Abstract
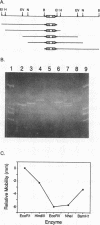
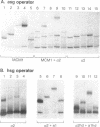
Structural and biochemical studies of monomer homeodomain-DNA complexes have not so far revealed any cases of pronounced DNA distortion. In this paper we show that multimeric complexes of the yeast homeodomain proteins a1 and alpha 2 induced significant bends in their operators upon binding. Based on a series of circular permutation experiments, we found that a dimer of alpha 2 bound to operator DNA produced a mild bend in the DNA, whereas the alpha 2-MCM1-DNA and the a1-alpha 2-DNA complexes exhibited much sharper bends. As these latter two complexes represent the in vivo form of DNA-bound a1 and alpha 2, we conclude that, in the cell, these homeodomain proteins are associated with pronounced bends in DNA. We discuss possible roles for these bends in transcriptional repression.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ceska T. A., Lamers M., Monaci P., Nicosia A., Cortese R., Suck D. The X-ray structure of an atypical homeodomain present in the rat liver transcription factor LFB1/HNF1 and implications for DNA binding. EMBO J. 1993 May;12(5):1805–1810. doi: 10.2210/pdb1lfb/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie-Martin F., Magasanik B. Positive and negative effects of DNA bending on activation of transcription from a distant site. J Mol Biol. 1992 Oct 20;227(4):996–1008. doi: 10.1016/0022-2836(92)90516-m. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Gartenberg M. R., Shrader T. E. DNA bending in protein-DNA complexes. Methods Enzymol. 1991;208:118–146. doi: 10.1016/0076-6879(91)08011-6. [DOI] [PubMed] [Google Scholar]
- Echols H. Nucleoprotein structures initiating DNA replication, transcription, and site-specific recombination. J Biol Chem. 1990 Sep 5;265(25):14697–14700. [PubMed] [Google Scholar]
- Goutte C., Johnson A. D. Yeast a1 and alpha 2 homeodomain proteins form a DNA-binding activity with properties distinct from those of either protein. J Mol Biol. 1993 Oct 5;233(3):359–371. doi: 10.1006/jmbi.1993.1517. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Giese K., Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994 Mar;10(3):94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- Gustafson T. A., Taylor A., Kedes L. DNA bending is induced by a transcription factor that interacts with the human c-FOS and alpha-actin promoters. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2162–2166. doi: 10.1073/pnas.86.7.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington R. E. DNA curving and bending in protein-DNA recognition. Mol Microbiol. 1992 Sep;6(18):2549–2555. doi: 10.1111/j.1365-2958.1992.tb01431.x. [DOI] [PubMed] [Google Scholar]
- Herschbach B. M., Arnaud M. B., Johnson A. D. Transcriptional repression directed by the yeast alpha 2 protein in vitro. Nature. 1994 Jul 28;370(6487):309–311. doi: 10.1038/370309a0. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Herskowitz I. A repressor (MAT alpha 2 Product) and its operator control expression of a set of cell type specific genes in yeast. Cell. 1985 Aug;42(1):237–247. doi: 10.1016/s0092-8674(85)80119-7. [DOI] [PubMed] [Google Scholar]
- Keleher C. A., Redd M. J., Schultz J., Carlson M., Johnson A. D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992 Feb 21;68(4):709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Komachi K., Redd M. J., Johnson A. D. The WD repeats of Tup1 interact with the homeo domain protein alpha 2. Genes Dev. 1994 Dec 1;8(23):2857–2867. doi: 10.1101/gad.8.23.2857. [DOI] [PubMed] [Google Scholar]
- Lobell R. B., Schleif R. F. DNA looping and unlooping by AraC protein. Science. 1990 Oct 26;250(4980):528–532. doi: 10.1126/science.2237403. [DOI] [PubMed] [Google Scholar]
- Mak A., Johnson A. D. The carboxy-terminal tail of the homeo domain protein alpha 2 is required for function with a second homeo domain protein. Genes Dev. 1993 Oct;7(10):1862–1870. doi: 10.1101/gad.7.10.1862. [DOI] [PubMed] [Google Scholar]
- Nash H. A. Bending and supercoiling of DNA at the attachment site of bacteriophage lambda. Trends Biochem Sci. 1990 Jun;15(6):222–227. doi: 10.1016/0968-0004(90)90034-9. [DOI] [PubMed] [Google Scholar]
- Patterton H. G., Simpson R. T. Nucleosomal location of the STE6 TATA box and Mat alpha 2p-mediated repression. Mol Cell Biol. 1994 Jun;14(6):4002–4010. doi: 10.1128/mcb.14.6.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. L., Stark M. R., Johnson A. D., Dahlquist F. W. Heterodimerization of the yeast homeodomain transcriptional regulators alpha 2 and a1 induces an interfacial helix in alpha 2. Biochemistry. 1994 Aug 9;33(31):9294–9302. doi: 10.1021/bi00197a033. [DOI] [PubMed] [Google Scholar]
- Pérez-Martín J., Espinosa M. Protein-induced bending as a transcriptional switch. Science. 1993 May 7;260(5109):805–807. doi: 10.1126/science.8387228. [DOI] [PubMed] [Google Scholar]
- Roth S. Y., Dean A., Simpson R. T. Yeast alpha 2 repressor positions nucleosomes in TRP1/ARS1 chromatin. Mol Cell Biol. 1990 May;10(5):2247–2260. doi: 10.1128/mcb.10.5.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. Y., Shimizu M., Johnson L., Grunstein M., Simpson R. T. Stable nucleosome positioning and complete repression by the yeast alpha 2 repressor are disrupted by amino-terminal mutations in histone H4. Genes Dev. 1992 Mar;6(3):411–425. doi: 10.1101/gad.6.3.411. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Smith D. L., Johnson A. D. Flexibility of the yeast alpha 2 repressor enables it to occupy the ends of its operator, leaving the center free. Genes Dev. 1988 Jul;2(7):807–816. doi: 10.1101/gad.2.7.807. [DOI] [PubMed] [Google Scholar]
- Schleif R. DNA looping. Annu Rev Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- Smith D. L., Johnson A. D. A molecular mechanism for combinatorial control in yeast: MCM1 protein sets the spacing and orientation of the homeodomains of an alpha 2 dimer. Cell. 1992 Jan 10;68(1):133–142. doi: 10.1016/0092-8674(92)90212-u. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjian R., Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994 Apr 8;77(1):5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- Travers A. A. Why bend DNA? Cell. 1990 Jan 26;60(2):177–180. doi: 10.1016/0092-8674(90)90729-x. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzamarias D., Struhl K. Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature. 1994 Jun 30;369(6483):758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- Verrijzer C. P., van Oosterhout J. A., van Weperen W. W., van der Vliet P. C. POU proteins bend DNA via the POU-specific domain. EMBO J. 1991 Oct;10(10):3007–3014. doi: 10.1002/j.1460-2075.1991.tb07851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershon A. K., Johnson A. D. A short, disordered protein region mediates interactions between the homeodomain of the yeast alpha 2 protein and the MCM1 protein. Cell. 1993 Jan 15;72(1):105–112. doi: 10.1016/0092-8674(93)90054-t. [DOI] [PubMed] [Google Scholar]
- Wolberger C., Vershon A. K., Liu B., Johnson A. D., Pabo C. O. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991 Nov 1;67(3):517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. Catabolite activator protein-induced DNA bending in transcription initiation. J Mol Biol. 1991 May 20;219(2):201–215. doi: 10.1016/0022-2836(91)90562-k. [DOI] [PubMed] [Google Scholar]
- Zwieb C., Kim J., Adhya S. DNA bending by negative regulatory proteins: Gal and Lac repressors. Genes Dev. 1989 May;3(5):606–611. doi: 10.1101/gad.3.5.606. [DOI] [PubMed] [Google Scholar]
- van de Putte P., Goosen N. DNA inversions in phages and bacteria. Trends Genet. 1992 Dec;8(12):457–462. doi: 10.1016/0168-9525(92)90331-w. [DOI] [PubMed] [Google Scholar]
- van der Vliet P. C., Verrijzer C. P. Bending of DNA by transcription factors. Bioessays. 1993 Jan;15(1):25–32. doi: 10.1002/bies.950150105. [DOI] [PubMed] [Google Scholar]