy S. Sorting through the cell biology of Alzheimer’s disease: intracellular pathway to pathogenesis. Neuron. 2006;52:15–31. doi: 10.1016/j.neuron.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, Arnim CAF, Breiderhoff T, Jansen P, Wu X, Bales KR, Cappai R, Masters CL, Gliemann J, Mufson EJ, Hyman BT, Paul SM, Nykjaer A, Willnow TE. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Willnow TE, Petersen CM, Nykjaer A. VPS10P-domain receptors - regulators of neuronal viability and function. Nat. Rev. Neurosci. 2008;9:899–909. doi: 10.1038/nrn2516. [DOI] [PubMed] [Google Scholar]
- [4].Esler WP, Wolfe MS. A protrait of Alzheimer secretases - new features and familiar faces. Science. 2001;293:1449–1454. doi: 10.1126/science.1064638. [DOI] [PubMed] [Google Scholar]
- [5].Vassar R, Citron M. Aβ-generating enzymes: recent advances in β- and γ-secretase research. Neuron. 2000;27:419–422. doi: 10.1016/s0896-6273(00)00051-9. [DOI] [PubMed] [Google Scholar]
- [6].Yan R, Bienkowski MJ, Shuck ME, Miao H, Tory MC, Pauley AM, Brashler JR, Stratman NC, Mathews WR, Buhl AE, Cater DB, Tomasselli A, Parodi LA, Heinrikson RL, Gurney ME. Membrane-anchored aspartyl protease with Alzheimer’s disease β-secretase activity. Nature. 1999;402:533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- [7].Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz E, Denis P, Teplow DB, Ross S, Amarante P, Leoeloff R, Lou Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AJ, Curran E, Burgess T, Louis J, Collins F, Treanor J, Rogers G, Citron M. β-Secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- [8].Hardy JA, Higgins GA. Alzheimer’s disease: The amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- [9].Tanzi RE, Bertram L. Twenty years of the Alzheimer’s disease amyloid hypothesis: A genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- [10].Selkoe DJ. The ups and downs of A beta. Nat. Med. 2006;12:758–759. doi: 10.1038/nm0706-758. [DOI] [PubMed] [Google Scholar]
- [11].Jellinger KA. Alzheimer 100 - highlights in the history of Alzheimer research. J. Neural Transm. 2006;113:1603–1623. doi: 10.1007/s00702-006-0578-3. [DOI] [PubMed] [Google Scholar]
- [12].Jakob-Roetne R, Jacobsen H. Alzheimer’s disease: From pathology to therapeutic approaches. Angew. Chem.-Int. Edit. 2009;48:3030–3059. doi: 10.1002/anie.200802808. [DOI] [PubMed] [Google Scholar]
- [13].Selkoe DJ. Alzheimer’s disease: genes, protein, and therapy. Physiol. Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- [14].Dodson SE, Gearing M, Lippa CF, Montine TJ, Levey AI, Lah JJ. LR11/SorLA expression is reduced in sporadic Alzheimer disease but not in familial Alzheimer disease. J. Neuropathol. Exp. Neurol. 2006;65:866–872. doi: 10.1097/01.jnen.0000228205.19915.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Offe K, Dodson SE, Shoemaker JT, Fritz JJ, Gearing M, Levey AI, Lah JJ. The lipoprotein receptor LR11 regulates amyloid β production and amyloid precursor protein traffic in endosomal compartments. J. Neurosci. 2006;26:1596–1603. doi: 10.1523/JNEUROSCI.4946-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rogaeva E, Meng Y, Lee JH, Gu YJ, Kawarai T, Zou FG, Katayama T, Baldwin CT, Cheng R, Hasegawa H, Chen FS, Shibata N, Lunetta KL, Pardossi-Piquard R, Bohm C, Wakutani Y, Cupples LA, Cuenco KT, Green RC, Pinessi L, Rainero I, Sorbi S, Bruni A, Duara R, Friedland RP, Inzelberg R, Hampe W, Bujo H, Song YQ, Andersen OM, Willnow TE, Graff-Radford N, Petersen RC, Dickson D, Der SD, Fraser PE, Schmitt-Ulms G, Younkin S, Mayeux R, Farrer LA, George-Hyslop PS. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 2007;2:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mayeux R, Hyslop PSG. Alzeimer’s disease: advances in trafficking. Neurology. 2008;7:2–3. doi: 10.1016/S1474-4422(07)70298-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Spoelgen R, Von Arnim CAF, Thomas AV, Peltan ID, Koker M, Deng A, Irizarry MC, Anderson DE, Willnow TE, Hyman BT. Interaction of the cytosolic domains of sorLA/LR11 with the amyloid precursor protein (APP) and beta-secretase beta-site APP-cleaving enzyme. J. Neurosci. 2006;26:418–428. doi: 10.1523/JNEUROSCI.3882-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jacob J, Madsen P, Jacobsen C, Nielsen MS, Gliemann J, Petersen CM. Activation and functional characterization of the mosaic receptor SorLA/LR11. J. Biol. Chem. 2001;276:222788–222796. doi: 10.1074/jbc.M100857200. [DOI] [PubMed] [Google Scholar]
- [20].Yamazaki H, Bujo H, Kusunoki J, Seimiya K, Kanaki T, Morisaki N, Schneider WJ, Saito Y. Elements of neural adhesion molecules and a yeast vacuolar protein sorting receptor are present in a novel mammalian low density lipoprotein receptor family member. J. Biol. Chem. 1996;271:24761–24768. doi: 10.1074/jbc.271.40.24761. [DOI] [PubMed] [Google Scholar]
- [21].Youker RT, Shinde U, Day R, Thomas G. At the crossroads of homoeostasis and disease: roles of the PACS proteins in membrane traffic and apoptosis. Biochem. J. 2009;421:1–15. doi: 10.1042/BJ20081016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nielsen MS, Gustafsen C, Madsen P, Nyengaard JR, Hermey G, Bakke O, Mari M, Schu P, Pohlmann R, Dennes A, Petersen CM. Sorting by the cytoplasmic domain of the amyloid precursoe protein binding receptor SorLA. Mol. Cell. Biol. 2007;27:6842–6851. doi: 10.1128/MCB.00815-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jacobsen L, Madsen P, Nielsen MS, Geraerts WPM, Gliemann J, Smit AB, Petersen CM. The sorLA cytoplasmic domain interacts with GGA1 and −2 and defines minimum requirements for GGA binding. FEBS Lett. 2002;511:155–158. doi: 10.1016/s0014-5793(01)03299-9. [DOI] [PubMed] [Google Scholar]
- [24].Schmidt V, Sporbert A, Rohe M, Reimer T, Rehm A, Andersen OM, Willnow TE. SorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. J. Biol. Chem. 2007;282:32956–32964. doi: 10.1074/jbc.M705073200. [DOI] [PubMed] [Google Scholar]
- [25].Andersen OM, Schmidt V, Spoelgen R, Gliemann J, Behlke J, Galatis D, McKinstry WJ, Parker MW, Masters CL, Hyman BT, Cappai R, Willnow TE. Molecular dissection of the interaction between amyloid precursor protein and its neuronal trafficking receptor SorLA/LR11. Biochemistry. 2006;45:2618–2628. doi: 10.1021/bi052120v. [DOI] [PubMed] [Google Scholar]
- [26].Bohm C, Seibel NM, Henkel B, Steiner H, Haass C, Hampe W. SorLA signaling by regulated intramembrane proteolysis. J. Biol. Chem. 2006;281:14547–14533. doi: 10.1074/jbc.M601660200. [DOI] [PubMed] [Google Scholar]
- [27].Nyborg AC, Ladd TB, Zwizinski CW, Golde TE. Sortilin, SorCS1b and SorLA Vps10p sorting receptors, are novel γ-secretase substrates. Mol. Neurodegener. 2006;1:3. doi: 10.1186/1750-1326-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Junge F, Schneider B, Reckel S, Schwarz D, Dotsch V, Bernhard F. Large-scale production of functional membrane proteins. Cell. Mol. Life Sci. 2008;65:1729–1755. doi: 10.1007/s00018-008-8067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Qin HJ, Hu J, Hua YZ, Challa SV, Cross TA, Gao FP. Construction of a series of vectors for high throughput cloning and expression screening of membrane proteins from Mycobacterium tuberculosis. BMC Biotechnol. 2008;8:51. doi: 10.1186/1472-6750-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Korepanova A, Moore JD, Nguyen HB, Hua Y, Cross TA, Gao F. Expression of membrane proteins from Mycobacterium tuberculosis in Escherichia coli as fusions with maltose binding protein. Protein Expres. Purif. 2007;53:24–30. doi: 10.1016/j.pep.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nietlispach D, Ito Y, Laue ED. A novel approach for the sequential backbone assignment of large proteins: Selective intra-HNCA and DQ-HNCA. J. Am. Chem. Soc. 2002;124:11199–11207. doi: 10.1021/ja025865m. [DOI] [PubMed] [Google Scholar]
- [32].Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- [33].Seimiya K, Moerwald S, Bujo H, Yamazaki H, Kusunoki J, Kanaki T, Morisaki N, Nimpf J, Schneider WJ, Saito Y. Identification and molecular characterization of LR11 from man and chicken. Atherosclerosis. 1997;134:362–362. [Google Scholar]
- [34].Cramer JF, Gustafsen C, Behrens MA, Oliveira CLP, Pedersen JS, Madsen P, Petersen CM, Thirup SS. GGA autoinhibition revisited. Traffic. 2010;11:259–273. doi: 10.1111/j.1600-0854.2009.01017.x. [DOI] [PubMed] [Google Scholar]
- [35].Wohnert J, Franz KJ, Nitz M, Imperiali B, Schwalbe H. Protein alignment by a coexpressed lanthanide-binding tag for the measurement of residual dipolar couplings. J. Am. Chem. Soc. 2003;125:13338–13339. doi: 10.1021/ja036022d. [DOI] [PubMed] [Google Scholar]
- [36].Nitz M, Franz KJ, Maglathlin RL, Imperiali B. A powerful combinatorial screen to identify high-affinity terbium(III)-binding peptides. ChemBioChem. 2003;4:272–276. doi: 10.1002/cbic.200390047. [DOI] [PubMed] [Google Scholar]
- [37].Franz KJ, Nitz M, Imperiali B. Lanthanide-binding tags as versatile protein coexpression probes. ChemBioChem. 2003;4:265–271. doi: 10.1002/cbic.200390046. [DOI] [PubMed] [Google Scholar]
- [38].Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres AO, Cahill SM, Hicks DB, Grinius L, Krulwich TA, Girvin ME. An evaluation of detergents for NMR structural studies of membrane proteins. J. Biomol. NMR. 2004;28:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- [39].Poget SF, Girvin ME. Solution NMR of membrane proteins in bilayer mimics: Small is beautiful, but sometimes bigger is better. Biochim. Biophys. Acta. 2007;1768:3098–3106. doi: 10.1016/j.bbamem.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Traaseth NJ, Verardi R, Torgersen KD, Karim CB, Thomas DD, Veglia G. Spectroscopic validation of the pentameric structure of phospholarnban. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14676–14681. doi: 10.1073/pnas.0701016104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kim HJ, Howell SC, Van Horn WD, Jeon YH, Sanders CR. Recent advances in the application of solution NMR spectroscopy to multi-span integral membrane proteins. Prog. NMR Spectrosc. 2009;55:335–360. doi: 10.1016/j.pnmrs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shenkarev ZO, Lyukmanova EN, Paramonov AS, Shingarova LN, Chupin VV, Kirpichnikov MP, Blommers MJJ, Arseniev AS. Lipid-protein nanodiscs as reference medium in detergent screening for high-resolution NMR studies of integral membrane proteins. J. Am. Chem. Soc. 2010;132:5628–5629. doi: 10.1021/ja9097498. [DOI] [PubMed] [Google Scholar]
- [43].Raschle T, Hiller S, Yu TY, Rice AJ, Walz T, Wagner G. Structural and functional characterization of the integral membrane protein VDAC-1 in lipid bilayer nanodiscs. J. Am. Chem. Soc. 2009;131:17777–17779. doi: 10.1021/ja907918r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yang J, Ma YQ, Page RC, Misra S, Plow EF, Qin J. Structure of an integrin alpha IIb beta 3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation. Proc. Natl. Acad. Sci. U.S.A. 2009;106:17729–17734. doi: 10.1073/pnas.0909589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Thompson JD, Higgins DG, Gibson TJ. CLUSTAL-W - Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.