Abstract
Systems biology uses experimental and computational approaches to characterize large sample populations systematically, process large datasets, examine and analyze regulatory networks, and model reactions to determine how components are joined to form functional systems. Systems biology technologies, data and knowledge are particularly useful in understanding disease processes and drug actions. An important area of integration between systems biology and drug discovery is the concept of polypharmacology: the treatment of diseases by modulating more than one target. Polypharmacology for complex diseases is likely to involve multiple drugs acting on distinct targets that are part of a network regulating physiological responses. This review discusses the current state of the systems-level understanding of diseases and both the therapeutic and adverse mechanisms of drug actions. Drug-target networks can be used to identify multiple targets and to determine suitable combinations of drug targets or drugs. Thus, the discovery of new drug therapies for complex diseases may be greatly aided by systems biology.
Keywords: Drug discovery, drug resistance, polypharmacology, regulatory network, systems biology, targeted therapy
Introduction
Drug discovery is a complex and expensive process. With increasing focus on diseases that involve many genes, identifying potential drugs has become more complicated. Over the next 5 years, the pharmaceutical industry will be in a compromised financial position as a result of upcoming patent expirations on many blockbuster drugs, estimated to be worth a total of approximately US $160 billion for generics manufacturers. Currently, the R&D effort required for a drug to progress from clinical trials to market involves 5 to 10 years and costs an average of US $1 billion. Both prior to and during clinical trials, substantial research is undertaken to gain a comprehensive understanding of the action of drugs and possible off-target effects. Methodologies to classify drugs according to their properties (eg, specificity, half-life and toxicity), molecular structure and SAR have been established over the past decade [1–3]. However, the high rate of drug attrition indicates that new approaches are required to improve and expedite the drug discovery process. In theory, the large amount of data obtained on drug action, metabolism, safety and efficacy in preclinical studies, as well as in phase I and II clinical trials, should ensure a high success rate in phase III trials. However, many drugs fail at the phase III stage, primarily because of a lack of efficacy (ie, no significant difference between placebo and drug treatment, or between the investigational drug and current treatment alternatives) [4]. The risk of failure is higher if the drug has a novel mechanism of action (ie, a newly identified target) [4]. This problematic evidence indicates that new approaches are necessary in drug-discovery strategies.
This review highlights the manner in which systems biology methodologies could enable a polypharmacology approach to drug discovery. The presented definition of polypharmacology encompasses both one drug binding to multiple targets and multiple drugs that bind to different targets within a network. Potential exists for polypharmacology to be co-integrated with systems biology. The impact of a systems-level understanding of complex diseases on polypharmacology and the potential impact on new drug discovery are discussed, as well as methods that have been used in systems biology experiments.
The use of systems biology to gain perspective on complex diseases
Defining the complex mechanisms underlying diseases is challenging. The viewpoint that diseases can be understood by a Mendelian perspective, or the ‘one gene-one disease’ theory, and treated with a ‘magic–bullet’ therapy, has proven to be unsuccessful. Many diseases are not associated with a single genetic determinant. Instead, a complex multiplicity of genetic determinants leads to a disease state, and a single genetic determinant can influence more than one disease. In addition, environmental factors, tissue type, hormone levels and age play a role in how genetic determinants dictate disease manifestation. This complexity in disease origins arises because the impact on protein function or expression level is controlled by the regulatory network within which a protein exists.
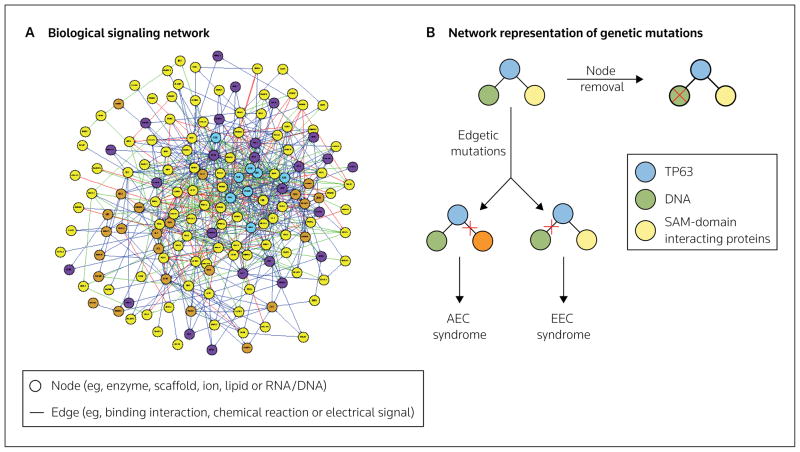
In order to gain a systems-level perspective of disease at the molecular level, a systems view of the relevant physiological function is essential. Each physiological process has an underlying signaling network of chemicals, hormones, protein receptors, ligands, enzymes, transcription factors, ions or DNA/RNA that modulate biochemical reactions, electrical signals, mRNA transcription and protein translation. These reactions can each occur with different kinetics, and simultaneously and/or at different timepoints, as well as at varying levels of magnitude. Therefore, each physiological function or phenotype is controlled by a complicated network of signals. Each physiological component of the network (eg, protein, ion or chemical) can be considered a ‘node’, and each interaction between two nodes (eg, binding or chemical reaction) is an ‘edge’ (Figure 1A) [5,6]. The scale-free and redundant properties of biological networks allow for network perturbation without a complete loss of function. Thus, in a disease system, the signaling networks underlying physiological symptoms are most likely perturbed at more than one point (node or edge).
Figure 1. Biological signaling networks and network representation of genetic mutations in disease.
(A) Biological signaling molecules and the interactions of these molecules are represented in biological signaling networks as nodes and edges, respectively. (B) In a network representation, genetic mutations associated with disease are either node removals or edgetic mutations. Node removals are truncated gene products and edgetic mutations disrupt the interaction between two nodes. For example, two different disorders, the ectrodactyly–ectodermal dysplasia–clefting (EEC) and ankyloblepharon-ectodermal dysplasia-clefting (AEC; Hay-Wells) syndromes are associated with two different edgetic mutations within one signaling motif that contains the interactions between the transcription factor TP63 (tumor protein 63) and DNA, and between TP63 and sterile α motif (SAM)-domain interacting proteins [15].
Numerous genomic, proteomic and phosphoproteomic studies have focused on discovering gene expression signatures and protein signaling pathways associated with a particular disease [7–11]. Many of these studies are hypothesis-driven; however, high-throughput studies (Table 1), such as genome-wide association studies (GWAS), proteomic experiments, or short hairpin (sh)RNA/siRNA experiments with a particular disease model, are also prominent in the literature. Disease-associated genetic data are assembled into several sources, including compilation studies [12] and databases of gene mutation data, such as the Online Mendelian Inheritance in Man (OMIM) database or the Catalogue of Somatic Mutations in Cancer (COSMIC). Such data provide a foundation for studies to examine genes that harbor mutations in disease as parts of a network, generating information on the network properties of these nodes (genes) [13–15].
Table 1.
Selected systems biology technologies used in drug discovery.
| Methodology | Description | Reference |
|---|---|---|
| Genomics and proteomics (eg, SILAC-MS, phospho-proteomics and reverse-phase protein microarrays) | Large-scale experimental analysis of signaling molecules. Cells under various conditions (eg, drug treatment, stress and conditions mimicking disease) are analyzed for response to treatment compared with untreated control cells. The expression level and status of phosphorylation and other post-translational modifications can be monitored. | [8–11, 65,66] |
| Large-scale molecular screening | Experimental and computational methods to identify both unknown targets and the promiscuity of drugs. These analyses can enable further characterization of targets that are inhibited by the same drugs or, conversely, drugs that inhibit the same targets. | [1,60,61,67] |
| Network and computational analyses | The identification and understanding of networks that surround a drug target. This analysis can lead to the identification of a novel drug target in the same pathway or the identification of mechanisms of drug resistance. | [5,68–70] |
| shRNA/siRNA screens | Silencing one gene at a time and analyzing the effect in cells or in vivo. These methods can be used to understand the function of a protein or the effect of inhibiting the protein, as a tool for drug target validation. These studies can lead to information on the signaling network involved and unforeseen effects of a potential drug on the signaling network. | [71–73] |
shRNA Short hairpin RNA, SILAC-MS stable isotope labeling with amino acids in cell culture-mass spectrometry
Network representation of genetic mutations in disease
A recent computational study of disease network properties demonstrated that the phenotypic manifestations of diseases are more likely to be caused by genetic mutations that perturb networks at an edge rather than at a node [15]. Edgetic perturbations (Figure 1B) are more commonly observed in mutations that span multiple diseases, and are caused by in-frame mutations and the expression of an almost full-length gene product that lacks an interaction with a neighboring node. In contrast, node mutations (Figure 1B) lead to the expression of truncated gene products, but do not necessarily affect the interaction of the node with other proteins in the respective signaling network. Edgetic mutations have potential consequences at more than one node in the network. Furthermore, different mutations of the same gene product may affect different edges, with each edgetic mutation being associated with a different disorder. In an example highlighted by Zhong et al [15], two different edgetic mutations of the transcription factor TP63 (tumor protein 63) are involved in two different developmental disorders, ectrodactyly-ectodermal dysplasia–clefting (EEC) and ankyloblepharon–ectodermal dysplasia–clefting (AEC; Hay-Wells) syndromes. Although both syndromes are associated with facial clefting, EEC is also associated with hand and foot deformities and AEC with complete or partial eyelid fusion. TP63 mutations associated with EEC syndrome disrupt the interaction between TP63 and DNA, whereas TP63 mutations associated with AEC syndrome disrupt protein-protein interactions involving the sterile α motif (SAM) domain of TP63 [15] (Figure 1B). Therefore, not only the identity of the mutated gene, but also the type of mutation, determines the role of genetic mutations in disease; these multiple components of mutations add complexity to identifying suitable new therapeutic drug targets.
Network analyses of the relationships between drugs and drug targets
Given that genetic mutations and respective signaling networks, symptom manifestation and disease progression have a complex relationship, achieving a therapeutic effect with drug intervention is a multifaceted process that depends strongly on the signaling network containing the therapeutically targeted node. The relationship between drugs and drug targets has been studied from a network perspective [16,17]. These studies analyzed the data available in drug-target databases such as DrugBank [18,19], the Therapeutic Targets Database (TTD) [20,21], World Molecular Bioactivity (WOMBAT) [22] and the Potential Drug Target Database (PDTD) [23]. A study by Yildrim et al organized all approved drugs reported by DrugBank into a drug-target network, in which the drugs are depicted as nodes that are connected if the drugs share a protein target [17]. A target-protein network, in which the proteins are nodes that are connected if the proteins are targeted by the same drug, was also generated. In both networks, the majority of nodes were connected to at least one other drug/target: more than half of the drugs in the drug-target network formed a ‘giant interconnected cluster’ (island); however, this island was smaller than the largest cluster in a comparable randomized network of interactions, and the largest cluster in the complementary target-protein network was also significantly smaller than the equivalent cluster in a random network. These trends indicate that many approved drugs are based on the same therapeutic targets. When investigational drugs were included in this analysis, the size of the largest cluster within the target-protein network increased, indicating a trend toward a more diversified pool of drug targets [17].
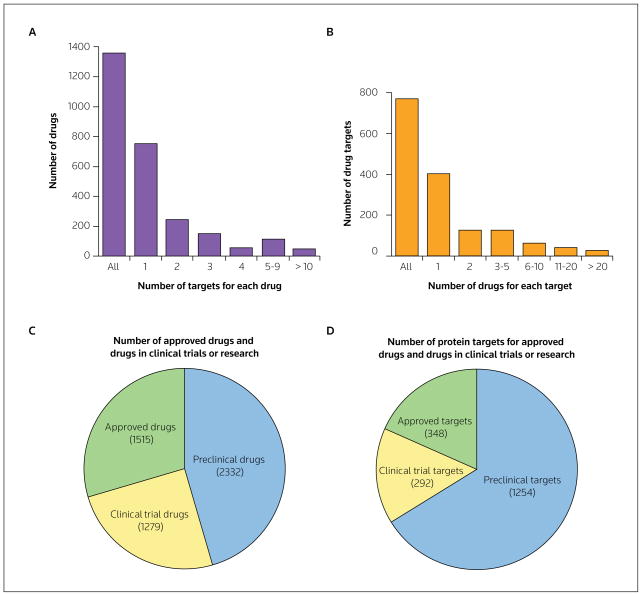
Data on the number of currently approved drugs and drug targets, extracted from DrugBank and TTD on March 23, 2010, are depicted in Figure 2. The number of unique targets in DrugBank increased from 349 in 2007 to 764 with an update implemented on January 1, 2008 [18]. Although the majority of approved drugs have been identified to have one to three targets (Figures 2A and 2B), the actual number of molecules to which the drugs bind is likely to be substantially higher, as Drugbank generally lists only the intended therapeutic targets. Furthermore, the affinity for each target is not specified, and the affinity of the drug is likely to differ for each target.
Figure 2. The analysis of drugs and drug targets.
Approved, non-illicit, non-nutraceutical drugs and drug targets were extracted from DrugBank 2.0, [18,19] on March 23, 2010. The number of unique drug targets was 764, and the total number of drugs was 1366. The number of drugs for each target (A) and the number of targets for each drug (B) are shown. A total of 5126 drugs and 1894 drug targets listed on the Therapeutic Target Database (TTD) [20,21] on March 23, 2010 were categorized by drug type (C) and drug target type (D), according to whether the drugs were approved, in clinical trials or in the preclinical phase [21].
The WOMBAT database compiles the affinities of drugs for any protein target published in the literature, including interactions other than the intended therapeutic interaction. According to a recent drug-network study, the average number of targets per drug is 1.8 when considering therapeutic targets only (ie, in DrugBank), increasing to 2.7 if the interactions reported in WOMBAT are also considered; if both the targets reported in the literature and any predicted targets are considered, this number increases to 6.3 [24]. The result is polypharmacology: drugs bind to more than one target, often including targets other than the intended therapeutic targets.
The drug-target statistics are indicative of the challenge of achieving specificity for one drug target over other targets. Specificity for a protein target of interest is only part of the overall complexity of the physiological effect of a drug treatment. Many pathways contain sequential enzymatic steps that exhibit a variety of kinetics, with first- or second-order reactions and different rate-limiting steps, creating a complex context in which the dosage of a drug must be optimized in order to achieve a therapeutic effect. In addition, other factors, such as the bioavailability, absorption rate, metabolism and half-life of a particular drug, will also affect how the drug interacts with multiple targets within a network.
Therapeutic polypharmacology in systematic drug combinations
As a description for the binding of a drug to more than one target, polypharmacology can lead to multiple outcomes, both beneficial and harmful. To consider both outcomes separately, polypharmacology can be divided into two types: therapeutic polypharmacology and adverse polypharmacology (Table 2). Therapeutic polypharmacology includes the concept of treating multigenic, complex diseases by targeting multiple targets with one or more drugs, in order to effectively reset the regulatory network processes that are altered in the disease state. Adverse polypharmacology comprises the scenarios in which the ‘off-target’ binding of drugs leads to adverse effects. Such interactions include binding to protein targets other than the therapeutic target and binding to the therapeutic target in non-target tissue.
Table 2.
Comparison of therapeutic and adverse polypharmacology.
| Type of polypharmacology | Description | Role of systems biology |
|---|---|---|
| Therapeutic polypharmacology | The treatment of multigenic, complex diseases based on the analysis of the signaling networks of the disease state and the systems-level effects of modulating multiple protein targets with one or more drug. | Enables drug design based on systems-level knowledge: involves the modulation of multiple nodes in one or more regulatory networks. |
| Adverse polypharmacology | The adverse, physiological effect caused by drug binding to protein targets other than the therapeutic target or binding to the therapeutic target in non-target tissue. | Enables an understanding of the adverse event, including the effects of an interaction with a non-target regulatory network or signal propagation within the regulatory network that lead to an adverse phenotype. |
The systematic treatment of a single disease at two or more targets (ie, critical nodes) requires a combination of the empirical knowledge of which drugs are effective against each pathophysiology and the knowledge gained from a systems approach to understanding how multiple nodes cooperate in a signaling network to produce the pathophysiology associated with the disease. Ideally, each disease would be treated by therapeutic polypharmacology – the combination of ‘targeted therapies’ that modulate multiple, specific signaling components or interactions that malfunction in a given disease [25]. The regulatory network surrounding the drug targets and/or disease would be analyzed, accounting for the modulations resulting from the targeted therapy treatment. Combining the building of interaction networks for various diseases with genome-wide analyses of changes, such as SNPs or copy-number variations, will allow a significant expansion of the list of possible drug targets in a physiologically relevant manner. Each gene product associated with disease is not necessarily ‘druggable’; however, an analysis of the druggability of the human genome, according to the structural and functional properties of each protein, supports the view that the current ‘drugome’ can be greatly expanded and diversified [26]. Combining a network perspective with the empirical knowledge of genetic changes associated with disease, in order to achieve an understanding of the consequences of targeting each node, contrasts with the establishment of drug targets through empirical knowledge, such as the use of trial-and-error approaches in prescribing drugs and drug combinations. For example, given the known genetic amplifications or mutations associated with cancer, there are several protocols that require a receptor expression test before drug use, including the requirement for HER2 expression in breast cancer prior to the prescription of trastuzumab [27,28], and EGFR expression in pancreatic cancer for enrollment in a cetuximab combination clinical trial [29]. However, these diagnostic tests and empirical studies do not offer a mechanistic insight into why drugs function or do not function, or what role a genetic change plays in disease progression. Such information may be obtained through careful analyses of the signaling networks involved. As discussed in subsequent sections, a therapeutic polypharmacology approach can introduce robustness in therapeutic targeting and has the potential to minimize the clinical failure of drugs resulting from lack of effectiveness, through targeting a combination of multiple, critical nodes.
The decision to treat a disease using a combination of drugs with different targets is often derived from insights based on genomic studies. For example, a gene expression or protein signal profiling experiment in cells treated with a drug can indicate which genes or signaling mechanisms are upregulated and interfere with drug action. In addition, GWAS on cohorts of patients with different responses to a drug can indicate which upregulated genes may be causing drug resistance. However, because of the variety of genetic backgrounds in disease, particularly cancer, a given combination of therapies designed to downregulate multiple targets will not be effective for every patient. Before designing combination therapies, analyzing the signaling network involved with a drug action within the context of a patient’s genetic background may be critical to the success of the therapy. Accounting for the effect of the combination of drug effects within a signaling network will be a distinguishing feature between currently used, empirical combination therapy and therapeutic polypharmacology. The following examples demonstrate how components of the signaling network surrounding a drug target can play important roles in drug action, and why possible drug combinations can be effective.
Therapeutic pharmacology in antibiotic therapy
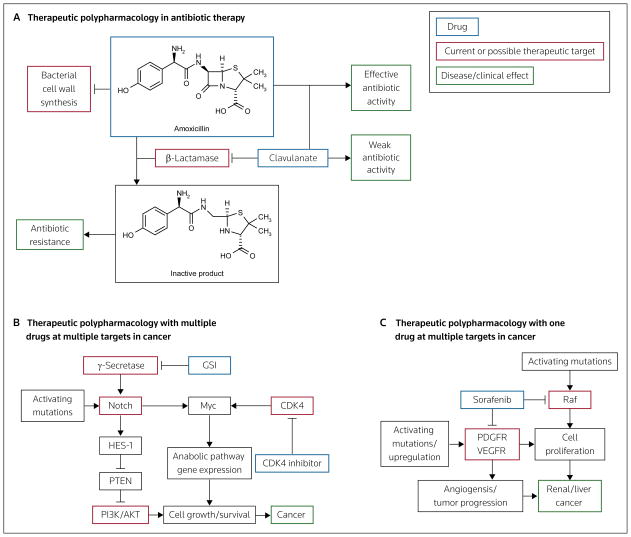
A well-understood example of therapeutic polypharmacology is resistance to β-lactam antibiotics, often caused by the bacterial enzyme β-lactamase. Bacteria produce various isoforms of β-lactamase, which degrades β-lactam antibiotics and renders these drugs inactive at inhibiting bacterial cell wall synthesis (Figure 3A). Combining β-lactamase inhibitors with β-lactam antibiotics overcomes this antibiotic resistance. This combination is now widely used as an effective antibiotic therapy. However, there are multiple mechanisms of antibiotic resistance, which continues to be a problem and has been the focus of many studies (reviewed in [30]).
Figure 3. Selected examples of therapeutic polypharmacology.
(A) Resistance to β-lactam antibiotics (eg, amoxicillin) is caused by degradation of the drugs by bacterial β-lactamase. Inhibitors of β-lactamase (eg, clavulanate) prevent the degradation of the antibiotics, thereby increasing the effectiveness of these drugs. (B) Notch mutations are associated with cancer. Several drugs have been designed that target γ-secretase, the enzyme upstream of Notch; however, resistance to these γ-secretase inhibitors (GSIs) can be caused by mutations in the MYC gene. A combination of drugs that target both Notch and Myc (through inhibition of CDK4) has been demonstrated to reverse Myc-induced GSI resistance and effectively treat cancer. (C) The anticancer agent sorafenib is an example of a drug with multiple targets (eg, Raf and PDGFR/VEGFR) that is used to treat a single disease (ie, renal or liver cancer).
Therapeutic polypharmacology for multiple drugs at multiple targets
Studies have demonstrated that certain mutations within a targeted molecule, or at a molecule in another pathway, can render a drug ineffective. In these scenarios, which are common in the treatment of cancer, a therapeutic polypharmacology approach is most likely to succeed. In a recent example, the discovery that Notch mutations are associated with T-cell acute lymphoblastic leukemia (T-ALL) led to the development of inhibitors of this pathway, such as γ-secretase inhibitors (GSIs; γ-secretase is an upstream regulator of Notch), for the treatment of cancer [31]. A stapled peptide that directly inhibits the transcriptional activity of Notch was recently described [32]. The enhanced signaling of the Notch pathway that results from Notch mutations leads to aberrant cell survival through anabolic gene expression. GSI treatment typically has a limited success rate because many patients with T-ALL exhibit other mutations that compensate for the inhibition of the γ-secretase pathway (eg, PTEN deletion) [31] (Figure 3B). This situation illustrates the potential utility of a therapeutic polypharmacology approach to target both Myc and Notch in diseases involving aberrant Notch signaling, which has also been observed in lung adenocarcinoma and other cancers [31]. In a recent study by Rao et al, a therapeutic polypharmacology approach was used to examine the properties that render cells sensitive or insensitive to GSI therapy [33]. The results prompted the design of a combination therapy of a GSI with an inhibitor of CDK4, which acts upstream of Myc (Figure 3B). The therapeutic polypharmacology downregulated the compensatory signal (ie, Myc) through CDK4 inhibition, allowing increased sensitivity to Notch inhibition, thereby producing a therapeutic effect (ie, inhibition of cell growth) [33].
Therapeutic polypharmacology for one drug at multiple targets
Therapeutic polypharmacology can also arise from one drug binding multiple targets that contribute to the overall effectiveness of a treatment. For example, sorafenib (Nexavar), a drug used to treat renal and liver cancers, was originally designed to inhibit Raf kinase isoforms, but was also demonstrated to inhibit the PDGF and VEGF receptor tyrosine kinases (RTKs) [34–36], both of which play a role in cancer progression. Thus, the ability of sorafenib to demonstrate activity against these receptors in conjunction with anti-Raf activity is advantageous. Sorafenib appears to inhibit cancer cell proliferation by inhibiting Raf, and prevent tumor progression and angiogenesis by inhibiting PDGFR and VEGFR (Figure 3C). In another example, lapatinib (Tykerb), which is used to treat metastatic breast cancer, inhibits ErbB1 and HER2; the upregulation of both of these molecules has been observed in breast cancer, as well as in several other types of cancer [37].
Therapeutic polypharmacology in designed combination therapy
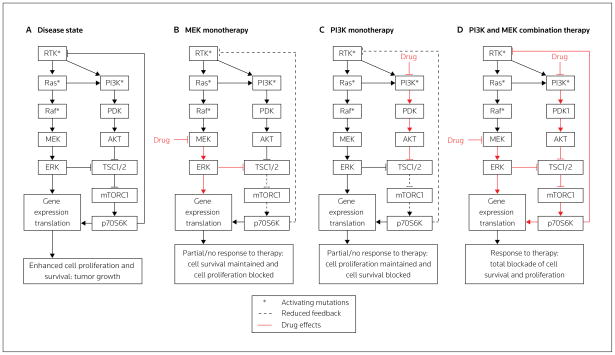
Another example of therapeutic polypharmacology is illustrated with the regulatory network surrounding the growth factor RTK in cancer (Figure 4A). Dysregulation of the RTK, MEK and PI3K pathways resulting from various mutations within these pathways have been observed in cancer. These altered signaling events have implications for the effectiveness of targeted therapies; for example, KRAS mutations are often associated with resistance to EGFR-targeted therapies [38]. The observed high frequency of KRAS mutations in cancer has led to the development of inhibitors of Raf and MEK, both of which act downstream of K-Ras [39,40] (Figure 4). BRAF mutations have also been observed in various cancers, leading to the development of B-Raf inhibitors [41]. Recent studies in cells containing mutated Ras have also demonstrated that B-Raf inhibitors can lead to enhanced MEK/ERK activation, caused by the inhibitors functioning as C-Raf activators because of the heterodimerization of B-Raf and C-Raf [42,43]. Resistance to EGFR therapy can also be coupled with the maintenance of PI3K signaling through a variety of mechanisms, including loss of PTEN (a phosphatase and negative regulator of AKT) activity, or ErbB3 or Met amplification [44]. The association of PI3K signaling with cancer and resistance to therapies has led to the development of mTOR and PI3K inhibitors [45].
Figure 4. The significance therapeutic polypharmacology in designing combination therapy.
(A) Many types of cancer involve mutations in, or aberrant expression of, Ras, Raf, PI3K or receptor tyrosine kinases (RTKs), such as EGFR, HER2, PDGFR and FGFR. Cell proliferation and survival require both the MAPK and PI3K pathways to be active. Compensatory mechanisms cause a limited or null response to monotherapies targeted at different nodes in the RTK signaling network, such as the reduction in the p70S6K/RTK negative feedback caused by either MEK (B) or PI3K inhibition (C). (D) A combination of therapies can overcome these compensatory mechanisms and could effectively cause disease regression.
Each targeted therapy affects the signaling network in a different manner, and downstream inhibitors produce a set of compensatory signals that differs from those involved in upstream inhibition. For example, the mechanisms of compensation in MEK and PI3K/AKT inhibition (downstream inhibition) differ from those of EGFR inhibition (upstream inhibition). MEK inhibition can cause an increase in PI3K/AKT activation (Figure 4B) and, conversely, PI3K/AKT inhibition can cause an increase in MAPK activation (Figure 4C) [46,47]. These paradoxical results are caused by a negative feedback loop in RTK signaling that is mediated by TSC2/p70S6K signaling and controlled by AKT and ERK. Thus, when MEK/ERK or PI3K/AKT is inhibited, the negative feedback loop is partially shut down (Figures 4B and 4C), causing signal amplification in the non-inhibited arm of the pathway that could enable cancer progression [46]. Such mechanisms of resistance are particularly common in cancer progression and underscore the importance of careful analysis of signal flow within the surrounding signaling network of drug targets, in order to understand the mechanisms of resistance and to design therapeutic polypharmacology appropriately to overcome this resistance.
Potential applications for therapeutic polypharmacology
The ideal strategy for therapeutic polypharmacology would be to couple a screen in each patient with the types of mutations and other genomic signatures that exist in the diseased tissue, examine the effects of the mutations in signaling networks (ie, edge versus node), and design a therapeutic approach to target the up- or downregulated signals that drive the pathophysiology specifically. There are many scenarios in which combinatorial treatments would be useful. The most widely used method for combining drugs involves either a decision by the prescribing physician or clinical trial design; this method is based on the available safety information in resources such as the drug label or the physician’s desk reference. Adding network analyses to estimate the potential improved efficacy of drug combinations compared with monotherapy could be useful in clinical trial design for specific disease conditions. In general, only a small fraction of clinical trials in the US focuses on therapeutic polypharmacology. According to ClinicalTrials.gov, in February 2010, the number of clinical trials for drug combinations in the US (6720) accounted for approximately 15% of the total number of clinical trials (44,832) [48].
A major issue with screening drug combinations on a large scale is the selection of an appropriate dose for each drug, as the combination of each physiologically relevant concentration would lead to a prohibitively high number of test conditions. Only recently, in a study by Lehar et al, have computational methodologies used to make predictions on drug combinations been improved [49]. These methodologies are based on the dose-response curves and network connectivity of the individual drugs. The study demonstrated a clear relationship between the shape of the drug-dose response surface and the connectivity of the drug targets [49], potentially enabling large-scale combinatorial studies of several compounds at selected concentrations. This modeling technology was the foundation used by the biotechnology company CombinatoRx Inc in the development of an experimental platform to test combinations of pre-approved, generic drugs for new indications [50,51]. The approach used by CombinatoRx is empirical with respect to targeting nodes within the disease signaling network systematically, in order to search for combinations of approved drugs that will have novel activity against a disease. However, the experimental methodology developed is also valuable for the future of systems pharmacology in medicine.
Systems biology and therapeutic polypharmacology approaches are likely to expand the druggable genome, through a comprehensive perspective on disease and drug-action mechanisms. Systems-level studies can extend the list of targets that would be mechanistically preferred drug targets for a particular disease. Network analyses of drugs and drug-target interactions have demonstrated that the currently used drug targets are limited in diversity, but that the drugs in current R&D pipelines are expanding the drug-target diversity [17]. As genomic knowledge increases and is combined with the knowledge of mechanistic pathophysiology, the list of drug targets will also increase. In addition, the chemistry and biology of drug-design technology has improved greatly. Innovations include the structure-based design and high-throughput discovery of small-molecule, active-site inhibitors of enzymes and biological therapeutics, including mAbs, peptides and antisense therapies. These innovations are complemented by the number of drug targets in R&D pipelines, the number of which increases substantially when research targets are included (Figures 2C and 2D). However, even with the addition of research targets, the total number of currently known targets is only a small fraction of the number of potential targets encoded by the human genome. Thus, the combination of new drug-discovery technologies and systems-level discovery should improve and expand the druggable genome.
In addition, the Pharmaceutical Assets Portal, a recent initiative by the Clinical and Translational Service Award (CTSA), was designed to encourage the use of drugs that were abandoned at a clinical stage for repositioning (ie, to further research on a drug molecule with a new target or indication) [52,53]. This portal allows academic clinicians and scientists, as well as researchers in the pharmaceutical industry, to access data on drugs that have failed at various phases of clinical trials. The toxicity and bioavailability of most of these compounds have already been tested in humans, providing an advantage in overcoming the barriers involved in initiating the progression of a novel compound as a potential commercial drug product. The majority of the drugs included in the portal have known targets, and the available information on the drugs includes gene-target data. Details of researchers who have been linked to the protein target and diseases through publications are accessible using the Foci of Expertise (FoX) tool [52]. This initiative provides opportunities not only for identifying new drug indications, but also for the use of combinations of drugs, designed with therapeutic polypharmacology and network-based logic.
Adverse polypharmacology: Off-target binding and on-target binding in non-target tissue
As noted, adverse polypharmacology is defined as the negative effects of drugs that bind multiple targets, such as toxicity or other detrimental physiological effects. For example, NSAIDs inhibit the COX-1 and COX-2 isoforms, which have differing physiological roles. While COX-2 inhibition leads to the targeted effect of a reduction in the levels of prostaglandins that are responsible for pain and inflammation, COX-1 inhibition causes a reduction in the levels of prostaglandins that protect the lining of the stomach, leading to stomach pain or ulcers. A more unexpected side effect of an increased risk of heart attack and stroke was observed with high-dose treatments of the COX-2-specific inhibitor rofecoxib (Vioxx), resulting in the withdrawal of the drug from the market in 2004 [54]. The biochemical reasons for these side effects are still not completely understood [55]. Toxicity or adverse effects can also arise when the drug has an unexpected effect in a tissue other than the diseased tissue or organ. For example, HER2 inhibitors used to treat metastatic breast cancer can have cardiac toxicity effects as a result of the inhibition of HER2 expressed in cardiac tissue [56,57].
Many adverse effects are not observed at a significant level until a drug has reached clinical trials or is on the market and widely used, indicating that the drug may first need to reach an expanded, genetically diverse population. The field of pharmacogenetics studies the genetic factors that influence different drug behavior in individuals [58,59]. Screening candidate drugs for binding against every protein encoded in the human genome is not currently possible. However, research is underway to build computational tools to predict drug-target binding and, therefore, predict off-target binding and possible adverse effects [1,60]. A substantial component of this research involves analyzing patterns in the molecular structures of drugs with respect to the macromolecules with which the drugs interact. For example, if a class of drugs binds to the serotonin receptor and the drugs contain a common structural feature, then that common chemical entity is coded. Unrelated structures are parsed with these codes to predict the protein targets with which these structures might interact. A recent study using this method predicted that paroxetine (Paxil) and fluoxetine (Prozac), both SSRIs, were also β-blockers [61]. The binding of these drugs to the β-adrenergic receptor was verified, and the researchers speculated that some of the observed adverse effects of the drugs, such as increased heart rate and sexual dysfunction, could be attributed to this off-target binding [61]. These methods can also be applied in the prediction of novel binders to drug targets [1,62].
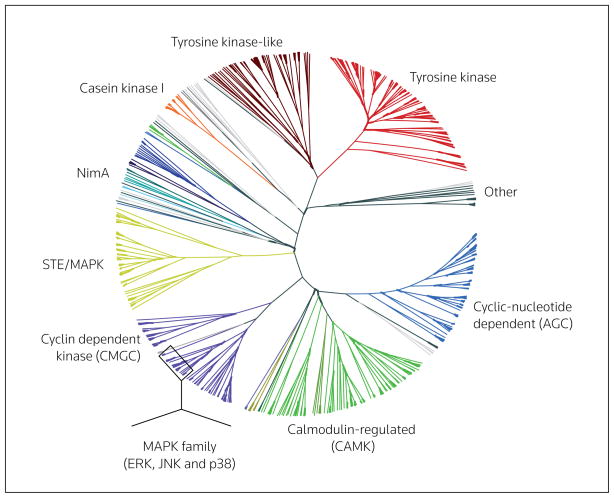
Predicting targets of drugs and drug candidates has become particularly important with the increased focus on kinase inhibitor drugs. The majority of targeted therapies involve kinase targets, and 13 different kinase inhibitors (both antibody and small-molecule) have been approved by the FDA for cancer treatment alone, with more inhibitors being pursued in other therapeutic areas and in clinical trials. Kinases have a common active site, but also have a large diversity of biological roles. The design of drugs that target the kinase domain relies heavily on the unique properties of the kinase target that distinguishes each kinase. In general, kinases with a similar function share the highest degree of sequence similarity, but there are exceptions in which kinases with high sequence similarity have diverse biological functions [63]. For example, the MAPK family, which includes the ERK and JNK isoforms, as well as p38, is responsible for a large diversity of physiological functions, including mediating cell proliferation and apoptosis [64]. However, the members of this family have highly related structures (Figure 5). Predictive studies have concluded that kinases with > 40 to 50% primary sequence identity are more likely to follow similar SARs with respect to small-molecule inhibitors [1]. The fact that kinase inhibitors tend to bind to kinases with similar structures is therefore not surprising, and inhibiting these kinases may have significantly diverse physiological implications.
Figure 5. A phylogenetic tree of the kinases coded for within the human genome.
Kinases are grouped and classified primarily by the sequence of the catalytic domains. Different groups are color-coded according to this classification [63,74].
Conclusion
Basic cell biological research continues to delineate the functions of each protein in the proteome in order to construct the ‘interactome’ (ie, how all cellular components interact), and to apply this perspective to the mechanisms underlying disease. The greater this understanding becomes, the greater the need to integrate this knowledge to enhance the discovery of new drugs. While drug-discovery platforms have progressed toward the identification of targeted therapies, attrition rates remain high, particularly for the therapies with novel mechanisms. A systems biology perspective provides an integrated basis for the understanding of the complex mechanisms of disease and targeted therapy action. The full integration of systems biology and drug discovery with polypharmacology will enable the logical design of targeted, combination therapies that are capable of efficiently restoring the equilibrium of an altered disease state. Methodologies in both fields are being developed that can aid this process; however, full implementation is still required.
Acknowledgments
The authors thank Seth Berger and Sherry Jenkins for help with analyzing the drug target data presented in Figure 2. The authors are supported by the National Institute of General Medical Sciences (NIGMS) Systems Biology Center Grants P50GM071558 and GM54508.
References
• of special interest
- 1.Bamborough P, Drewry D, Harper G, Smith GK, Schneider K. Assessment of chemical coverage of kinome space and its implications for kinase drug discovery. J Med Chem. 2008;51 (24):7898–7914. doi: 10.1021/jm8011036. [DOI] [PubMed] [Google Scholar]
- 2.Frye SV. Structure-activity relationship homology (SARAH): A conceptual framework for drug discovery in the genomic era. Chem Biol. 1999;6(1):R3–R7. doi: 10.1016/S1074-5521(99)80013-1. [DOI] [PubMed] [Google Scholar]
- 3.Lovering F, Bikker J, Humblet C. Escape from flatland: Increasing saturation as an approach to improving clinical success. J Med Chem. 2009;52(21):6752–6756. doi: 10.1021/jm901241e. [DOI] [PubMed] [Google Scholar]
- 4.Gordian M, Singh N, Zemmel R, Elias T. Why products fail in phase III. In Vivo. 2006:2006800066. [Google Scholar]
- 5•.Berger SI, Iyengar R. Network analyses in systems pharmacology. Bioinformatics. 2009;25(19):2466–2472. doi: 10.1093/bioinformatics/btp465. Provides an in-depth review of network analyses in disease and pharmacology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411(6833):41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 7.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18(4):644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf-Yadlin A, Sevecka M, MacBeath G. Dissecting protein function and signaling using protein microarrays. Curr Opin Chem Biol. 2009;13(4):398–405. doi: 10.1016/j.cbpa.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6(2):95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 10.Speer R, Wulfkuhle J, Espina V, Aurajo R, Edmiston KH, Liotta LA, Petricoin EF., 3rd Molecular network analysis using reverse phase protein microarrays for patient tailored therapy. Adv Exp Med Biol. 2008;610:177–186. doi: 10.1007/978-0-387-73898-7_13. [DOI] [PubMed] [Google Scholar]
- 11.Macek B, Mann M, Olsen JV. Global and site-specific quantitative phosphoproteomics: Principles and applications. Annu Rev Pharmacol Toxicol. 2009;49:199–221. doi: 10.1146/annurev.pharmtox.011008.145606. [DOI] [PubMed] [Google Scholar]
- 12.Jimenez-Sanchez G, Childs B, Valle D. The effect of Mendelian disease on human health. In: Scriver CR, Beaudet AR, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; New York, NY, USA: 2001. pp. 167–174. [Google Scholar]
- 13.Feldman I, Rzhetsky A, Vitkup D. Network properties of genes harboring inherited disease mutations. Proc Natl Acad Sci USA. 2008;105(11):4323–4328. doi: 10.1073/pnas.0701722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci USA. 2007;104(21):8685–8690. doi: 10.1073/pnas.0701361104. The first global network perspective study on how disease genes are connected with respect to their protein signaling networks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong Q, Simonis N, Li QR, Charloteaux B, Heuze F, Klitgord N, Tam S, Yu H, Venkatesan K, Mou D, Swearingen V, et al. Edgetic perturbation models of human inherited disorders. Mol Syst Biol. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Ma’ayan A, Jenkins SL, Goldfarb J, Iyengar R. Network analysis of FDA approved drugs and their targets. Mt Sinai J Med. 2007;74(1):27–32. doi: 10.1002/msj.20002. One of the first examples, along with reference [17], of network analyses of drugs and the proteins these drugs target, providing a global perspective of the properties of approved drugs and drugs in research pipelines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nat Biotechnol. 2007;25(10):1119–1126. doi: 10.1038/nbt1338. One of the first examples, along with reference [18], of network analyses of drugs and the proteins targeted by these drugs, providing a global perspective of the properties of approved drugs and the drugs in research pipelines. [DOI] [PubMed] [Google Scholar]
- 18.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(Database issue):D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: A knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(Database issue):D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Ji ZL, Chen YZ. TTD: Therapeutic Target Database. Nucleic Acids Res. 2002;30(1):412–415. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu F, Han B, Kumar P, Liu X, Ma X, Wei X, Huang L, Guo Y, Han L, Zheng C, Chen Y. Update of TTD: Therapeutic Target Database. Nucleic Acids Res. 2010;38(Database issue):D787–D791. doi: 10.1093/nar/gkp1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olah M, Rad R, Ostopovici L, Bora A, Hadaruga N, Hadaruga D, Moldovan R, Fulias A, Mracec M, Oprea TI. WOMBAT and WOMBAT-PK: Bioactivity databases for lead and drug discovery. In: Schreiber SL, Kapoor TM, Wess G, editors. Chemical Biology. Wiley-VCH; Weinheim, Germany: 2007. pp. 760–786. [Google Scholar]
- 23.Gao Z, Li H, Zhang H, Liu X, Kang L, Luo X, Zhu W, Chen K, Wang X, Jiang H. PDTD: A web-accessible protein database for drug target identification. BMC Bioinformatics. 2008;9:104. doi: 10.1186/1471-2105-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24•.Mestres J, Gregori-Puigjane E, Valverde S, Sole RV. Data completeness – The Achilles heel of drug-target networks. Nat Biotechnol. 2008;26(9):983–984. doi: 10.1038/nbt0908-983. Examines the possibilities of how drug-target databases underestimate the number of therapeutic targets for each drug. Incorporating all targets is important when considering and predicting adverse effects or new therapeutic effects of existing drugs. [DOI] [PubMed] [Google Scholar]
- 25•.Wist AD, Berger SI, Iyengar R. Systems pharmacology and genome medicine: A future perspective. Genome Med. 2009;1(1):11. doi: 10.1186/gm11. Describes the genomic basis of disease, systems pharmacology and the integration of these processes in the future of personalized medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1(9):727–730. doi: 10.1038/nrd892. Analyzes the druggability of the proteins expressed in the human genome. Although not all of the defined druggable proteins are associated with disease, this assessment provides a perspective on how drug pipelines may evolve in the future. [DOI] [PubMed] [Google Scholar]
- 27.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC., Jr American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25 (33):5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 28.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman PA, Swain SM, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353(16):1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 29.Cascinu S, Berardi R, Labianca R, Siena S, Falcone A, Aitini E, Barni S, Di Costanzo F, Dapretto E, Tonini G, Pierantoni C, et al. Cetuximab plus gemcitabine and cisplatin compared with gemcitabine and cisplatin alone in patients with advanced pancreatic cancer: A randomised, multicentre, phase II trial. Lancet Oncol. 2008;9(1):39–44. doi: 10.1016/S1470-2045(07)70383-2. [DOI] [PubMed] [Google Scholar]
- 30.Kong KF, Schneper L, Mathee K. β-Lactam antibiotics: From antibiosis to resistance and bacteriology. APMIS. 2010;118(1):1–36. doi: 10.1111/j.1600-0463.2009.02563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizzo P, Osipo C, Foreman K, Golde T, Osborne B, Miele L. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27 (38):5124–5131. doi: 10.1038/onc.2008.226. [DOI] [PubMed] [Google Scholar]
- 32.Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE. Direct inhibition of the Notch transcription factor complex. Nature. 2009;462(7270):182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao SS, O’Neil J, Liberator CD, Hardwick JS, Dai X, Zhang T, Tyminski E, Yuan J, Kohl NE, Richon VM, Van der Ploeg LH, et al. Inhibition of Notch signaling by γ secretase inhibitor engages the Rb pathway and elicits cell cycle exit in T-cell acute lymphoblastic leukemia cells. Cancer Res. 2009;69 (7):3060–3068. doi: 10.1158/0008-5472.CAN-08-4295. [DOI] [PubMed] [Google Scholar]
- 34.Wilhelm SM, Adnane L, Newell P, Villanueva A, Llovet JM, Lynch M. Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling. Mol Cancer Ther. 2008;7(10):3129–3140. doi: 10.1158/1535-7163.MCT-08-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, Chen C, Zhang X, Vincent P, McHugh M, Cao Y, et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the Raf/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64(19):7099–7109. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- 36.Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, Negrier S, Chevreau C, Solska E, Desai AA, Rolland F, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356(2):125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 37.Xia W, Mullin RJ, Keith BR, Liu LH, Ma H, Rusnak DW, Owens G, Alligood KJ, Spector NL. Anti-tumor activity of GW572016: A dual tyrosine kinase inhibitor blocks EGF activation of EGFR/ErbB2 and downstream ERK1/2 and AKT pathways. Oncogene. 2002;21(41):6255–6263. doi: 10.1038/sj.onc.1205794. [DOI] [PubMed] [Google Scholar]
- 38.Lopez-Chavez A, Carter CA, Giaccone G. The role of KRAS mutations in resistance to EGFR inhibition in the treatment of cancer. Curr Opin Investig Drugs. 2009;10(12):1305–1314. [PubMed] [Google Scholar]
- 39.Friday BB, Adjei AA. Advances in targeting the Ras/Raf/MEK/ERK mitogen-activated protein kinase cascade with MEK inhibitors for cancer therapy. Clin Cancer Res. 2008;14(2):342–346. doi: 10.1158/1078-0432.CCR-07-4790. [DOI] [PubMed] [Google Scholar]
- 40.Young A, Lyons J, Miller AL, Phan VT, Alarcon IR, McCormick F. Ras signaling and therapies. Adv Cancer Res. 2009;102:1–17. doi: 10.1016/S0065-230X(09)02001-6. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Chen Y, Rao SS, Chen XM, Liu HC, Qin JH, Tang WF, Yue W, Zhou X, Lu T. Recent advances in the research and development of B-Raf inhibitors. Curr Med Chem. 2010 doi: 10.2174/092986710791111242. [DOI] [PubMed] [Google Scholar]
- 42•.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, Marais R. Kinase-dead BRaf and oncogenic Ras cooperate to drive tumor progression through CRaf. Cell. 2010;140(2):209–221. doi: 10.1016/j.cell.2009.12.040. This study, along with reference [43], reveals a counterintuitive result of inhibiting B-Raf in BRAF wild-type and KRAS mutant cancers. Specifically, inhibiting B-Raf in cancers with certain mutational backgrounds causes tumor growth. This effect has serious implications for the target patient populations of several B-Raf drugs that are either marketed or in clinical trials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, Morales T, et al. Raf inhibitors prime wild-type Raf to activate the MAPK pathway and enhance growth. Nature. 2010;464(7287):431–435. doi: 10.1038/nature08833. This study, along with reference [42], reveals a counterintuitive result of inhibiting B-Raf in BRAF wild-type and KRAS mutant cancers. Specifically, inhibiting B-Raf in cancers with certain mutational backgrounds causes tumor growth. This effect has serious implications for the target patient populations of several B-Raf drugs that are either marketed or in clinical trials. [DOI] [PubMed] [Google Scholar]
- 44.Klein S, Levitzki A. Targeting the EGFR and the PKB pathway in cancer. Curr Opin Cell Biol. 2009;21(2):185–193. doi: 10.1016/j.ceb.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 45.Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28(6):1075–1083. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sos ML, Fischer S, Ullrich R, Peifer M, Heuckmann JM, Koker M, Heynck S, Stuckrath I, Weiss J, Fischer F, Michel K, et al. Identifying genotype-dependent efficacy of single and combined PI3K- and MAPK-pathway inhibition in cancer. Proc Natl Acad Sci USA. 2009;106(43):18351–18356. doi: 10.1073/pnas.0907325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, Papa A, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118(9):3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.ClinicalTrials.gov. US National Library of Medicine; Bethesda, MD, USA: 2010. clinicaltrials.gov/ [Google Scholar]
- 49.Lehar J, Krueger AS, Avery W, Heilbut AM, Johansen LM, Price ER, Rickles RJ, Short GF, 3rd, Staunton JE, Jin X, Lee MS, et al. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat Biotechnol. 2009;27 (7):659–666. doi: 10.1038/nbt.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolfson W. Speed dating for molecules CombinatoRx looks for that special synergy. Chem Biol. 2006;13(5):461–462. doi: 10.1016/j.chembiol.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 51.CombinatoRx drug discovery engine (cHTS) CombinatoRx; Cambridge, MA, USA: 2010. www.combinatorx.com/discovery_cHTS_technology.php. [Google Scholar]
- 52.Clinical and Translational Science Awards (CTSA) Pharmaceutical Assets Portal Foci of Expertise (FoX) NIH; Bethesda, MD, USA: 2010. www.ctsapharmaportal.org/fociofexpertise.html. [Google Scholar]
- 53.Schubert C. Matchmaking service links up researchers to wallflower drugs. Nat Med. 2010;16(1):7. doi: 10.1038/nm0110-7b. [DOI] [PubMed] [Google Scholar]
- 54.Merck & Co Inc. Merck announces voluntary worldwide withdrawal of VIOXX. Press Release. 2004 September 30; [Google Scholar]
- 55.Bombardier C, Laine L, Reicin A, Shapiro D, Burgos-Vargas R, Davis B, Day R, Ferraz MB, Hawkey CJ, Hochberg MC, Kvien TK, et al. Comparison of upper gastrointestinal toxicity of rofecoxib and naproxen in patients with rheumatoid arthritis. VIGOR Study Group. N Engl J Med. 2000;343(21):1520–1528. doi: 10.1056/NEJM200011233432103. 2 p following 1528. [DOI] [PubMed] [Google Scholar]
- 56.Tan-Chiu E, Yothers G, Romond E, Geyer CE, Jr, Ewer M, Keefe D, Shannon RP, Swain SM, Brown A, Fehrenbacher L, Vogel VG, et al. Assessment of cardiac dysfunction in a randomized trial comparing doxorubicin and cyclophosphamide followed by paclitaxel, with or without trastuzumab as adjuvant therapy in node-positive, human epidermal growth factor receptor 2-overexpressing breast cancer: NSABP B-31. J Clin Oncol. 2005;23(31):7811–7819. doi: 10.1200/JCO.2005.02.4091. [DOI] [PubMed] [Google Scholar]
- 57.Perez EA. Cardiac toxicity of ErbB2-targeted therapies: What do we know? Clin Breast Cancer. 2008;8(Suppl 3):S114–S120. doi: 10.3816/cbc.2008.s.007. [DOI] [PubMed] [Google Scholar]
- 58.Court MH. A pharmacogenomics primer. J Clin Pharmacol. 2007;47(9):1087–1103. doi: 10.1177/0091270007303768. [DOI] [PubMed] [Google Scholar]
- 59.Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429(6990):464–468. doi: 10.1038/nature02626. [DOI] [PubMed] [Google Scholar]
- 60•.Ong SE, Schenone M, Margolin AA, Li X, Do K, Doud MK, Mani DR, Kuai L, Wang X, Wood JL, Tolliday NJ, et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci USA. 2009;106(12):4617–4622. doi: 10.1073/pnas.0900191106. Experimental methodology, combining quantitative proteomics and affinity enrichment methods, was used to identify proteins that bind to small molecules or drugs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61•.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, Whaley R, et al. Predicting new molecular targets for known drugs. Nature. 2009;462(7270):175–181. doi: 10.1038/nature08506. One of several studies, including reference [62] that used structure-based algorithms to search for commonalities among drugs or small-molecule ligands and predict new protein targets to which these drugs/ligands bind. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62•.Fuller JC, Burgoyne NJ, Jackson RM. Predicting druggable binding sites at the protein-protein interface. Drug Discov Today. 2009;14(3–4):155–161. doi: 10.1016/j.drudis.2008.10.009. One of several studies, including reference [61] that used structure-based algorithms to search for commonalities among drugs or small-molecule ligands and predict new protein targets to which these drugs/ligands bind. [DOI] [PubMed] [Google Scholar]
- 63.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 64.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK and p38 protein kinases. Science. 2002;298(5600):1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 65.Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G, Mann M, Daub H. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics. 2009;8(7):1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.VanMeter A, Signore M, Pierobon M, Espina V, Liotta LA, Petricoin EF., 3rd Reverse-phase protein microarrays: Application to biomarker discovery and translational medicine. Expert Rev Mol Diagn. 2007;7(5):625–633. doi: 10.1586/14737159.7.5.625. [DOI] [PubMed] [Google Scholar]
- 67.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25(9):1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 68.Iyengar R. Why we need quantitative dynamic models. Sci Signal. 2009;2(64):eg3. doi: 10.1126/scisignal.264eg3. [DOI] [PubMed] [Google Scholar]
- 69.Stites EC, Trampont PC, Ma Z, Ravichandran KS. Network analysis of oncogenic Ras activation in cancer. Science. 2007;318(5849):463–467. doi: 10.1126/science.1144642. [DOI] [PubMed] [Google Scholar]
- 70.Kreeger PK, Lauffenburger DA. Cancer systems biology: A network modeling perspective. Carcinogenesis. 2010;31(1):2–8. doi: 10.1093/carcin/bgp261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3(9):715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 72.Gaither A, Iourgenko V. RNA interference technologies and their use in cancer research. Curr Opin Oncol. 2007;19(1):50–54. doi: 10.1097/CCO.0b013e328011a8b0. [DOI] [PubMed] [Google Scholar]
- 73.Chang K, Elledge SJ, Hannon GJ. Lessons from Nature: MicroRNA-based shRNA libraries. Nat Methods. 2006;3(9):707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- 74.Kinase.com. Salk Institute; La Jolla, CA, USA: 2009. www.kinase.com/human/kinome. [Google Scholar]