Abstract
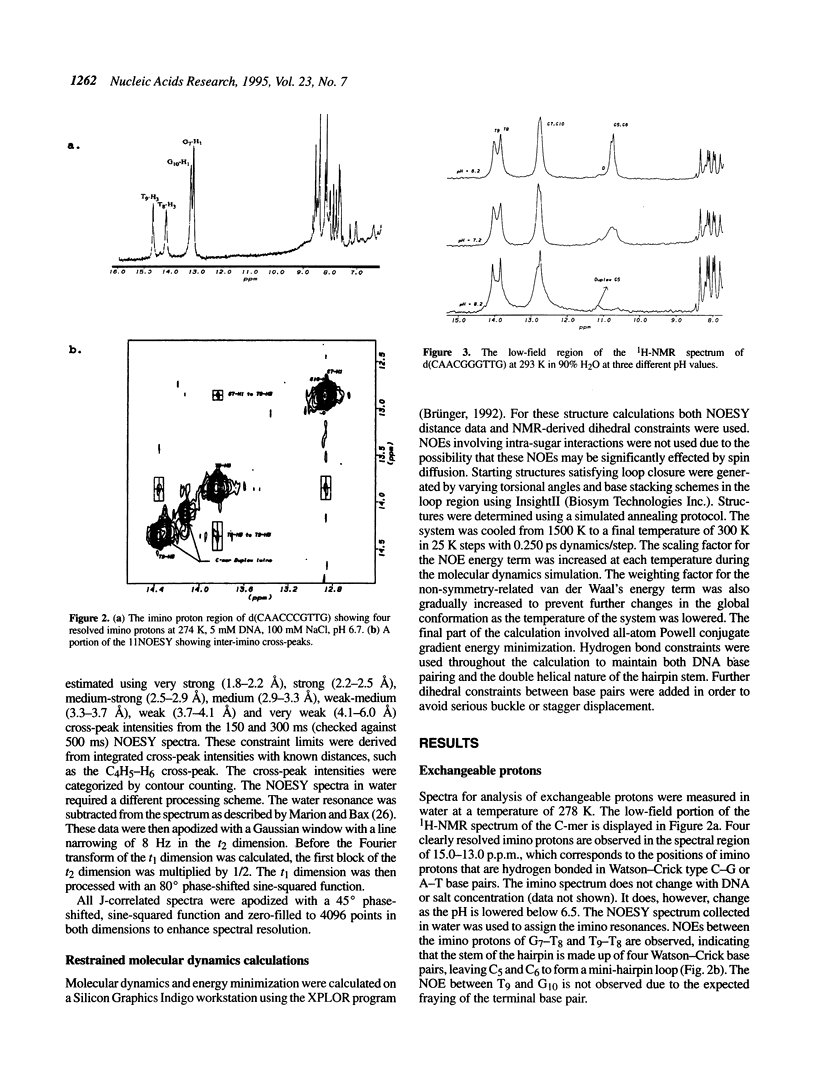
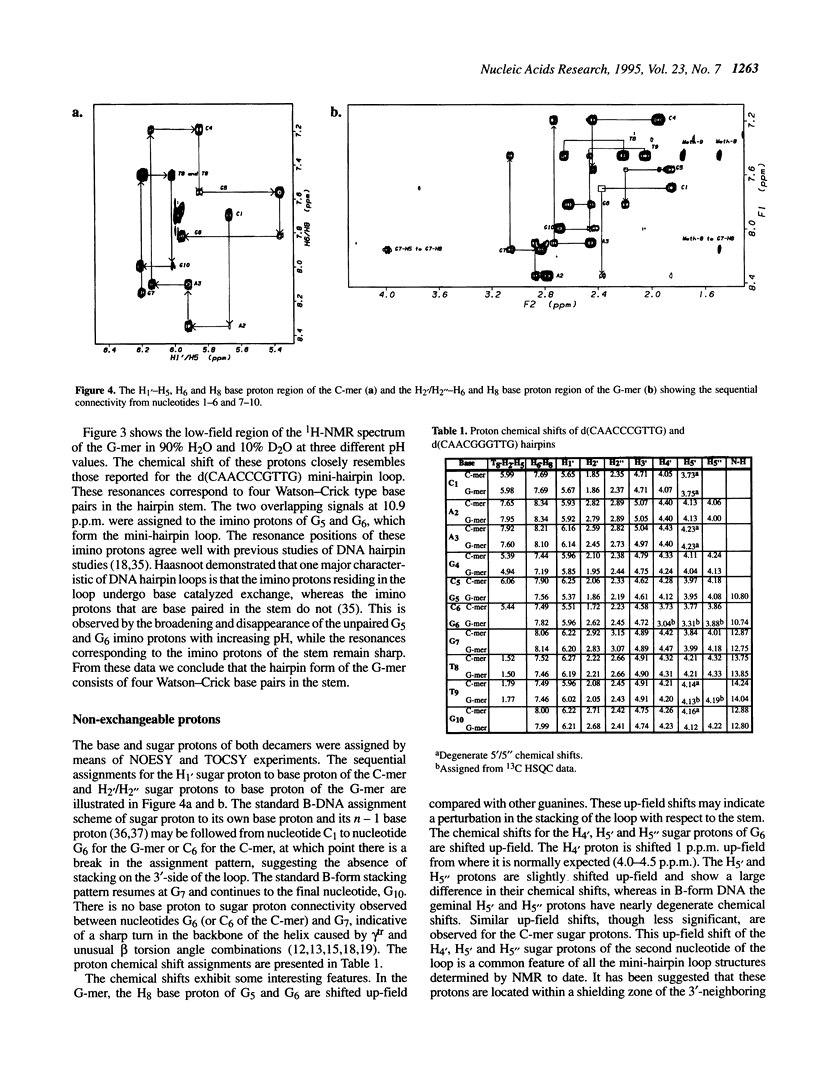
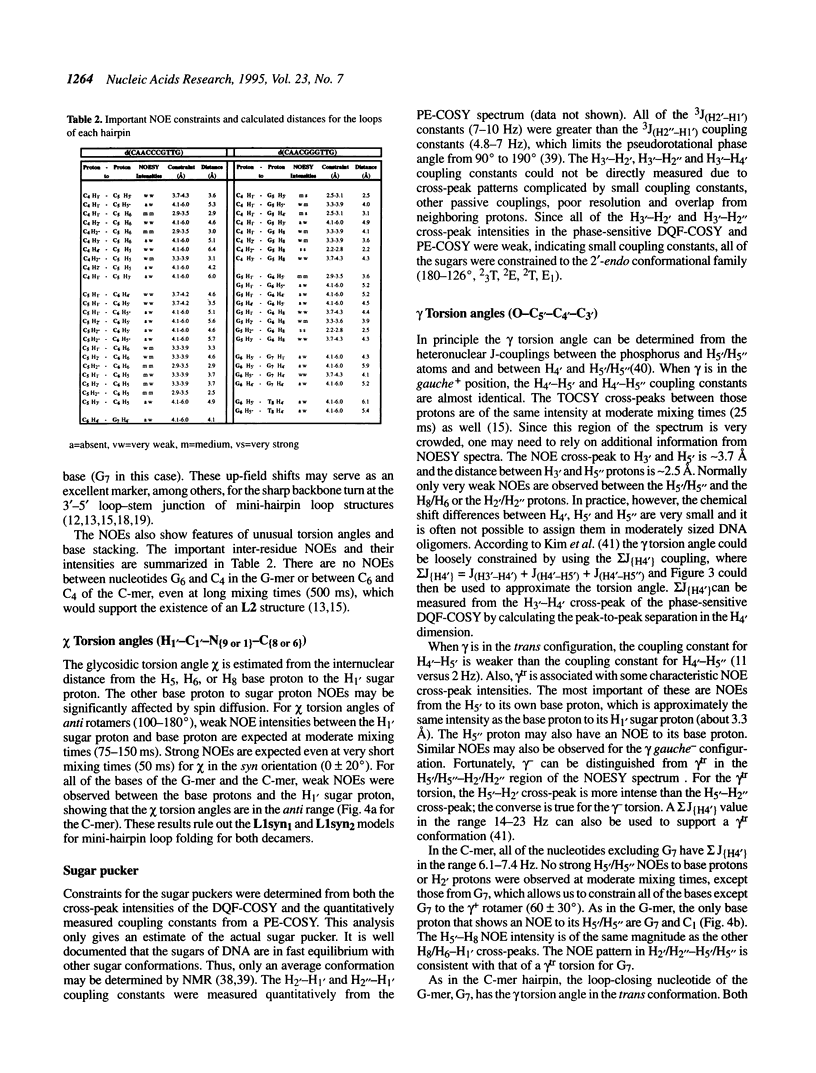
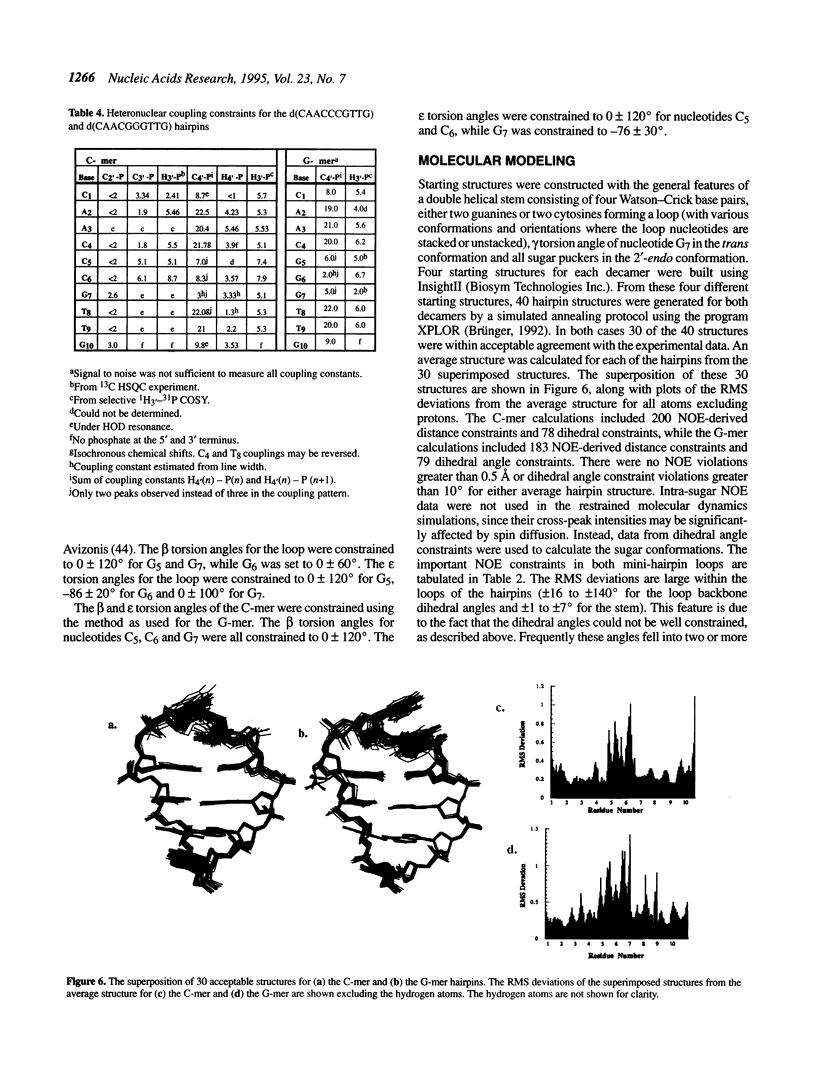
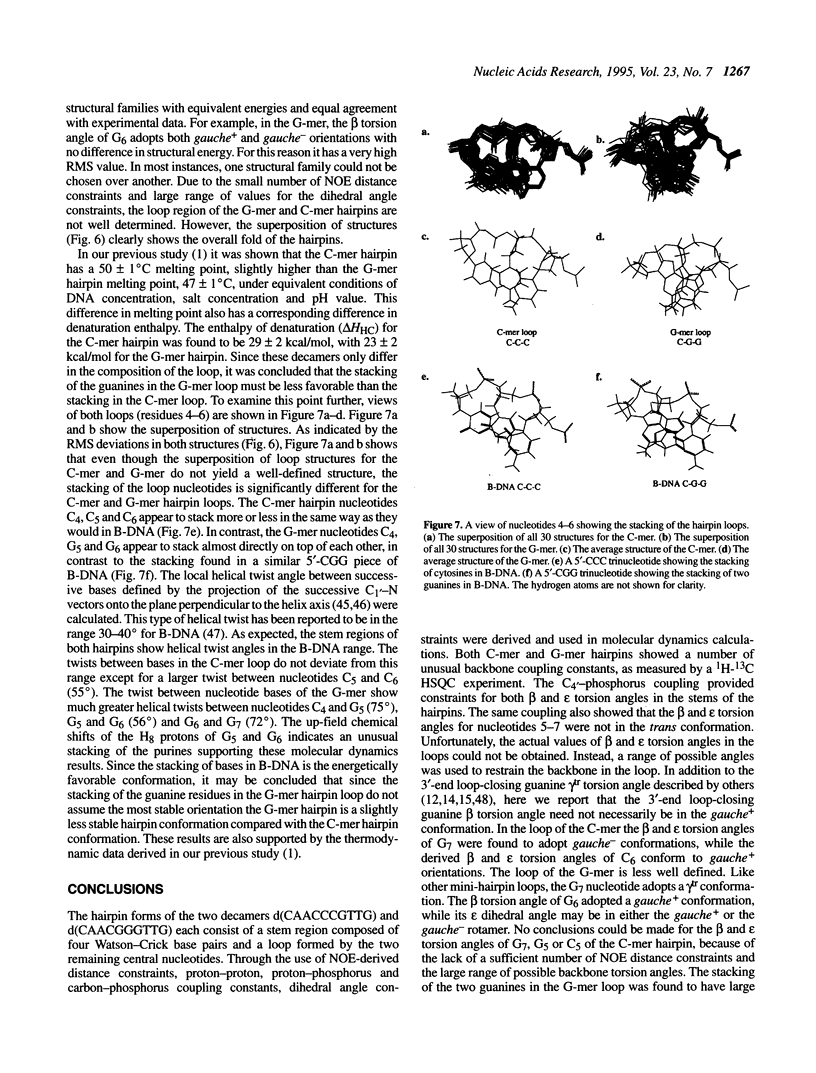
The DNA decamers, d(CAACCCGTTG) and d(CAACGGGTTG) were studied in solution by proton and heteronuclear NMR. Under appropriate conditions of pH, temperature, salt concentration and DNA concentration, both decamers form hairpin conformations with similar stabilities [Avizonis and Kearns (1995) Biopolymers, 35, 187-200]. Both decamers adopt mini-hairpin loops, where the first and last four nucleotides are involved in Watson-Crick hydrogen bonding and the central two nucleotides, CC or GG respectively, form the loop. Through the use of proton-proton, proton-phosphorus and natural abundance proton-carbon NMR experiments, backbone torsion angles (beta, gamma and epsilon), sugar puckers and interproton distances were measured. The nucleotides forming the loops of these decamers were found to stack upon one another in an L1 type of loop conformation. Both show gamma tr and unusual beta torsion angles in the loop-closing nucleotide G7, as expected for mini-hairpin loop formation. Our results indicate that the beta and epsilon torsion angles of the fifth and sixth nucleotides that form the loop and the loop-closing nucleotide G7 are not in the standard trans conformation as found in B-DNA. Although the loop structures calculated from NMR-derived constraints are not well defined, the stacking of the bases in the two different hairpins is different. This difference in the base stacking of the loop may provide an explanation as to why the cytosine-containing hairpin is thermodynamically more stable than the guanine-containing hairpin.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avizonis D. Z., Kearns D. R. Kinetic and thermodynamic characterization of DNA duplex-hairpin interconversion for two DNA decamers: d(CAACGGGTTG) and d(CAACCCGTTG). Biopolymers. 1995 Feb;35(2):187–200. doi: 10.1002/bip.360350207. [DOI] [PubMed] [Google Scholar]
- Blommers M. J., Walters J. A., Haasnoot C. A., Aelen J. M., van der Marel G. A., van Boom J. H., Hilbers C. W. Effects of base sequence on the loop folding in DNA hairpins. Biochemistry. 1989 Sep 5;28(18):7491–7498. doi: 10.1021/bi00444a049. [DOI] [PubMed] [Google Scholar]
- Definitions and nomenclature of nucleic acid structure parameters. EMBO J. 1989 Jan;8(1):1–4. doi: 10.1002/j.1460-2075.1989.tb03339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Germann M. W., Kalisch B. W., Lundberg P., Vogel H. J., van de Sande J. H. Perturbation of DNA hairpins containing the EcoRI recognition site by hairpin loops of varying size and composition: physical (NMR and UV) and enzymatic (EcoRI) studies. Nucleic Acids Res. 1990 Mar 25;18(6):1489–1498. doi: 10.1093/nar/18.6.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot C. A., Hilbers C. W. Effective water resonance suppression in 1D- and 2D-FT-1H-NMR spectroscopy of biopolymers in aqueous solution. Biopolymers. 1983 May;22(5):1259–1266. doi: 10.1002/bip.360220502. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., Hilbers C. W., van der Marel G. A., van Boom J. H., Singh U. C., Pattabiraman N., Kollman P. A. On loop folding in nucleic acid hairpin-type structures. J Biomol Struct Dyn. 1986 Apr;3(5):843–857. doi: 10.1080/07391102.1986.10508468. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., Westerink H. P., van der Marel G. A., van Boom J. H. Conformational analysis of a hybrid DNA-RNA double helical oligonucleotide in aqueous solution: d(CG)r(CG)d(CG) studied by 1D- and 2D-1H NMR spectroscopy. J Biomol Struct Dyn. 1983 Oct;1(1):131–149. doi: 10.1080/07391102.1983.10507430. [DOI] [PubMed] [Google Scholar]
- Hilbers C. W., Haasnoot C. A., de Bruin S. H., Joordens J. J., van der Marel G. A., van Boom J. H. Hairpin formation in synthetic oligonucleotides. Biochimie. 1985 Jul-Aug;67(7-8):685–695. doi: 10.1016/s0300-9084(85)80156-5. [DOI] [PubMed] [Google Scholar]
- Howard F. B., Chen C. Q., Ross P. D., Miles H. T. Hairpin formation in the self-complementary dodecamer d-GGTACGCGTACC and derivatives containing GA and IA mispairs. Biochemistry. 1991 Jan 22;30(3):779–782. doi: 10.1021/bi00217a030. [DOI] [PubMed] [Google Scholar]
- Ikuta S., Chattopadhyaya R., Ito H., Dickerson R. E., Kearns D. R. NMR study of a synthetic DNA hairpin. Biochemistry. 1986 Aug 26;25(17):4840–4849. doi: 10.1021/bi00365a018. [DOI] [PubMed] [Google Scholar]
- Ippel J. H., Lanzotti V., Galeone A., Mayol L., van den Boogaart J. E., Pikkemaat J. A., Altona C. An NMR study of the conformation and thermodynamics of the circular dumbbell d [formula: see text] Slow exchange between two- and four-membered hairpin loops. J Biomol Struct Dyn. 1992 Apr;9(5):821–836. doi: 10.1080/07391102.1992.10507961. [DOI] [PubMed] [Google Scholar]
- Kim S. G., Lin L. J., Reid B. R. Determination of nucleic acid backbone conformation by 1H NMR. Biochemistry. 1992 Apr 14;31(14):3564–3574. doi: 10.1021/bi00129a003. [DOI] [PubMed] [Google Scholar]
- Lankhorst P. P., Haasnoot C. A., Erkelens C., Altona C. Carbon-13 NMR in conformational analysis of nucleic acid fragments. 2. A reparametrization of the Karplus equation for vicinal NMR coupling constants in CCOP and HCOP fragments. J Biomol Struct Dyn. 1984 Jun;1(6):1387–1405. doi: 10.1080/07391102.1984.10507527. [DOI] [PubMed] [Google Scholar]
- Lavery R., Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J Biomol Struct Dyn. 1988 Aug;6(1):63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- Marion D., Wüthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem Biophys Res Commun. 1983 Jun 29;113(3):967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- Nadeau J. G., Gilham P. T. Anomalous hairpin formation in an oligodeoxyribonucleotide. Nucleic Acids Res. 1985 Nov 25;13(22):8259–8274. doi: 10.1093/nar/13.22.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonowicz E. P., Gorenstein D. G. Two-dimensional 1H and 31P NMR spectra and restrained molecular dynamics structure of a mismatched GA decamer oligodeoxyribonucleotide duplex. Biochemistry. 1990 Sep 18;29(37):8845–8858. doi: 10.1021/bi00489a048. [DOI] [PubMed] [Google Scholar]
- Orbons L. P., van Beuzekom A. A., Altona C. Conformational and model-building studies of the hairpin form of the mismatched DNA octamer d(m5C-G-m5C-G-T-G-m5C-G). J Biomol Struct Dyn. 1987 Jun;4(6):965–987. doi: 10.1080/07391102.1987.10507692. [DOI] [PubMed] [Google Scholar]
- Orbons L. P., van der Marel G. A., van Boom J. H., Altona C. An NMR study of polymorphous behaviour of the mismatched DNA octamer d(m5C-G-m5C-G-A-G-m5C-G) in solution. The B-duplex and hairpin forms. Eur J Biochem. 1987 Dec 30;170(1-2):225–239. doi: 10.1111/j.1432-1033.1987.tb13690.x. [DOI] [PubMed] [Google Scholar]
- Orbons L. P., van der Marel G. A., van Boom J. H., Altona C. An NMR study of the polymorphous behavior of the mismatched DNA octamer d(m5C-G-m5C-G-T-G-m5C-G) in solution. The B, Z, and hairpin forms. J Biomol Struct Dyn. 1987 Jun;4(6):939–963. doi: 10.1080/07391102.1987.10507691. [DOI] [PubMed] [Google Scholar]
- Orbons L. P., van der Marel G. A., van Boom J. H., Altona C. Hairpin and duplex formation of the DNA octamer d(m5C-G-m5C-G-T-G-m5C-G) in solution. An NMR study. Nucleic Acids Res. 1986 May 27;14(10):4187–4196. doi: 10.1093/nar/14.10.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters J. M., de Vroom E., van der Marel G. A., van Boom J. H., Koning T. M., Kaptein R., Altona C. Hairpin structures in DNA containing arabinofuranosylcytosine. A combination of nuclear magnetic resonance and molecular dynamics. Biochemistry. 1990 Jan 23;29(3):788–799. doi: 10.1021/bi00455a029. [DOI] [PubMed] [Google Scholar]
- Raghunathan G., Jernigan R. L., Miles H. T., Sasisekharan V. Conformational feasibility of a hairpin with two purines in the loop. 5'-d-GGTACIAGTACC-3'. Biochemistry. 1991 Jan 22;30(3):782–788. doi: 10.1021/bi00217a031. [DOI] [PubMed] [Google Scholar]
- Roy S., Weinstein S., Borah B., Nickol J., Appella E., Sussman J. L., Miller M., Shindo H., Cohen J. S. Mechanism of oligonucleotide loop formation in solution. Biochemistry. 1986 Nov 18;25(23):7417–7423. doi: 10.1021/bi00371a025. [DOI] [PubMed] [Google Scholar]
- Sakata T., Hiroaki H., Oda Y., Tanaka T., Ikehara M., Uesugi S. Studies on the structure and stabilizing factor of the CUUCGG hairpin RNA using chemically synthesized oligonucleotides. Nucleic Acids Res. 1990 Jul 11;18(13):3831–3839. doi: 10.1093/nar/18.13.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder P., Ippel J. H., van den Elst H., van der Marel G. A., van Boom J. H., Altona C., Kessler H. Heteronuclear NMR of DNA with the heteronucleus in natural abundance: facilitated assignment and extraction of coupling constants. Nucleic Acids Res. 1992 Sep 25;20(18):4747–4751. doi: 10.1093/nar/20.18.4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior M. M., Jones R. A., Breslauer K. J. Influence of loop residues on the relative stabilities of DNA hairpin structures. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6242–6246. doi: 10.1073/pnas.85.17.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers M. F., Byrd R. A., Gallo K. A., Samson C. J., Zon G., Egan W. Nuclear magnetic resonance and circular dichroism studies of a duplex--single-stranded hairpin loop equilibrium for the oligodeoxyribonucleotide sequence d(CGCGATTCGCG). Nucleic Acids Res. 1985 Sep 11;13(17):6375–6386. doi: 10.1093/nar/13.17.6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G., Cheong C., Tinoco I., Jr Structure of an unusually stable RNA hairpin. Biochemistry. 1991 Apr 2;30(13):3280–3289. doi: 10.1021/bi00227a016. [DOI] [PubMed] [Google Scholar]
- Wemmer D. E., Chou S. H., Hare D. R., Reid B. R. Duplex-hairpin transitions in DNA: NMR studies on CGCGTATACGCG. Nucleic Acids Res. 1985 May 24;13(10):3755–3772. doi: 10.1093/nar/13.10.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstrom E., Tinoco I., Jr The stability of RNA hairpin loops containing A-U-G: An-U-G-Um. Biopolymers. 1974 Nov;13(11):2367–2383. doi: 10.1002/bip.1974.360131116. [DOI] [PubMed] [Google Scholar]
- Williamson J. R., Boxer S. G. Multinuclear NMR studies of DNA hairpins. 1. Structure and dynamics of d(CGCGTTGTTCGCG). Biochemistry. 1989 Apr 4;28(7):2819–2831. doi: 10.1021/bi00433a012. [DOI] [PubMed] [Google Scholar]
- Williamson J. R., Boxer S. G. Multinuclear NMR studies of DNA hairpins. 2. Sequence-dependent structural variations. Biochemistry. 1989 Apr 4;28(7):2831–2836. doi: 10.1021/bi00433a013. [DOI] [PubMed] [Google Scholar]
- Xodo L. E., Manzini G., Quadrifoglio F., van der Marel G. A., van Boom J. H. Oligodeoxynucleotide folding in solution: loop size and stability of B-hairpins. Biochemistry. 1988 Aug 23;27(17):6321–6326. doi: 10.1021/bi00417a018. [DOI] [PubMed] [Google Scholar]
- Xodo L. E., Manzini G., Quadrifoglio F., van der Marel G., van Boom J. H. Hairpin structures in synthetic oligodeoxynucleotides: sequence effects on the duplex-to-hairpin transition. Biochimie. 1989 Jul;71(7):793–803. doi: 10.1016/0300-9084(89)90042-4. [DOI] [PubMed] [Google Scholar]


