Abstract
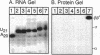
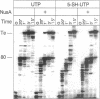
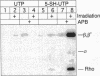
We report a modified synthesis for 5-mercapto-UTP (5-SH-UTP) and its use for analysis of protein-RNA interactions utilizing Escherichia coli and T7 RNA polymerases and yeast RNA polymerases I and III. 5-SH-UTP did not affect transcriptional pausing, Rho-independent termination or recognition of the E. coli transcription complex by NusA. RNA containing 5-SH-UMP did not crosslink to polymerase when irradiation was 302 or 337 nm. Transcription complexes containing RNA substituted with 5-SH-UMP were post-transcriptionally modified to attach a photocross-linking group to thiol-tagged nucleotides in the RNA on the surface of the polymerase of free in solution. The pKa for 5-SH-UTP was determined to be 5.6, so modification of the thiol groups in the RNA with p-azidophenacyl bromide could be carried out at pH 7. Addition of the transcription termination factor Rho, a RNA binding protein, to E. coli transcription complexes resulted in RNA crosslinking to Rho and to the beta and beta' subunits of polymerase. Using 5-SH-UTP, one can distinguish RNA binding domains on the surface of RNA polymerases or other RNA binding proteins from those buried within the protein.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bardos T. J., Kalman T. I. Spectrophotometric and chemical studies of 5-mercaptouracil, 5-mercaptodeoxyuridine, and their S-substituted derivatives. J Pharm Sci. 1966 Jun;55(6):606–610. doi: 10.1002/jps.2600550615. [DOI] [PubMed] [Google Scholar]
- Bowser C. A., Hanna M. M. Sigma subunit of Escherichia coli RNA polymerase loses contacts with the 3' end of the nascent RNA after synthesis of a tetranucleotide. J Mol Biol. 1991 Jul 20;220(2):227–239. doi: 10.1016/0022-2836(91)90009-u. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. J. The selectivity of transcription. Annu Rev Biochem. 1974;43(0):721–775. doi: 10.1146/annurev.bi.43.070174.003445. [DOI] [PubMed] [Google Scholar]
- Das A. Control of transcription termination by RNA-binding proteins. Annu Rev Biochem. 1993;62:893–930. doi: 10.1146/annurev.bi.62.070193.004333. [DOI] [PubMed] [Google Scholar]
- Dissinger S., Hanna M. M. Active site labeling of Escherichia coli transcription elongation complexes with 5-[4-azidophenacyl)thio)uridine 5'-triphosphate. J Biol Chem. 1990 May 5;265(13):7662–7668. [PubMed] [Google Scholar]
- Dissinger S., Hanna M. M. RNA-protein interactions in a Nus A-containing Escherichia coli transcription complex paused at an RNA hairpin. J Mol Biol. 1991 May 5;219(1):11–25. doi: 10.1016/0022-2836(91)90853-x. [DOI] [PubMed] [Google Scholar]
- Erie D. A., Yager T. D., von Hippel P. H. The single-nucleotide addition cycle in transcription: a biophysical and biochemical perspective. Annu Rev Biophys Biomol Struct. 1992;21:379–415. doi: 10.1146/annurev.bb.21.060192.002115. [DOI] [PubMed] [Google Scholar]
- Friedman D. I. Interaction between bacteriophage lambda and its Escherichia coli host. Curr Opin Genet Dev. 1992 Oct;2(5):727–738. doi: 10.1016/s0959-437x(05)80133-9. [DOI] [PubMed] [Google Scholar]
- Gonzalez N., Wiggs J., Chamberlin M. J. A simple procedure for resolution of Escherichia coli RNA polymerase holoenzyme from core polymerase. Arch Biochem Biophys. 1977 Aug;182(2):404–408. doi: 10.1016/0003-9861(77)90521-5. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Nodwell J. R., Mason S. W. Transcriptional antitermination. Nature. 1993 Jul 29;364(6436):401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- Hanna M. M., Dissinger S., Williams B. D., Colston J. E. Synthesis and characterization of 5-[(4-Azidophenacyl)thio]uridine 5'-triphosphate, a cleavable photo-cross-linking nucleotide analogue. Biochemistry. 1989 Jul 11;28(14):5814–5820. doi: 10.1021/bi00440a017. [DOI] [PubMed] [Google Scholar]
- Hanna M. M. Photoaffinity cross-linking methods for studying RNA-protein interactions. Methods Enzymol. 1989;180:383–409. doi: 10.1016/0076-6879(89)80113-2. [DOI] [PubMed] [Google Scholar]
- Hanna M. M. Photocrosslinking analysis of protein-RNA interactions in E. coli transcription complexes. Cell Mol Biol Res. 1993;39(4):393–399. [PubMed] [Google Scholar]
- Hanna M. M., Zhang Y., Reidling J. C., Thomas M. J., Jou J. Synthesis and characterization of a new photocrosslinking CTP analog and its use in photoaffinity labeling E. coli and T7 RNA polymerases. Nucleic Acids Res. 1993 May 11;21(9):2073–2079. doi: 10.1093/nar/21.9.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A. Protein-protein communication within the transcription apparatus. J Bacteriol. 1993 May;175(9):2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K., Benkovic S. J., Modrich P. Glu-111 is required for activation of the DNA cleavage center of EcoRI endonuclease. J Biol Chem. 1989 Jul 15;264(20):11807–11815. [PubMed] [Google Scholar]
- Nudler E., Goldfarb A., Kashlev M. Discontinuous mechanism of transcription elongation. Science. 1994 Aug 5;265(5173):793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- Pavco P. A., Steege D. A. Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J Biol Chem. 1990 Jun 15;265(17):9960–9969. [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sentenac A. Eukaryotic RNA polymerases. CRC Crit Rev Biochem. 1985;18(1):31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- Tanner N. K., Hanna M. M., Abelson J. Binding interactions between yeast tRNA ligase and a precursor transfer ribonucleic acid containing two photoreactive uridine analogues. Biochemistry. 1988 Nov 29;27(24):8852–8861. doi: 10.1021/bi00424a025. [DOI] [PubMed] [Google Scholar]
- Thayer G. C., Brosius J. In vivo transcription from deletion mutations introduced near Escherichia coli ribosomal RNA promoter P2. Mol Gen Genet. 1985;199(1):55–58. doi: 10.1007/BF00327509. [DOI] [PubMed] [Google Scholar]
- Wright D. J., King K., Modrich P. The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J Biol Chem. 1989 Jul 15;264(20):11816–11821. [PubMed] [Google Scholar]
- Zhang Y., Hanna M. M. NusA changes the conformation of Escherichia coli RNA polymerase at the binding site for the 3' end of the nascent RNA. J Bacteriol. 1994 Mar;176(6):1787–1789. doi: 10.1128/jb.176.6.1787-1789.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]