Abstract
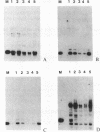
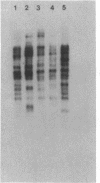
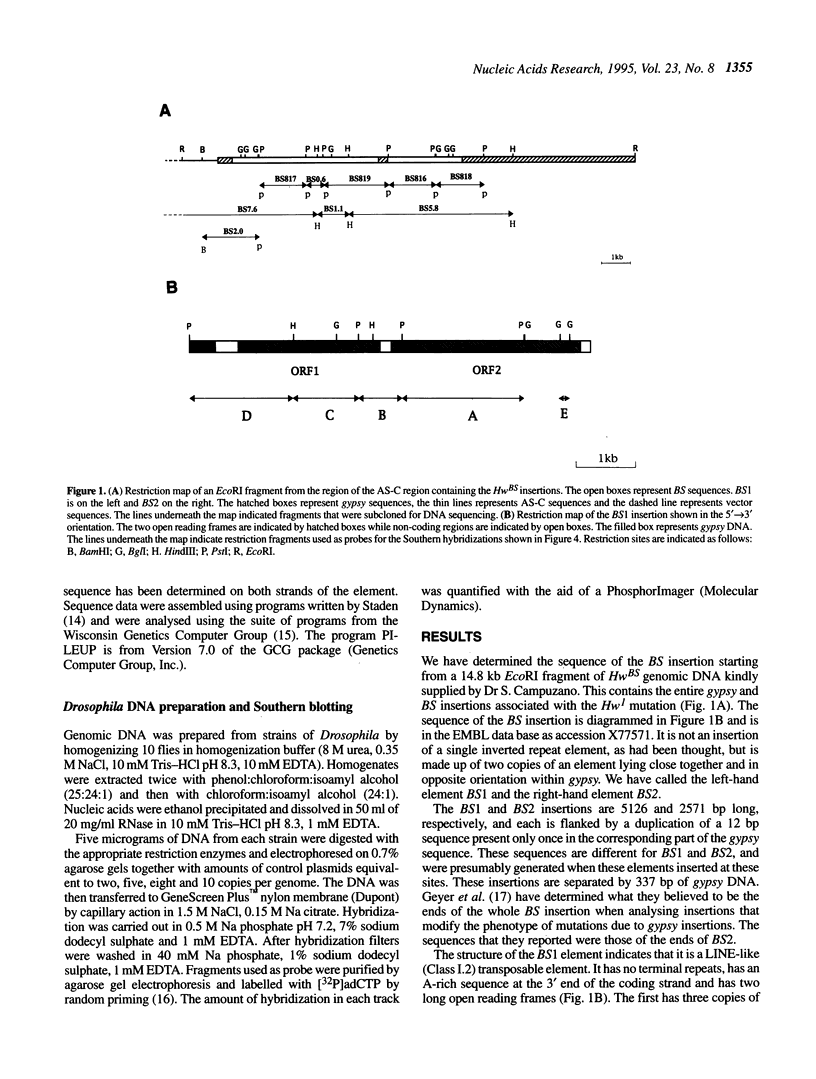
Transposable elements with long terminal inverted repeats are rare and only one family of elements of this sort has been identified in the genome of Drosophila melanogaster. An insertion associated with the HSBS mutation of the achaete-scute complex has been reported to be a second element of this type. We have determined the complete sequence of this insertion and have shown that it is in fact two copies of a new LINE-like transposable element, that we have called BS, inserted in opposite orientation 337 bp apart. Like other elements of this type, BS has two open reading frames that appear to encode a gag-like polypeptide and a reverse transcriptase. There are few complete BS elements in the five strains of D.melanogaster that we have tested and they appear to transpose infrequently. The events that may have lead to the double BS insertion are discussed in terms of the supposed mechanism of transposition of LINE-like elements.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brierley H. L., Potter S. S. Distinct characteristics of loop sequences of two Drosophila foldback transposable elements. Nucleic Acids Res. 1985 Jan 25;13(2):485–500. doi: 10.1093/nar/13.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheton A. I transposable elements and I-R hybrid dysgenesis in Drosophila. Trends Genet. 1990 Jan;6(1):16–21. doi: 10.1016/0168-9525(90)90044-7. [DOI] [PubMed] [Google Scholar]
- Campuzano S., Balcells L., Villares R., Carramolino L., García-Alonso L., Modolell J. Excess function hairy-wing mutations caused by gypsy and copia insertions within structural genes of the achaete-scute locus of Drosophila. Cell. 1986 Jan 31;44(2):303–312. doi: 10.1016/0092-8674(86)90764-6. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nocera P. P. Close relationship between non-viral retroposons in Drosophila melanogaster. Nucleic Acids Res. 1988 May 11;16(9):4041–4052. doi: 10.1093/nar/16.9.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nocera P. P., Graziani F., Lavorgna G. Genomic and structural organization of Drosophila melanogaster G elements. Nucleic Acids Res. 1986 Jan 24;14(2):675–691. doi: 10.1093/nar/14.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J. P., Palmiter R. D. Retrotransposition of a mouse L1 element. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8792–8795. doi: 10.1073/pnas.88.19.8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett D. H., Lister C. K., Kellett E., Finnegan D. J. Transposable elements controlling I-R hybrid dysgenesis in D. melanogaster are similar to mammalian LINEs. Cell. 1986 Dec 26;47(6):1007–1015. doi: 10.1016/0092-8674(86)90815-9. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Geyer P. K., Corces V. G. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992 Oct;6(10):1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- Geyer P. K., Green M. M., Corces V. G. Mutant gene phenotypes mediated by a Drosophila melanogaster retrotransposon require sequences homologous to mammalian enhancers. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8593–8597. doi: 10.1073/pnas.85.22.8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goman M., Mons B., Scaife J. The complete sequence of a Plasmodium malariae SSUrRNA gene and its comparison to other plasmodial SSUrRNA genes. Mol Biochem Parasitol. 1991 Apr;45(2):281–288. doi: 10.1016/0166-6851(91)90096-o. [DOI] [PubMed] [Google Scholar]
- Harden N., Ashburner M. Characterization of the FB-NOF transposable element of Drosophila melanogaster. Genetics. 1990 Oct;126(2):387–400. doi: 10.1093/genetics/126.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison D. A., Geyer P. K., Spana C., Corces V. G. The gypsy retrotransposon of Drosophila melanogaster: mechanisms of mutagenesis and interaction with the suppressor of Hairy-wing locus. Dev Genet. 1989;10(3):239–248. doi: 10.1002/dvg.1020100313. [DOI] [PubMed] [Google Scholar]
- Jakubczak J. L., Burke W. D., Eickbush T. H. Retrotransposable elements R1 and R2 interrupt the rRNA genes of most insects. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3295–3299. doi: 10.1073/pnas.88.8.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubczak J. L., Xiong Y., Eickbush T. H. Type I (R1) and type II (R2) ribosomal DNA insertions of Drosophila melanogaster are retrotransposable elements closely related to those of Bombyx mori. J Mol Biol. 1990 Mar 5;212(1):37–52. doi: 10.1016/0022-2836(90)90303-4. [DOI] [PubMed] [Google Scholar]
- Jensen S., Heidmann T. An indicator gene for detection of germline retrotransposition in transgenic Drosophila demonstrates RNA-mediated transposition of the LINE I element. EMBO J. 1991 Jul;10(7):1927–1937. doi: 10.1002/j.1460-2075.1991.tb07719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare K., Alley M. R., Cullingford T. E., Driver A., Sanderson M. J. DNA sequence of the Doc retroposon in the white-one mutant of Drosophila melanogaster and of secondary insertions in the phenotypically altered derivatives white-honey and white-eosin. Mol Gen Genet. 1991 Jan;225(1):17–24. doi: 10.1007/BF00282637. [DOI] [PubMed] [Google Scholar]
- Pittler S. J., Davis R. L. A new family of the poly-deoxyadenylated class of Drosophila transposable elements identified by a representative member at the dunce locus. Mol Gen Genet. 1987 Jun;208(1-2):325–328. doi: 10.1007/BF00330460. [DOI] [PubMed] [Google Scholar]
- Potter S. S. DNA sequence analysis of a Drosophila foldback transposable element rearrangement. Mol Gen Genet. 1982;188(1):107–110. doi: 10.1007/BF00333002. [DOI] [PubMed] [Google Scholar]
- Potter S. S. DNA sequence of a foldback transposable element in Drosophila. Nature. 1982 May 20;297(5863):201–204. doi: 10.1038/297201a0. [DOI] [PubMed] [Google Scholar]
- Priimägi A. F., Mizrokhi L. J., Ilyin Y. V. The Drosophila mobile element jockey belongs to LINEs and contains coding sequences homologous to some retroviral proteins. Gene. 1988 Oct 30;70(2):253–262. doi: 10.1016/0378-1119(88)90197-7. [DOI] [PubMed] [Google Scholar]
- Pritchard M. A., Dura J. M., Pélisson A., Bucheton A., Finnegan D. J. A cloned I-factor is fully functional in Drosophila melanogaster. Mol Gen Genet. 1988 Nov;214(3):533–540. doi: 10.1007/BF00330491. [DOI] [PubMed] [Google Scholar]
- Pélisson A., Finnegan D. J., Bucheton A. Evidence for retrotransposition of the I factor, a LINE element of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4907–4910. doi: 10.1073/pnas.88.11.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiha H., Miller J. R., Woods L. C., Glover D. M. Arrangements and rearrangements of sequences flanking the two types of rDNA insertion in D. melanogaster. Nature. 1981 Apr 30;290(5809):749–753. doi: 10.1038/290749a0. [DOI] [PubMed] [Google Scholar]
- Roiha H., Rubin G. M., O'Hare K. P element insertions and rearrangements at the singed locus of Drosophila melanogaster. Genetics. 1988 May;119(1):75–83. doi: 10.1093/genetics/119.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Leclercq L., Göbel E., Saedler H. Cin4, an insert altering the structure of the A1 gene in Zea mays, exhibits properties of nonviral retrotransposons. EMBO J. 1987 Dec 20;6(13):3873–3880. doi: 10.1002/j.1460-2075.1987.tb02727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana C., Harrison D. A., Corces V. G. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 1988 Nov;2(11):1414–1423. doi: 10.1101/gad.2.11.1414. [DOI] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton N. S., Potter S. S. Complete foldback transposable elements encode a novel protein found in Drosophila melanogaster. EMBO J. 1989 Jun;8(6):1887–1894. doi: 10.1002/j.1460-2075.1989.tb03585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H., Kikuno R., Hayashida H., Miyata T., Kugimiya W., Inouye S., Yuki S., Saigo K. Close structural resemblance between putative polymerase of a Drosophila transposable genetic element 17.6 and pol gene product of Moloney murine leukaemia virus. EMBO J. 1985 May;4(5):1267–1272. doi: 10.1002/j.1460-2075.1985.tb03771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J., Karpen G. H., Craig N., Spradling A. C. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics. 1993 Feb;133(2):347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truett M. A., Jones R. S., Potter S. S. Unusual structure of the FB family of transposable elements in Drosophila. Cell. 1981 Jun;24(3):753–763. doi: 10.1016/0092-8674(81)90101-x. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990 Oct;9(10):3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]