Abstract
Tensor-based morphometry (TBM) is an analysis technique where anatomical information is characterized by means of the spatial transformations mapping a customized template with the observed images. Therefore, accurate inter-subject non-rigid registration is an essential prerequisite for both template estimation and image warping. Subsequent statistical analysis on the spatial transformations is performed to highlight voxel-wise differences. Most of previous TBM studies did not explore the influence of the registration parameters, such as the parameters defining the deformation and the regularization models. In this work performance evaluation of TBM using stationary velocity field (SVF) diffeomorphic registration was performed in a subset of subjects from Alzheimer’s Disease Neuroimaging Initiative (ADNI) study. A wide range of values of the registration parameters that define the transformation smoothness and the balance between image matching and regularization were explored in the evaluation. The proposed methodology provided brain atrophy maps with very detailed anatomical resolution and with a high significance level compared with results recently published on the same data set using a non-linear elastic registration method.
Keywords: Alzheimer’s disease, Tensor-based morphometry, Diffeomorphic registration
Introduction
Alzheimer’s disease (AD) is the most common form of age-related dementia and one of the most serious health problems in the industrialized world. It manifests with progressive cognitive decline initially shown as memory loss and then spreads to affect all other cognitive faculties and the patients’ ability to conduct an independent lifestyle. Mild cognitive impairment (MCI) is a relatively recent concept introduced to recognize the intermediate cognitive state where patients are neither cognitively intact nor demented (Petersen et al., 2001; Petersen, 2004; Winblad et al., 2004). Some MCI patients harbor an alternative pathological diagnosis such as dementia with Lewy bodies, vascular dementia, hippocampal sclerosis, frontotemporal dementia and even some MCI cases can also be attributed to non-degenerative pathology.
In spite of recent advances in understanding the genetics, neuropathology and neuropsychology of AD, we are still lacking sensitive and specific biological markers useful in the preclinical stages. AD-associated brain changes can be clinically evaluated in-vivo with the help of neuroimaging, using either structural technique such as magnetic resonance imaging (MRI) and diffusion tensor imaging or functional approaches such as positron emission tomography (Mosconi, 2005; Nordberg, 2008), functional MRI (Dickerson and Sperling, 2008), arterial spin labeling (Du et al., 2006) and spectroscopy (Kantarci et al., 2002; Modrego et al., 2005). Reliable biomarkers of the underlying pathology that can also predict disease progression in MCI are needed and several candidate brain measures have been examined in a wealth of cross-sectional and longitudinal neuroimaging studies.
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) (Mueller et al., 2005a,b) is a large multi-site longitudinal structural MRI and fluorodeoxyglucose positron emission tomography (FDG-PET) study of 800 adults, ages 55 to 90, including 200 elderly controls, 400 subjects with mild cognitive impairment, and 200 patients with AD. The ADNI was launched in 2003 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public–private partnership. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. Several brain morphometry studies on ADNI data have been already published (Fan et al., 2008a; Hua et al., 2008a,b, 2009; Morra et al., 2008; Leow et al., 2009; Misra et al., 2009; Qiu et al., 2009; Schuff et al., 2009).
Nowadays several techniques for analysis of brain anatomy are available. The oldest approach is the region of interest (ROI) technique which measures the volume of specific brain structures. It relies on delineation of the regions of interest. Volumetry is a powerful and intuitive technique that has yielded a wealth of findings, but has some drawbacks. ROI analysis requires an accurate a priori hypothesis, so analyses often tend to be limited to one or two structures of interest. This limitation is important when complex and dynamic atrophy patterns are sought, which is the case of AD. Hippocampus and entorhinal cortex are the regions more frequently analyzed in this pathology (Laakso et al., 1995; Krasuski et al., 1998; Jack et al., 1999; Du et al., 2001; Pennanen et al., 2004). In addition, when using manual delineation, the ROI method is operator-dependent, susceptible to bias and time consuming (Barnes et al., 2009).
More specific and subtle shape information of particular regions or structures, such as the hippocampus, has been analyzed by means of statistical shape analysis. Different shape features have been used, such as landmark coordinates (Csernansky et al., 2000, 2004), thickness or radial atrophy maps (Thompson et al., 2007; Querbes et al., 2009; Qiu et al., 2009), and medial representations (Styner et al., 2003). However, these methods share some limitations with the ROI analysis because an a priori hypothesis about the target structure is required together with the task of accurate delineation.
A different paradigm is to perform voxel-wise statistical analysis of anatomical information for the whole brain volume. One of the techniques belonging to this paradigm is tensor-based morphometry (TBM), which identifies regional structural differences in the brain, across groups or over time, from the gradients of the deformation fields that align images to a common anatomical template (Frackowiak, 2004). The anatomical information is encoded in the spatial transformation. Therefore, accurate inter-subject non-rigid registration is an essential tool. Many different registration approaches have been proposed, all having several tuning parameters, including parameters defining the deformation model, the regularization model, the optimization technique and the interpolation approach. With the new advent of recent and powerful non-rigid registration algorithms based on the large deformation paradigm (Leow et al., 2007; Lepore et al., 2008; Brun et al., 2009), TBM is being increasingly used. Subsequent statistical analysis is performed on the spatial transformations to highlight statistical differences between groups (Chiang et al., 2007a,b), or to classify individuals into diagnostic labels (Fan et al., 2008a,b; Duchesne et al., 2008). One of the simplest and most common TBM features is the determinant of the Jacobian matrix which can be interpreted as a local atrophy/expansion factor (Leow et al., 2006; Lepore et al., 2007; Chiang et al., 2007a; Lee et al., 2007). More complete descriptors can be also used, such as the complete Jacobian matrix J, or rotation-invariant features, such as the strain tensor (Lepore et al., 2006, 2008; Ridgway et al., 2008).
One of the main limitations of the TBM is the non-uniform distribution of the variance of the warpings, which is typically larger at cortical folds than in subcortical regions. This variance may be due to anatomical variability and possible misregistration errors. Accordingly, subtle anatomical differences between groups may be unnoticed especially in these regions.
Even though many different non-rigid registration methods could be considered as potentially suitable for TBM studies, the methods belonging to the large deformation paradigm have the advantage of offering a large flexibility required to characterize the anatomical variability in cross-sectional studies of elderly subjects and dementia patients. Some of these methods are fluid registration (Christensen et al., 1996; D’Agostino et al., 2003), the large deformation diffeomorphic metric mapping (LDDMM) (Csernansky et al., 2000; Beg et al., 2005; Wang et al., 2007), diffeomorphic demons (Vercauteren et al., 2007) and stationary velocity field (SVF) diffeomorphic methods (Ashburner, 2007; Hernandez et al., 2007, 2009; Vercauteren et al., 2008). The warping in all previous methods is a diffeomorphism, which is an invertible and differentiable mapping obtained by integrating a smooth velocity vector field.
SVF diffeomorphic registration has been recently proposed as a simplified version of the LDDMM algorithm, by constraining the parameterization to a stationary velocity field. With this simplified characterization, the forward and backward integration of the velocity field are identified with the group exponential and can be computed using fast methods (Arsigny et al., 2006; Bossa et al., 2008) with smaller memory requirements than in the LDDMM method. To our knowledge, two diffeomorphic registration algorithms with SVF parameterization were proposed at about the same time (Hernandez et al., 2007; Ashburner, 2007). Both can be fitted in the same variational framework, with some differences in the optimization technique. At the same time an extension of the demons registration method to diffeomorphic transformations was proposed in Vercauteren et al. (2007) where a Lie group optimization technique was used. The regularization in Vercauteren et al. (2007, 2009) was externally imposed by means of Gaussian smoothing.
The aim of this paper is twofold. Firstly, to illustrate that SVF diffeomorphic registration may be a good choice for TBM studies because it allows large deformations and offers a good accuracy/complexity trade-off. In particular, the SVF diffeomorphic registration method is used on the same data set analyzed in a recent TBM study using non-linear elastic registration (Hua et al., 2008a). Secondly, to quantify and illustrate the effect of using different values of the registration parameters in a TBM study. In addition to SVF diffeomorphic registration, diffeomorphic demons 2 was also explored.
Materials and methods
Subjects
In this study we selected the same subset of 120 subjects from ADNI database as in Hua et al. (2008a) in order to make an easier comparison. To summarize, MRI baseline scans, divided into 3 groups: 40 healthy elderly individuals (denoted as Nor), 40 individuals with amnestic MCI, and 40 individuals with probable AD. Each group of 40 subjects was well matched in terms of gender and age. Likewise (Hua et al., 2008a), an independent second group of normal subjects (denoted as Nor2), age- and gender-matched to the first group of controls, was selected to test whether analysis techniques correctly detect no differences when comparing the two independent groups of normal subjects.
All subjects underwent clinical/cognitive assessment at the time of scan acquisition. As part of each subject’s cognitive evaluation, the Mini-Mental State Examination (MMSE) was performed to provide a global measure of mental status based on evaluation of five cognitive domains. The Clinical Dementia Rating (CDR) was also assessed as a measure of dementia severity. The elderly normal subjects had MMSE scores between 28 and 30 (inclusive), a global CDR of 0, and no symptoms of depression, MCI, or other forms of dementia. The MCI subjects had MMSE scores in the range of 24 to 28, a global CDR of 0.5, and mild memory complaints, with memory impairment assessed via education-adjusted scores on the Wechsler Memory Scale – Logical Memory II. All AD patients met NINCDS/ADRDA criteria for probable AD with an MMSE score between 20 and 23. As such, these subjects would be considered as having mild to moderate, but not severe, AD. Table 1 shows a summary of demographic and clinical data. More details about criteria for patient selection and exclusion can be found in Hua et al. (2008a) and in the ADNI protocol (Mueller et al., 2005a,b).
Table 1.
Demographic data. The format of the values is average (min–max).
| Group |
AD |
MCI |
Nor |
|||
|---|---|---|---|---|---|---|
| Sex | Male | Female | Male | Female | Male | Female |
| N | 21 | 19 | 21 | 19 | 21 | 19 |
| Age | 76.4 (56–87) | 75.6 (56–87) | 76.5 (57–88) | 75.2 (55–86) | 76.6 (63–85) | 75.8 (62–89) |
| MMSE | 21.7 (20–23) | 22.1(20–23 ) | 27.1 (26–28) | 27.1 (26–28) | 29.2 (28–30) | 29.3 (28–30) |
MRI acquisition, image correction and pre-processing
High-resolution structural brain MRI scans were acquired at multiple ADNI sites with 1.5T MRI scanners using the standard ADNI MRI protocol. For each subject, two T1-weighted MRI scans were collected using a sagittal 3D magnetization-prepared rapid acquisition with gradient echo (MP-RAGE) sequence with voxel size of 0.94×0.94×1.2 mm3. The images were calibrated with phantom-based geometric corrections to ensure consistency among scans acquired at different sites. Additional image corrections included geometric distortion correction, bias field correction and geometrical scaling. See Hua et al. (2008a) for more details. The pre-processed images are available to the scientific community and were downloaded from the ADNI website.
Brain images were intensity-normalized by means of histogram matching with a linear mapping that aligned the 95-th percentile of the intensity histogram to an intensity value of 95. To adjust for global differences in brain positioning and scale across individuals, all scans were linearly registered to the stereotaxic space defined by the International Consortium for Brain Mapping (ICBM-53) (Mazziotta et al., 2001) with an affine transformation (12 degrees of freedom). Aligned images were resampled in an isotropic space of 220 voxels along each axis (x, y, and z) with a final voxel size of 1 mm3.
Stationary velocity field (SVF) diffeomorphic registration
The registration method can be formulated as a variational problem, where the cost function to be minimized contains an image matching term E1 between a template image T and a target image I and a regularization term E2 in order to guarantee the smoothness of the transformation,
| (1) |
where the weight σ (regularization parameter) balances the relative importance between image matching and regularization, and ϕ is the template warping parameterized as
| (2) |
being v a stationary velocity vector field and the group exponential mapping is defined as exp(tv) ≡ φt. In this work we selected the Sum of Squared Differences (SSD) as matching criteria E1 and the regularization term as E2(v) = ∫ (Lv)2dx being L a linear invertible differential operator. The L operator was chosen as in Beg et al. (2005), L = Id − αΔ, where Δ is the Laplacian operator and the parameter α penalizes up to second-order derivatives of the velocity field. All in all, the cost function is given by
| (3) |
The optimization was performed with a non-linear conjugate-gradient strategy (Nocedal and Wright, 1999; Hager and Zhang, 2006), i.e. the search direction is a linear combination of the negative gradient direction and the search direction from the previous iteration. The gradient of Eq. (3) was computed as
| (4) |
being H = L†L, L†, the adjoint operator of L and Dφ the Jacobian matrix. Note that H is a smoothing kernel with a tuning parameter α. The amount of spatial correlation increases with larger values of the smoothing parameter α.
Additional implementation details are the following ones: a multi-scale pyramidal approach with 4 levels was used for computational savings and avoiding local minima; the exponential mapping was implemented as a forward Euler integration with 50 steps because this standard evolution method offered a good trade-off between accuracy and computational time (Bossa et al., 2008); the Laplacian operator was a centered-stencil; the filter H and its inverse were applied in the Fourier domain inducing periodic boundary conditions.
Unbiased average template
An average template is one of the key components of TBM studies. It provides a coordinate system where all image samples are registered. In order to make automatic registration easier and more robust, the template must represent common intensity and geometric features from the group of images. A common solution found in the literature is the estimation of an unbiased average template image by minimizing the deformations (Joshi et al., 2004; Hua et al., 2008a). When the registration method is not accurate enough to match anatomical structures, the unbiased template becomes smooth. This lack of sharp anatomical details in the template may reduce the sensitivity of a TBM study to detect subtle brain volume changes (Studholme et al., 2004).
In this work the unbiased template T was estimated from images of the Nor group, likewise in Hua et al. (2008a) because we assume that the disease process is one of structural removal and the morphometry analyses would be limited to those structures remaining in the disease group. An initial affine average atlas was estimated by means of voxel-wise averaging of all intensity- and spatial-normalized Nor group images. Next, an iterative process was used to estimate the template, including three stages for each iteration: non-linear registration of the affine-aligned images to the current estimated template; computing the bi-invariant mean φ̄ = exp(v̄) (Arsigny, 2006) of all warpings φi = exp(vi), and finally image intensities are averaged after subtracting the mean warping T = 1/NΣiIi(φi ◦ exp(−v̄)). Convergence is obtained after a few (typically less than 5) iterations.
As the particular values of the registration parameters have a strong impact in the final registration result, we estimated the control group atlas using a set of values of the parameters {α, σ} defined in Eq. (3). The average template better representing the anatomical details of the Nor group was selected for all subsequent analysis using visual criteria.
Brain atrophy statistical maps
To quantify the spatial distribution of brain atrophy3 in MCI and AD groups compared to the Nor group, the template was non-linearly registered to all individual brains (N = 120). The Jacobian map shows the spatial distribution of the Jacobian matrix determinant of the mapping and reflects the local brain volume change relative to the template. These Jacobian maps share a common anatomical coordinate system. Hypothesis testing was performed at each voxel to assess mean difference between patient groups. Voxel-wise two sample Student’s t-test with unequal variance on the log of Jacobian determinants was used. The log transformation helps to make the distribution of Jacobian determinants closer to a Gaussian distribution, which is the main assumption for the statistical test. This spatial distribution is denoted here as brain atrophy statistical map.
Regression maps between brain atrophy and clinical measurements
Any quantitative measure or surrogate marker estimated from MRI, such as local brain atrophy, has greater value if it can be shown to correlate with established measures of cognitive or clinical decline, or with future outcome measures. At each voxel, linear regressions were assessed between the log Jacobian determinant from all subjects (N = 120) and some clinical variables. The spatial distribution of the relations between local brain atrophy and clinical variables may provide valuable information to interpret the clinical effect of brain atrophy.
Informative regression parameters or features potentially useful for statistical maps are regression strength, usually quantified by the correlation coefficient r, regression significance, typically measured as a p-value, and the regression coefficients. All these measures were explored by means of statistical maps using parametric techniques. Voxel-wise regression F-test was used to assess significance of the linear model.
Student’s t-statistic supra-threshold volume (STV) plots
While in probability theory and statistics, the definition of cumulative distribution function (CDF) involves integration of a probability density function, in some recent neuroimaging studies (Leow et al., 2007; Lepore et al., 2008; Hua et al., 2008a) CDF has been used to quantify the number of voxels from a statistical map that achieve a significance level p. In these works, CDF plots were used to compare the statistical power of detecting significant effects using different experimental conditions or even different methods in TBM studies.
A small variant is proposed here: instead of p-values, the Student’s t-statistic is used. The first advantage of using t-statistic is that sign information (either atrophy or expansion) is preserved. The second one comes from the fact that while p-values can be estimated using several methods, either parametric or non-parametric, providing different results, Student’s t-statistic is a much simpler measurement. Therefore, supra-threshold volume (STV) plots illustrate the number of voxels in a statistical map having a Student’s t-statistic larger than a given t-threshold.
Correction for multiple comparisons
In order to correct for multiple comparisons false positive rate must be controlled. There are several false positive measures in the multiple testing problem. The standard measure is the familywise error rate (FWE) which quantifies the probability of observing at least one false positive (Hochberg and Tamhane, 1987; Nichols and Hayasaka, 2003). False discovery rate (FDR), defined as the expected fraction of false positives under the null hypothesis, was proposed later as a less conservative measure than FWE (Benjamini and Yosef, 1995). In this work both FWE- and FDR-based methods were used.
An omnibus test in order to control FDR was used as in previous neuroimaging studies (Chiang et al., 2007b; Hua et al., 2008a; Lepore et al., 2008; Leow et al., 2009). The null distribution was built using random permutations of the diagnostic labels. The number of voxels with larger significance than a p-threshold was computed in the real experiment and in the random assignments. The overall p-value for the significance of the map was obtained as the proportion of events with larger number of voxels for the randomized maps than for the original labeling.
A different alternative is to control FWE. Strong control of the FWE requires that false positives are controlled for each voxel in the statistical map where the null hypothesis holds, allowing localization of the particular significant voxels. This localization is essential to neuroimaging. FWE is usually analyzed by means of the distribution of the maximum statistic (Nichols and Hayasaka, 2003). In this work random permutations were used to empirically estimate the distribution of the maximum statistic. The 100(1 − p)-th percentile of this distribution defines a threshold tp for the statistical map that controls FWE at a level p.
Region of interest statistical analysis
In order to summarize the statistical map information from the voxel level to the ROI level, a scalar descriptor of the ROI is often computed. Many authors use the average Jacobian determinant which is a feature with a very intuitive interpretation: relative volume change of the ROI. The subsequent statistical analysis can be performed with univariate hypothesis testing. The results from this analysis could be directly compared with a rich list of manual volumetry studies performed on AD/MCI neuroimaging studies (Apostolova and Thompson, 2008). The main difference between both approaches is the consideration of either automatic or manual methods. In this work we used the average Jacobian determinant as ROI feature and statistical group analysis was performed by means of Student’s t-test.
Several subcortical regions of interest (ROIs) were automatically delineated at the template: hippocampus, amygdala, caudate nucleus, thalamus, putamen, pallidum and nucleus accumbens. These subcortical nuclei were segmented using the tool FIRST (Patenaude, 2007) from FSL package (Smith et al., 2004). Brain extraction tool, also from FSL package was also used in order to define a whole brain mask. All segmentations were visually checked. Only the brain mask was manually edited.
Results
Unbiased template
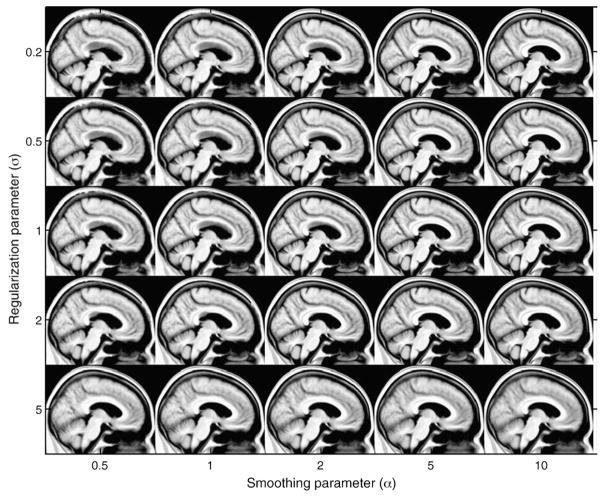
A wide range of different unbiased templates from the control group images were obtained using different values of the registration parameters {α, σ} in Eq. (3) that define the amount of smoothness and the balance between intensity matching and regularization respectively. Fig. 1 illustrates a sagittal view of the Nor group template estimated using the following values of the registration parameters α = [0.5, 1, 2, 5, 10] and σ = [0.2, 0.5, 1, 2, 5].
Fig. 1.
Illustration of sagittal views of the unbiased template of the Nor group with different values of the registration parameters {α, σ}.
Large values of the regularization parameter, i.e., σ = 5, produce an important blur in the templates for all values of the smoothing parameter α. On the other hand, unrealistic structures can be seen in most of the templates using α≤2 (see corpus callosum–lateral ventricle boundary). A possible reason can be that small values of the smoothing parameter α yield many local minima in the energy function to be minimized by the registration algorithm.
The values of the registration parameters in the interval {α = [5, 10], σ = [1, 2]} provide a good trade-off between regularization and smoothing. We visually checked that these templates preserve most of the anatomical details of the normal brain anatomy. For the rest of the study, the template was chosen as the one obtained with the values {α = 5, σ = 1}.
Student’s t-statistic STV plots
In order to compute non-rigid registration from the template to all brain images, the range of values of the registration parameters {α, σ} were slightly adjusted according to the results shown in Fig. 1. The value of σ = 5 was disregarded because the corresponding template did not show enough anatomical detail due to poor image matching; additionally a larger value of the smoothness parameter was considered. The new set of values of the registration parameters were α = [0.5, 1, 2, 5, 10, 20] and σ = [0.2, 0.5, 1, 2].
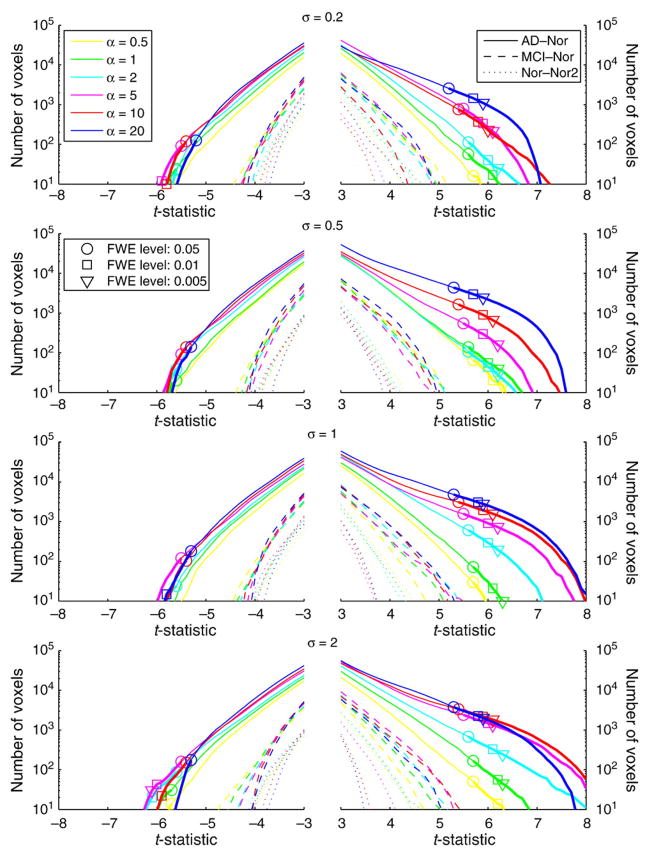
The STV curves of the Student’s t-statistic in Fig. 2 illustrate the sensitivity to detect significant brain volume changes between AD–Nor and MCI–Nor groups when using different values of the registration parameters {α, σ}. The STV curves corresponding to the null distribution were also computed comparing the two independent normal groups (Nor–Nor2). As only large values of the t-statistic are of interest, either positive for atrophy or negative for expansion, the horizontal axis shows values |t|≥3.
Fig. 2.
STV plot of the Student’s t-statistic in the brain mask for different values of the registration parameters {α, σ}. For each curve, there are marks showing the tp-threshold controlling FWE at level p = [0.05, 0.01, 0.005] (horizontal axis) as well as the number of voxels in the statistical map where t>tp-threshold (vertical axis).
An important asymmetry between atrophy and expansion can be observed in Fig. 2. For large enough values of the smoothing parameter α, the number of voxels with significant atrophy is larger than for expansion with the same significance level.
Most of the STV curves for AD–Nor group comparison show an increasing sensitivity to detect brain volume changes when increasing the value of the smoothing parameter α. The values of the registration parameters yielding voxels with larger t-statistic are {α = [5, 10], σ = 2}.
For each curve, a random permutation test with 10,000 permutations was performed to estimate the tp-threshold that controls FWE with significance level p. The values of tp are illustrated in Fig. 2 for p = [0.05, 0.01, 0.005]. All STV curves of the AD–Nor group comparison showed FWE-corrected significant voxels at level p = 0.05.
The optimal pattern for a STV curve would be the one that maximizes the number of voxels with higher significance, i.e. larger values of |t|. As the regularization is an extra penalty term to ensure smoothness of the mapping, a reasonable criterion could be to select the lowest value of α among the values that achieve a similar pattern of the STV curve. Accordingly, the values of the registration parameters {α = [5, 10], σ = 2} would be a good choice.
Brain atrophy statistical maps
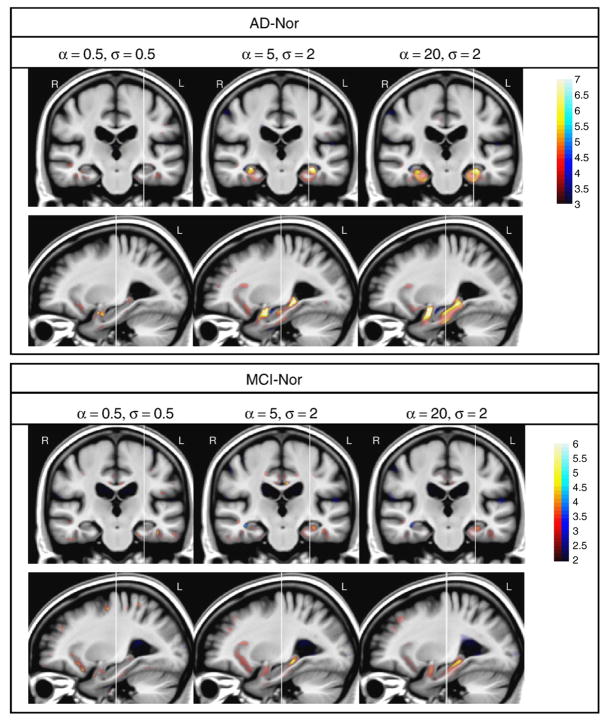
In order to illustrate the effect of using different values of the registration parameters in the spatial distribution of the brain atrophy, three sets of values were selected to represent different conditions: low-level smoothing with small regularization {α = 0.5, σ = 0.5}, large smoothing with large regularization {α = 20, σ = 2} and a point with intermediate smoothing {α = 5, σ = 2}. These working conditions are a representative sample of the different performance of STV curves illustrated in Fig. 2. Student’s t-statistic maps are shown in Fig. 3.
Fig. 3.
One coronal and sagittal view of brain atrophy statistical maps of AD–Nor and MCI–Nor groups with different values of {α, σ}. Color-bar values denote Student’s t-statistic. Red/blue color denotes atrophy/expansion respectively. Note that different color map scales are used in AD–Nor and MCI–Nor comparisons. Vertical lines define slice locations.
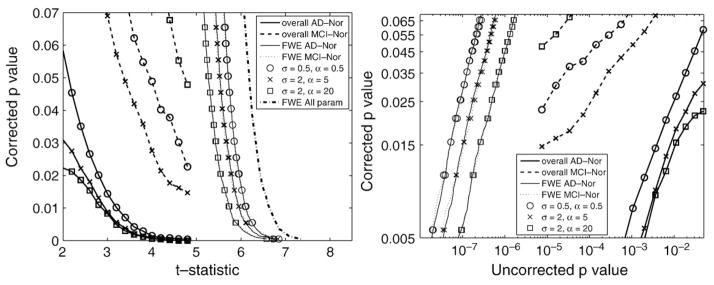
Assessment of statistical significance corrected for multiple comparisons is required in order to compare and to give an interpretation to Student’s t-maps. For each value of the registration parameters {α, σ}, 10,000 random permutations were used to correct for multiple comparisons with FWE- and FDR-based methods. Fig. 4 illustrates the corrected p-values for the three set of values of the registration parameters shown in Fig. 3. As statistical maps are typically shown with either t- or uncorrected p-value maps, two panels were used to illustrate the dependence of the corrected p-values on both measures. This information is redundant due to the known mapping between t-statistic and uncorrected p-values, but it may be helpful for comparison purposes. Note that while a t-threshold is used to control FWE, uncorrected p-value thresholds are used to estimate the overall significance.
Fig. 4.
Illustration of corrected p-value (either FWE p or overall p) versus Student’s t-statistic (left) and uncorrected p-value (right) for several values of the registration parameters {α, σ}. FWE p-values when controlling multiple comparisons taking into account the complete set of parameters are also shown in the left panel (FWE all param).
Using different values of the registration parameters {α, σ} provide atrophy maps with different amount of spatial correlation, and therefore the severity of the correction for multiple comparison will change. However, the values of the t-threshold tp controlling for FWE at level p for all values of {α, σ} differ in less than 0.5 units (see Figs. 2 and 4). This difference is difficult to appreciate in the Student’s t-statistic maps in Fig. 3.
Due to the fact that several values of the registration parameters were explored, an additional correction for multiple comparisons can be performed. For strong control of FWE, the distribution of the maximum statistic under the null hypothesis must be estimated. Accordingly, the maximum is computed not only across the voxels but also across the whole set of parameters {α, σ}. The mapping between t-threshold and this corrected p-value which takes into account the whole set of comparisons is also shown in the left panel of Fig. 4.
Brain atrophy statistical maps are strongly influenced by the values of the registration parameters {α, σ} used during the estimation of the warping between each subject and the template. In general, larger regions with significant differences between groups are obtained for larger values of the smoothing parameter α. However, too large values of α may produce smoothed statistical maps. For example, the statistical maps of the intermediate point {α = 5, σ = 2} in Fig. 3 show regions with sharp boundaries in agreement with anatomical structures affected by dementia, while the corresponding maps when using {α = 20, σ = 2} are blurred. See for example the boundaries of the parahippocampal gyrus in the AD–Nor comparison. Other structures with significant atrophy, such as the frontal part of the insula, are better represented when using {α = 5, σ = 2} than {α = 20, σ = 2}. When comparing AD–Nor versus MCI–Nor patient groups, AD group showed larger areas with stronger significance affected by brain atrophy.
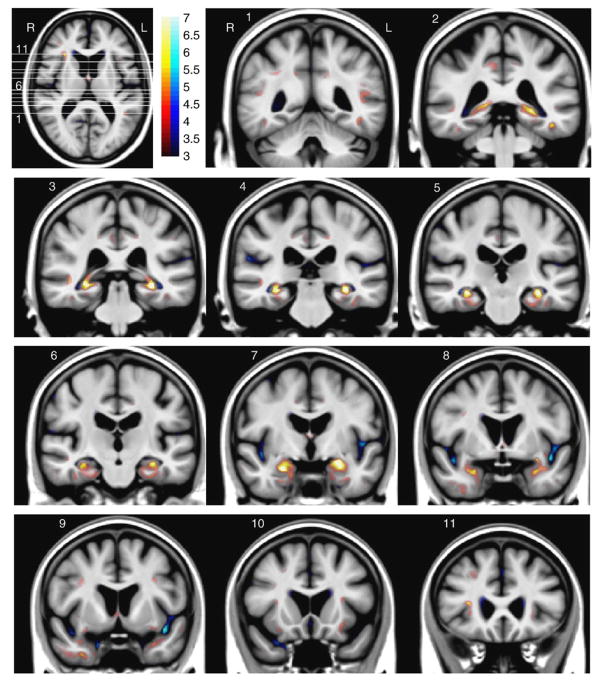
Fig. 5 shows in more detail the AD–Nor brain atrophy map for the intermediate point, i.e., the values of the registration parameters are {α = 5, σ = 2}. The following brain structures showed atrophy with a strong significance: left (see slice 1) and right (slices 3–4) superior temporal sulcus; bilateral posterior part of the cingulate gyrus (precuneus region) at slices 1–5; bilateral temporo-occipital sulcus at slices 1–2, with larger significance at the left side; bilateral hippocampus at slices 2–6, mainly affecting subiculum and CA1 regions; bilateral entorhinal cortex and parahippocampal gyrus at slices 4–7; bilateral amygdala at slice 7; temporal pole, more pronounced at right side (slice 9); anterior part of the right insula at slice 11 and axial slice, with a lower significance at the left insula (slice 10).
Fig. 5.
AD–Nor brain atrophy statistical map with registration parameter values {α = 5, σ = 2}. The white lines in the axial slice specify slice locations of the coronal views. Color-bar values denote Student’s t-statistic (and significance quantified as −log10 p). Red/blue color denotes atrophy/expansion respectively.
Regression analysis maps
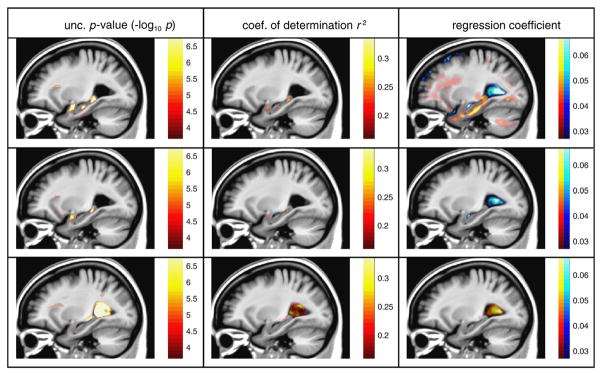
Voxel-wise linear regression analysis was performed with the following clinical variables: MMSEbaseline, MMSE12month and age. The interest here is not to discuss deeply the clinical interpretation of the relationship between brain atrophy and clinical measurements, but to illustrate the performance of the regression maps obtained with SVF diffeomorphic registration. Fig. 6 shows the spatial distribution of some regression features, such as the coefficient of determination r2, regression significance (uncorrected p-value) and regression coefficient. These statistical maps were obtained with registration parameters {α = 5, σ = 2}. It can be seen that Jacobian determinants at the hippocampus and amygdala showed a positive (right panel in Fig. 6) and significant (left panel in Fig. 6) relation with MMSEbaseline, because smaller values of the Jacobian determinants were related to lower MMSE scores. Note that the p-value regression map with MMSEbaseline is similar to the AD–Nor atrophy statistical map in Fig. 3. This result was expected because the clinical variable MMSEbaseline is closely related to the diagnostic label that defines patient groups. It can be noted that the atrophy-age regression maps have a completely different pattern: the most significant correlation was found in the lateral ventricles, which was positive, i.e. an increase in age was linearly related to expansion of the ventricles. In contrast, the regions showing a stronger linear relation between brain atrophy and cognitive status, either MMSEbaseline or MMSE12month, were located at structures known to be affected by dementia, such as hippocampus and amygdala.
Fig. 6.
A sagittal view of the statistical maps of regression between log Jacobian values and the following variables: MMSEbaseline (top), MMSE12month (middle) and age (bottom). Red/blue color denotes positive/negative values.
Region of interest analysis
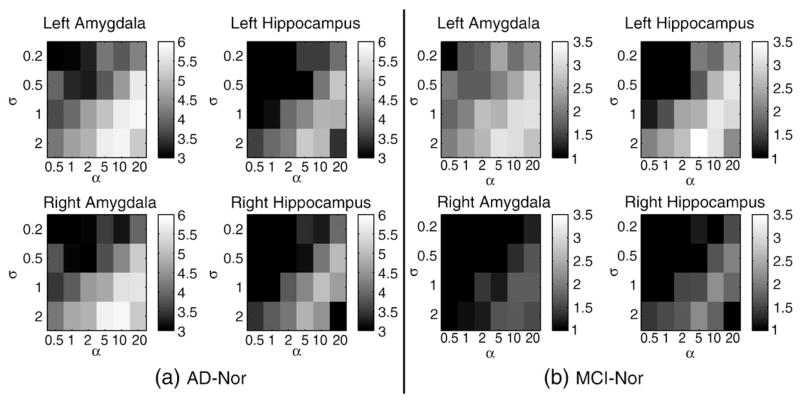
In order to assess statistical differences in the volume of subcortical regions across patient groups, univariate hypothesis testing was performed on the ROI-average Jacobian determinant of the mappings. Among the analyzed structures, only amygdalae and hippocampi presented significant volume differences, both in AD–Nor and MCI–Nor group comparisons. Fig. 7 shows the values of the Student’s t-statistic for the whole set of values of the registration parameters {α, σ}. It can be noted that the magnitude of the t-statistic in the ROI is smaller than the voxel-wise brain statistical maps due to the spatial averaging performed in the ROI analysis, especially at those structures with a heterogeneous atrophy. In our case, while the atrophy distribution at the amygdala was roughly homogeneous, an important heterogeneity was found in the hippocampus. Again, a good candidate of the registration parameter values is {α = 5, σ = 2} because it yields large differences between patient groups.
Fig. 7.
Student’s t-statistic on volume difference between patient groups using different values of the registration parameters {α, σ}. The ROIs are left/right amygdala and hippocampus.
Discussion
Two main contributions can be highlighted from this study. First, stationary velocity field (SVF) diffeomorphic registration seems to be an appropriate method for TBM studies on Alzheimer’s disease patients for the following reasons: it allows large deformations while preserving smoothness of the mapping, the computational requirements are not very high (typical computation time between 1 h and 2 h in a 64-bit 2.33 GHz processor for an image volume of 220×220×220) and more importantly because it provides brain atrophy maps with excellent spatial resolution. The second contribution is a thorough description of the effects of using different values of non-rigid registration parameters at several stages of a TBM study: template estimation, brain atrophy statistical maps and ROI analysis.
Selection of registration parameters
Even though the idea of exploring the values of the registration parameters is very old and recognized by many authors, the piece of information presented here is relevant because it provides criteria to select reasonable values. In this work we only explored two parameters: the coefficient α that specifies smoothness properties of the regularizer (in particular it penalizes up to second-order derivatives of the velocity field), and the relative weight 1/σ2 between image matching and regularization (see Eq. (3)). We illustrated the effect of varying these tuning parameters on the two most important stages of a TBM study: the template estimation and the statistical analysis of the warpings. In our experiments the parameter selection was performed in two stages. First, a reasonable template was visually selected after exploring tuning parameters. Secondly, statistical analysis for different values of the registration parameters was performed using a fixed template. We found that the effect of registration parameters in the performance of statistical analysis is much stronger than in the template estimation. Look for example at the differences in STV curves for parameter values {α = 5, σ = 2} and {α = 5, σ = 0.5} (Fig. 2) and ROI analysis (Fig. 7) compared to the small differences between the corresponding templates (Fig. 1).
Interestingly, the parameter values, {α = [5, 10], σ = 2}, obtained roughly the best performance under most criteria: its corresponding template showed sharp details of the brain anatomy and does not contain artificial structures (see Fig. 1); the STV curves for these values of the registration parameters showed the largest number of voxels with highest magnitude of t-statistic (see Fig. 2) and a very low rate of volume change detections when comparing the two independent normal groups (see Fig. 2); the brain atrophy statistical maps when comparing AD–Nor and MCI–Nor groups with {α = 5, σ = 2} showed significant regions with anatomically-defined boundaries and located at structures known to be affected by dementia (see Figs. 3–5); the ROI analysis, which can be interpreted as a volumetry analysis where delineation of the region is automatically performed with an atlas-based segmentation approach, showed that the same set of parameter values is a good choice for maximizing the statistical significance of volume difference of the hippocampus and amygdala between patient groups (see Fig. 7).
Regarding the selection of the template, future studies will consider quantitative measures for performance evaluation. For example, a common performance measure of the template is the variance (Allassonniere et al., 2007), i.e. distance between observed images and the template.
STV curves
Previous studies have used CDF plots of the uncorrected p-value in linear scale in order to assess statistical power for group analysis in TBM studies (Chiang et al., 2007a; Leow et al., 2007; Hua et al., 2008a; Leow et al., 2009). Log-scale representation has been used to focus on the most significant p-values (Ridgway et al., 2008). Taking into account that at distribution’s tail there is an almost linear relationship between t-statistic and log(p), the Student’s t-statistic STV plot is roughly equivalent to a CDF plot in log scale, but with the additional advantage that the atrophy/expansion information is preserved.
In most of the STV curves the sensitivity to detect volume changes in the AD–Nor group comparison increases with the smoothing and regularization parameters, α and σ respectively (see Fig. 2). However, in the curve with the largest values of regularization and smoothing parameters there is an important reduction of the number of voxels with largest t-statistic. This results shows that too much spatial correlation in the warpings degrades the sensitivity.
Regarding to the sign information in the STV curves, it can be noted from Fig. 2 that brain atrophy regions are larger and present higher significance than expansion regions for large enough values of the smoothing parameter α. This asymmetry is more pronounced in the AD–Nor group comparison but also visible when comparing MCI–Nor groups. This result is in agreement with the fact that the main reported sign of AD observed on MRI images is brain tissue atrophy of particular structures, starting at the temporal lobes.
Brain atrophy statistical maps
Comparing to previous whole brain morphometry studies, including voxel-based morphometry and TBM (Apostolova and Thompson, 2008, and references therein), the statistical maps illustrated in this work showed a much higher spatial resolution. In particular, when comparing AD–Nor groups, the following regions showed significant atrophy bilaterally: superior temporal sulcus, posterior part of the cingulate gyrus (precuneus region), temporo-occipital sulcus, hippocampus mainly affecting subiculum and CA1 regions, entorhinal cortex and parahippocampal gyrus, amygdala, the temporal pole, and the anterior part of the insula. When comparing MCI and normal groups, the regions with significant brain atrophy were smaller than in the AD case, but most of them presented again sharp anatomical boundaries of structures known to be affected by the dementia (Braak and Braak, 1995).
In our opinion, a good criterion for selecting the values of the registration parameters is the anatomical resolution of the brain atrophy maps. While the anatomical knowledge of pathology-induced changes in some brain disorders is relatively small, AD pathology is well-known to affect several specific structures (Braak and Braak, 1995). STV (and CDF) curves are compact descriptions of a brain atrophy map where the anatomical information is lost. Therefore they are not suitable for using such a priori information, unless the STV curve is computed within a pathology-related region.
Regression analysis
Regression analysis allowed to find linear relations between brain atrophy and clinical measurements. For example, brain tissue atrophy of elderly normal subjects, i.e. due to normal aging, is a global process affecting many different structures of the brain. In this case the atrophy is typically manifested as a lateral ventricle expansion because it is a compensatory effect while the tissue atrophy has a much disperse spatial distribution. Accordingly, the regions with the largest significance in the atrophy-age regression map were at the lateral ventricles. In contrast, AD manifests as brain tissue atrophy at specific structures in a known time-course, starting at the medial temporal lobe. Consequently, regression maps with a clinical variable of cognitive status showed that there was a significant linear relation between brain atrophy of hippocampus and amygdala with current cognitive status, i.e., MMSEbaseline and also even with future cognitive status, MMSE12month. This latter behavior is in agreement with previous hypothesis considering that brain atrophy could be used as an early marker of cognitive decline (Davatzikos et al., 2008).
Registration methods for TBM studies
Non-rigid registration is one of the key techniques in a TBM study and aims at defining anatomical correspondences between different brains. The strategies used to ensure the smoothness of the mapping by most of the registration methods belonging to the small deformation paradigm are based on either a parametric characterization of the mapping (Good et al., 2001; Studholme et al., 2004) or regularization of the displacement field (Thirion, 1998; Modersitzki, 2004; Hua et al., 2008a). In both cases the spatial frequency of the mapping is smoothed or band-limited, introducing a lower bound of the spatial resolution in a TBM study. In contrast, the regularization of the registration methods belonging to the large deformation paradigm is usually achieved by smoothing the velocity field instead of using an explicit smoothing of the mapping. As a consequence, there is no explicit bound of the spatial resolution of the mapping apart from the spatial sampling of the images.
In order to illustrate the effect of the values of the registration parameters in other registration methods we performed the same analysis using diffeomorphic demons (Vercauteren et al., 2007, 2009. We selected this method because it is available online,4 it allows large deformations while preserving topology, and at the same time it is based on a quite different strategy for regularization compared to SVF. Two smoothing kernels need to be defined in diffeomorphic demons: kdiff and kfluid, which are governed by the scale parameters s and g, respectively. The following set of the parameter values was used, s = [0.5, 1, 2, 4] and g = [0.5, 1, 2, 4, 6, 8], where a wide range of performances is observed with an ‘optimal’ STV curve inside the interval.
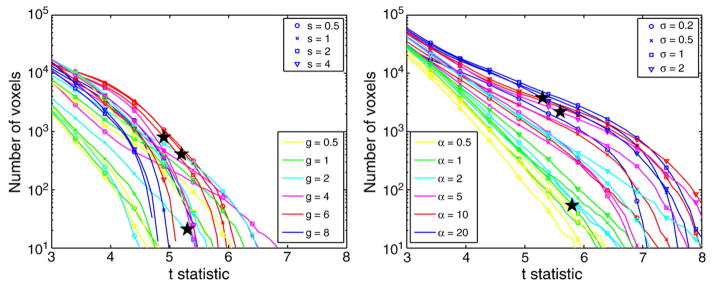
Fig. 8 illustrates the STV curves corresponding to brain atrophy for both registration methods, diffeomorphic demons and SVF diffeomorphic registration, when comparing AD and Nor groups. It is clearly shown that the number of voxels and the significance level strongly depend on the values of the registration parameters for both methods. Likewise in the SVF registration method, extreme values (either too small or large) of the diffeomorphic demons registration parameters produced STV curves far from the ‘optimal’ pattern. Even though SVF diffeomorphic registration obtained a larger sensitivity than diffeomorphic demons for detecting statistical differences between Nor and AD groups, one should be cautious before extrapolating this behavior to other performance measures and application domains, such as atlas-based segmentation, and even on a different set of images.
Fig. 8.
STV curves of the Student’s t-statistic in the brain mask for different values of the registration parameters for diffeomorphic demons (left) and SVF diffeomorphic (right) registration methods. Only brain atrophy for the Nor–AD group comparison is shown. The star marks illustrate the tp-threshold controlling FWE0.05 for a few selected values of the parameters corresponding to the brain atrophy statistical maps shown in Figs. 3 and 9.
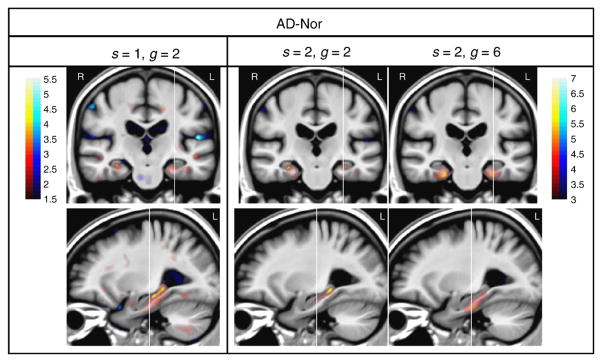
A few examples of the brain atrophy statistical maps obtained with diffeomorphic demons are shown in Fig. 9; they can be directly compared with the results obtained with SVF diffeomorphic registration (see Fig. 3). The brain atrophy statistical map with parameter values {s = 2, g = 6} lacks anatomical details probably due to a high level of smoothing. In contrast, the parameter values {s = 1, g = 2} yielded a map with higher spatial resolution but with a much lower significance level (note the different scale of the color map). The intermediate point {s = 2, g = 2} shows a compromise between resolution and significance.
Fig. 9.
One coronal and sagittal view of brain atrophy statistical maps of AD–Nor with different values of {s, g} in diffeomorphic demons. Color-bar values denote Student’s t-statistic. Red/blue color denotes atrophy/expansion respectively. Note that a different color map scale is used in the left column.
Recent TBM studies on ADNI data
In two previous cross-sectional TBM studies on ADNI data (Hua et al., 2008a,b) with population size N = 120 and 676 subjects respectively as well as in a longitudinal study with 100 subjects (Leow et al., 2009), the brain atrophy statistical maps had a poor spatial resolution compared with their template. Brain atrophy was found at regions without anatomically-driven boundaries providing larger volumes of brain atrophy at white matter tissue than at gray matter. Moreover, the tissue (gray and white matter) close to CSF showed Jacobian determinants larger than one. In our opinion this observed tissue expansion is mainly artificial due to the limited spatial resolution of the non-rigid registration method as pointed out in Hua et al. (2008a,b) and Leow et al. (2009). In contrast, in this work SVF diffeomorphic registration yielded brain atrophy statistical maps with significant regions in gray matter tissue delimited by sharp anatomical boundaries in the same data set as in Hua et al. (2008a). For example, entorhinal cortex and parahippocampal gyrus showed a very significant atrophy in Figs. 3–5. These thin cortical regions are especially relevant because they are affected at the early stages of the disease (Braak and Braak, 1995).
In this work a small subset of baseline images, N = 120, from ADNI database was used for two main reasons: to be able to make a fair and more direct comparison with a recent TBM study based on a nonlinear elastic registration method (Hua et al., 2008a) as well as to allow a feasible computation time when exploring several values of the registration parameters. Ongoing work in our group is focused on a TBM study with the complete data set from ADNI database with values of registration parameters learnt from this work. We hypothesize that with a larger data set the brain atrophy maps obtained with SVF diffeomorphic registration will show an improved anatomical resolution of the structures affected by atrophy.
Acknowledgments
This work was partially funded by research grants TEC2006-13966-C03-02 and TEC2009-14587-C03-01 from CICYT, TSI-020110-2009-362 from MITC and PI100/08 from DGA, Spain.
We thank X. Hua for providing the list of subjects analyzed in Hua et al. (2008a) in order to make an easier performance comparison. We also thank to the reviewers who helped to improve the manuscript.
Data collection and sharing for this project were funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI; Principal Investigator: Michael Weiner; NIH grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and through generous contributions from the following: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck and Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, Alzheimer’s Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging, with participation from the U.S. Food and Drug Administration. Industry partnerships are coordinated through the Foundation for the National Institutes of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory of NeuroImaging at the University of California, Los Angeles.
Footnotes
Diffeomorphic demons is available online at http://www.insight-journal.org/browse/publication/154.
As cross-sectional data is used in this work, brain atrophy/expansion refers to the volume change factor compared to the normal group, and not the usual concept of volume change along time.
References
- Allassonniere S, Amit Y, Trouve A. Towards a coherent statistical framework for dense deformable template estimation. J R Stat Soc B Stat Methodol. 2007;69:3–29. [Google Scholar]
- Apostolova LG, Thompson PM. Mapping progressive brain structural changes in early Alzheimer’s disease and mild cognitive impairment. Neuropsychologia. 2008;46(6):1597–1612. doi: 10.1016/j.neuropsychologia.2007.10.026. ( http://www.dx.doi.org/10.1016/j.neuropsychologia.2007.10.026) [DOI] [PMC free article] [PubMed]
- Arsigny V. PhD Thesis, École polytechnique. 2006. Processing data in Lie groups: An algebraic approach. application to non-linear registration and diffusion tensor MRI. [Google Scholar]
- Arsigny V, Commowick O, Pennec X, Ayache N. A log-Euclidean framework for statistics on diffeomorphisms. In: Larsen R, MN, Sporring J, editors. LNCS; Proc. MICCAI’06; Springer-Verlag; 2006. pp. 924–931. [DOI] [PubMed] [Google Scholar]
- Ashburner J. A fast diffeomorphic image registration algorithm. NeuroImage. 2007;38(1):95–113. doi: 10.1016/j.neuroimage.2007.07.007. (10/15) [DOI] [PubMed] [Google Scholar]
- Barnes J, Bartlett JW, van de Pol LA, Loy CT, Scahill RI, Frost C, Thompson P, Fox NC. A meta-analysis of hippocampal atrophy rates in Alzheimer’s disease. Neurobiol Aging. 2009 Nov;30(11):1711–1723. doi: 10.1016/j.neurobiolaging.2008.01.010. http://www.dx.doi.org/10.1016/j.neurobiolaging.2008.01.010. [DOI] [PMC free article] [PubMed]
- Beg MF, Miller MI, Trouvé A, Younes L. Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int J Comput Vis. 2005;61 (2):139–157. [Google Scholar]
- Benjamini Y, Yosef H. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Bossa M, Zacur E, Olmos S. Algorithms for computing the group exponential of diffeomorphisms: performance evaluation. MMBIA Workshop at Comp Vision Pattern Recog; 2008. Jun, pp. 1–8. [Google Scholar]
- Braak H, Braak E. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol Aging. 1995;16(3):271–278. doi: 10.1016/0197-4580(95)00021-6. (discussion 278–84) [DOI] [PubMed] [Google Scholar]
- Brun CC, Leporé N, Pennec X, Lee AD, Barysheva M, Madsen SK, Avedissian C, Chou Y-Y, de Zubicaray GI, McMahon KL, Wright MJ, Toga AW, Thompson PM. Mapping the regional influence of genetics on brain structure variability – a tensor-based morphometry study. Neuroimage. 2009 Oct;48(1):37–49. doi: 10.1016/j.neuroimage.2009.05.022. http://www.dx.doi.org/10.1016/j.neuroimage.2009.05.022. [DOI] [PMC free article] [PubMed]
- Chiang M-C, Dutton RA, Hayashi KM, Lopez OL, Aizenstein HJ, Toga AW, Becker JT, Thompson PM. 3D pattern of brain atrophy in HIV/AIDS visualized using tensor-based morphometry. Neuroimage. 2007a;34(1):44–60. doi: 10.1016/j.neuroimage.2006.08.030. ( http://www.dx.doi.org/10.1016/j.neuroimage.2006.08.030) [DOI] [PMC free article] [PubMed]
- Chiang M-C, Reiss AL, Lee AD, Bellugi U, Galaburda AM, Korenberg JR, Mills DL, Toga AW, Thompson PM. 3D pattern of brain abnormalities in Williams syndrome visualized using tensor-based morphometry. Neuroimage. 2007b;36(4):1096–1109. doi: 10.1016/j.neuroimage.2007.04.024. ( http://www.dx.doi.org/10.1016/j.neuroimage.2007.04.024) [DOI] [PMC free article] [PubMed]
- Christensen G, Rabbitt R, Miller M. Deformable templates using large deformation kinematics. IEEE Trans Image Process. 1996;5 (10):1435–1447. doi: 10.1109/83.536892. [DOI] [PubMed] [Google Scholar]
- Csernansky JG, Wang L, Joshi S, Miller JP, Gado M, Kido D, McKeel D, Morris JC, Miller MI. Early DAT is distinguished from aging by high-dimensional mapping of the hippocampus. Dementia of the Alzheimer type. Neurology. 2000;55 (11):1636–1643. doi: 10.1212/wnl.55.11.1636. [DOI] [PubMed] [Google Scholar]
- Csernansky JG, Wang L, Joshi SC, Ratnanather JT, Miller MI. Computational anatomy and neuropsychiatric disease: probabilistic assessment of variation and statistical inference of group difference, hemispheric asymmetry, and time-dependent change. Neuroimage. 2004;23(Suppl 1):S56–S68. doi: 10.1016/j.neuroimage.2004.07.025. ( http://www.dx.doi.org/10.1016/j.neuroimage.2004.07.025) [DOI] [PubMed]
- D’Agostino E, Maes F, Vandermeulen D, Suetens P. A viscous fluid model for multimodal non-rigid image registration using mutual information. Med Image Anal. 2003;7 (4):565–575. doi: 10.1016/s1361-8415(03)00039-2. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Fan Y, Wu X, Shen D, Resnick SM. Detection of prodromal Alzheimer’s disease via pattern classification of magnetic resonance imaging. Neurobiol Aging. 2008 Apr;29(4):514–523. doi: 10.1016/j.neurobiolaging.2006.11.010. http://www.dx.doi.org/10.1016/j.neurobiolaging.2006.11.010. [DOI] [PMC free article] [PubMed]
- Dickerson BC, Sperling RA. Functional abnormalities of the medial temporal lobe memory system in mild cognitive impairment and Alzheimer’s disease: insights from functional MRI studies. Neuropsychologia. 2008;46(6):1624–1635. doi: 10.1016/j.neuropsychologia.2007.11.030. ( http://www.dx.doi.org/10.1016/j.neuropsychologia.2007.11.030) [DOI] [PMC free article] [PubMed]
- Du AT, Jahng GH, Hayasaka S, Kramer JH, Rosen HJ, Gorno-Tempini ML, Rankin KP, Miller BL, Weiner MW, Schuff N. Hypoperfusion in frontotemporal dementia and Alzheimer disease by arterial spin labeling MRI. Neurology. 2006 Oct;67(7):1215–1220. doi: 10.1212/01.wnl.0000238163.71349.78. http://www.dx.doi.org/10.1212/01.wnl.0000238163.71349.78. [DOI] [PMC free article] [PubMed]
- Du AT, Schuff N, Amend D, Laakso MP, Hsu YY, Jagust WJ, Yaffe K, Kramer JH, Reed B, Norman D, Chui HC, Weiner MW. Magnetic resonance imaging of the entorhinal cortex and hippocampus in mild cognitive impairment and Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 2001;71 (4):441–447. doi: 10.1136/jnnp.71.4.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchesne S, Caroli A, Geroldi C, Barillot C, Frisoni GB, Collins DL. MRI-based automated computer classification of probable ad versus normal controls. IEEE Trans Med Imaging. 2008 Apr;27(4):509–520. doi: 10.1109/TMI.2007.908685. http://www.dx.doi.org/10.1109/TMI.2007.908685. [DOI] [PubMed]
- Fan Y, Batmanghelich N, Clark CM, Davatzikos C Alzheimer’s Disease Neuroimaging Initiative. Spatial patterns of brain atrophy in MCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. Neuroimage. 2008a Feb;39(4):1731–1743. doi: 10.1016/j.neuroimage.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Resnick SM, Wu X, Davatzikos C. Structural and functional biomarkers of prodromal Alzheimer’s disease: a high-dimensional pattern classification study. Neuroimage. 2008b;41(2):277–285. doi: 10.1016/j.neuroimage.2008.02.043. ( http://www.dx.doi.org/10.1016/j.neuroimage.2008.02.043) [DOI] [PMC free article] [PubMed]
- Frackowiak RSJ. Morphometry. Ch. 36. Academic Press; 2004. Human Brain Function; pp. 707–724. [Google Scholar]
- Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14(1 Pt 1):21–36. doi: 10.1006/nimg.2001.0786. ( http://www.dx.doi.org/10.1006/nimg.2001.0786) [DOI] [PubMed]
- Hager WW, Zhang H. A survey of non-linear conjugate gradient methods. Pac J Optim. 2006;2:35–58. [Google Scholar]
- Hernandez M, Bossa M, Olmos S. Registration of anatomical images using paths of diffeomorphisms parameterized with stationary vector field flows. Int J Comput Vis. 2009;85 (3):291–306. [Google Scholar]
- Hernandez M, Bossa MN, Olmos S. Registration of anatomical images using geodesic paths of diffeomorphisms parameterized with stationary vector fields. MMBIA Workshop at Int Conf Computer Vision; 2007. pp. 1–8. [Google Scholar]
- Hochberg Y, Tamhane A. Multiple Comparison Procedures. John Wiley & Sons; 1987. [Google Scholar]
- Hua X, Lee S, Yanovsky I, Leow AD, Chou Y-Y, Ho AJ, Gutman B, Toga AW, Jack CR, Bernstein MA, Reiman EM, Harvey DJ, Kornak J, Schuff N, Alexander GE, Weiner MW, Thompson PM, Initiative ADN. Optimizing power to track brain degeneration in Alzheimer’s disease and mild cognitive impairment with tensor-based morphometry: an ADNI study of 515 subjects. Neuroimage. 2009 Dec;48(4):668–681. doi: 10.1016/j.neuroimage.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Leow AD, Lee S, Klunder AD, Toga AW, Lepore N, Chou Y-Y, Brun C, Chiang M-C, Barysheva M, Jack CR, Bernstein MA, Britson PJ, Ward CP, Whitwell JL, Borowski B, Fleisher AS, Fox NC, Boyes RG, Barnes J, Harvey D, Kornak J, Schuff N, Boreta L, Alexander GE, Weiner MW, Thompson PM Alzheimer’s Disease Neuroimaging Initiative. 3D characterization of brain atrophy in Alzheimer’s disease and mild cognitive impairment using tensor-based morphometry. Neuroimage. 2008a;41(1):19–34. doi: 10.1016/j.neuroimage.2008.02.010. ( http://www.dx.doi.org/10.1016/j.neuroimage.2008.02.010) [DOI] [PMC free article] [PubMed]
- Hua X, Leow AD, Parikshak N, Lee S, Chiang MC, Toga AW, Jack CR, Weiner MW, Thompson PM Alzheimer’s Disease Neuroimaging Initiative. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer’s disease: an MRI study of 676 AD, MCI, and normal subjects. Neuroimage. 2008b;43 (3):458–469. doi: 10.1016/j.neuroimage.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Petersen RC, Xu YC, O’Brien PC, Smith GE, Ivnik RJ, Boeve BF, Waring SC, Tangalos EG, Kokmen E. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology. 1999;52 (7):1397–1403. doi: 10.1212/wnl.52.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. NeuroImage. 2004;23:151–160. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- Kantarci K, Xu Y, Shiung MM, O’Brien PC, Cha RH, Smith GE, Ivnik RJ, Boeve BF, Edland SD, Kokmen E, Tangalos EG, Petersen RC, Jack CR. Comparative diagnostic utility of different MR modalities in mild cognitive impairment and Alzheimer’s disease. Dement Geriatr Cogn Disord. 2002;14 (4):198–207. doi: 10.1159/000066021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasuski JS, Alexander GE, Horwitz B, Daly EM, Murphy DG, Rapoport SI, Schapiro MB. Volumes of medial temporal lobe structures in patients with Alzheimer’s disease and mild cognitive impairment (and in healthy controls) Biol Psychiatry. 1998 Jan;43(1):60–68. doi: 10.1016/s0006-3223(97)00013-9. [DOI] [PubMed] [Google Scholar]
- Laakso MP, Soininen H, Partanen K, Helkala EL, Hartikainen P, Vainio P, Hallikainen M, Hänninen T, Riekkinen PJ. Volumes of hippocampus, amygdala and frontal lobes in the MRI-based diagnosis of early Alzheimer’s disease: correlation with memory functions. J Neural Trans Park Dis Dement Sect. 1995;9 (1):73–86. doi: 10.1007/BF02252964. [DOI] [PubMed] [Google Scholar]
- Lee AD, Leow AD, Lu A, Reiss AL, Hall S, Chiang M-C, Toga AW, Thompson PM. 3D pattern of brain abnormalities in Fragile X syndrome visualized using tensor-based morphometry. Neuroimage. 2007 Feb;34(3):924–938. doi: 10.1016/j.neuroimage.2006.09.043. http://www.dx.doi.org/10.1016/j.neuroimage.2006.09.043. [DOI] [PMC free article] [PubMed]
- Leow AD, Klunder AD, Jack CR, Toga AW, Dale AM, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Whitwell JL, Borowski BJ, Fleisher AS, Fox NC, Harvey D, Kornak J, Schuff N, Studholme C, Alexander GE, Weiner MW, Thompson PM, Study ADNIPP. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage. 2006;31 (2):627–640. doi: 10.1016/j.neuroimage.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leow AD, Yanovsky I, Chiang M-C, Lee AD, Klunder AD, Lu A, Becker JT, Davis SW, Toga AW, Thompson PM. Statistical properties of Jacobian maps and the realization of unbiased large-deformation nonlinear image registration. IEEE Trans Med Imaging. 2007;26(6):822–832. doi: 10.1109/TMI.2007.892646. ( http://www.dx.doi.org/10.1109/TMI.2007.892646) [DOI] [PubMed]
- Leow AD, Yanovsky I, Parikshak N, Hua X, Lee S, Toga AW, Jack CR, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Borowski B, Shaw LM, Trojanowski JQ, Fleisher AS, Harvey D, Kornak J, Schuff N, Alexander GE, Weiner MW, Thompson PM Alzheimer’s Disease Neuroimaging Initiative. Alzheimer’s disease neuroimaging initiative: a one-year follow up study using tensor-based morphometry correlating degenerative rates, biomarkers and cognition. Neuroimage. 2009;45 (3):645–655. doi: 10.1016/j.neuroimage.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepore N, Brun C, Chou YY, Chiang MC, Dutton RA, Hayashi KM, Luders E, Lopez OL, Aizenstein HJ, Toga AW, Becker JT, Thompson PM. Generalized tensor-based morphometry of HIV/AIDS using multivariate statistics on deformation tensors. IEEE Trans Med Imaging. 2008;27(1):129–141. doi: 10.1109/TMI.2007.906091. http://www.dx.doi.org/10.1109/TMI.2007.906091. [DOI] [PMC free article] [PubMed]
- Lepore N, Brun C, Pennec X, Chou Y-Y, Lopez OL, Aizenstein HJ, Becker JT, Toga AW, Thompson PM. Mean template for tensor-based morphometry using deformation tensors. In: Ayache N, Ourselin S, Maeder A, editors. Proc MICCAI’07: LNCS. Vol. 4792. 2007. pp. 826–833. [DOI] [PubMed] [Google Scholar]
- Lepore N, Brun CA, Chiang M-C, Chou Y-Y, Dutton RA, Hayashi KM, Lopez OL, Aizenstein HJ, Toga AW, Becker JT, Thompson PM. Multivariate statistics of the Jacobian matrices in tensor based morphometry and their application to HIV/AIDS. In: Larsen R, Nielsen M, Sporring J, editors. LNCS; Proc. MICCAI’06; Springer-Verlag; 2006. pp. 191–198. [DOI] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Goualher GL, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond B Biol Sci. 2001 Aug;356(1412):1293–1322. doi: 10.1098/rstb.2001.0915. http://www.dx.doi.org/10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed]
- Misra C, Fan Y, Davatzikos C. Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: results from ADNI. Neuroimage. 2009;44(4):1415–1422. doi: 10.1016/j.neuroimage.2008.10.031. ( http://www.dx.doi.org/10.1016/j.neuroimage.2008.10.031) [DOI] [PMC free article] [PubMed]
- Modersitzki J. Numerical Methods for Image Registration. Oxford University Press; 2004. [Google Scholar]
- Modrego PJ, Fayed N, Pina MA. Conversion from mild cognitive impairment to probable Alzheimer’s disease predicted by brain magnetic resonance spectroscopy. Am J Psychiatry. 2005;162(4):667–675. doi: 10.1176/appi.ajp.162.4.667. ( http://www.dx.doi.org/10.1176/appi.ajp.162.4.667) [DOI] [PubMed]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR, Weiner MW, Thompson PM Alzheimer’s Disease Neuroimaging Initiative. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer’s disease mild cognitive impairment, and elderly controls. Neuroimage. 2008;43 (1):59–68. doi: 10.1016/j.neuroimage.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosconi L. Brain glucose metabolism in the early and specific diagnosis of Alzheimer’s disease. FDG-PET studies in MCI and AD Eur J Nucl Med Mol Imaging. 2005;32(4):486–510. doi: 10.1007/s00259-005-1762-7. ( http://www.dx.doi.org/10.1007/s00259-005-1762-7) [DOI] [PubMed]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, Trojanowski JQ, Toga AW, Beckett L. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin N Am. 2005a Nov;15(4):869–877. xi–xii. doi: 10.1016/j.nic.2005.09.008. http://www.dx.doi.org/10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005b;1(1):55–66. doi: 10.1016/j.jalz.2005.06.003. ( http://www.dx.doi.org/10.1016/j.jalz.2005.06.003) [DOI] [PMC free article] [PubMed]
- Nichols T, Hayasaka S. Controlling the familywise error rate in functional neuroimaging: a comparative review. Stat Methods Med Res. 2003 Oct;12(5):419–446. doi: 10.1191/0962280203sm341ra. [DOI] [PubMed] [Google Scholar]
- Nocedal J, Wright S. Springer Series in Operations Research. Springer-Verlag; New York: 1999. Numerical optimization. [Google Scholar]
- Nordberg A. Amyloid plaque imaging in vivo: current achievement and future prospects. Eur J Nucl Med Mol Imaging. 2008;35(Suppl 1):S46–S50. doi: 10.1007/s00259-007-0700-2. ( http://www.dx.doi.org/10.1007/s00259-007-0700-2) [DOI] [PubMed]
- Patenaude B. PhD thesis. University of Oxford; 2007. Bayesian statistical models of shape and appearance for subcortical brain segmentation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennanen C, Kivipelto M, Tuomainen S, Hartikainen P, Hänninen T, Laakso MP, Hallikainen M, Vanhanen M, Nissinen A, Helkala E-L, Vainio P, Vanninen R, Partanen K, Soininen H. Hippocampus and entorhinal cortex in mild cognitive impairment and early AD. Neurobiol Aging. 2004;25(3):303–310. doi: 10.1016/S0197-4580(03)00084-8. ( http://www.dx.doi.org/10.1016/S0197-4580(03)00084-8) [DOI] [PubMed]
- Petersen RC. Mild cognitive impairment as a diagnostic entity. J Intern Med. 2004;256(3):183–194. doi: 10.1111/j.1365-2796.2004.01388.x. ( http://www.dx.doi.org/10.1111/j.1365-2796.2004.01388.x) [DOI] [PubMed]
- Petersen RC, Doody R, Kurz A, Mohs RC, Morris JC, Rabins PV, Ritchie K, Rossor M, Thal L, Winblad B. Current concepts in mild cognitive impairment. Arch Neurol. 2001;58 (12):1985–1992. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- Qiu A, Fennema-Notestine C, Dale AM, Miller MI Alzheimer’s Disease Neuroimaging Initiative. Regional shape abnormalities in mild cognitive impairment and Alzheimer’s disease. Neuroimage. 2009;45 (3):656–661. doi: 10.1016/j.neuroimage.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querbes O, Aubry F, Pariente J, Lotterie J-A, Démonet J-F, Duret V, Puel M, Berry I, Fort J-C, Celsis P, Initiative ADN. Early diagnosis of Alzheimer’s disease using cortical thickness: impact of cognitive reserve. Brain. 2009 Aug;132(Pt 8):2036–2047. doi: 10.1093/brain/awp105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway G, Whitcher B, Nichols T, Ourselin S, Hill D, Fox N. Longitudinal multivariate tensor- and searchlight-based morphometry using permutation testing. Proc. 14th Org Human Brain Mapping.2008. [Google Scholar]
- Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, Thompson PM, Jack CR, Weiner MW Alzheimer’s Disease Neuroimaging Initiative. MRI of hippocampal volume loss in early Alzheimer’s disease in relation to ApoE genotype and biomarkers. Brain. 2009;132 (Pt 4):1067–1077. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, Bannister PR, Luca MD, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, Stefano ND, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. ( http://www.dx.doi.org/10.1016/j.neuroimage.2004.07.051) [DOI] [PubMed]
- Studholme C, Cardenas V, Blumenfeld R, Schuff N, Rosen HJ, Miller B, Weiner M. Deformation tensor morphometry of semantic dementia with quantitative validation. Neuroimage. 2004 Apr;21(4):1387–1398. doi: 10.1016/j.neuroimage.2003.12.009. http://www.dx.doi.org/10.1016/j.neuroimage.2003.12.009. [DOI] [PubMed]
- Styner M, Gerig G, Lieberman J, Jones D, Weinberger D. Statistical shape analysis of neuroanatomical structures based on medial models. Med Image Anal. 2003;7 (3):207–220. doi: 10.1016/s1361-8415(02)00110-x. [DOI] [PubMed] [Google Scholar]
- Thirion JP. Image matching as a diffusion process: an analogy with Maxwell’s demons. Med Image Anal. 1998;2 (3):243–260. doi: 10.1016/s1361-8415(98)80022-4. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, Dutton RA, Chiang M-C, Leow AD, Sowell ER, Zubicaray GD, Becker JT, Lopez OL, Aizenstein HJ, Toga AW. Tracking Alzheimer’s disease. Ann N Y Acad Sci. 2007;1097:183–214. doi: 10.1196/annals.1379.017. ( http://www.dx.doi.org/10.1196/annals.1379.017) [DOI] [PMC free article] [PubMed]
- Vercauteren T, Pennec X, Perchant A, Ayache N. Non-parametric diffeomorphic image registration with the demons algorithm. In: Ayache N, SO, Maeder A, editors. Proc MICCAI Vol 4792 of LNCS. Vol. 10. Springer-Verlag; Brisbane, Australia: Oct, 2007. pp. 319–326. [DOI] [PubMed] [Google Scholar]
- Vercauteren T, Pennec X, Perchant A, Ayache N. Proc MICCAI’08 Vol 5241 LNCS. Vol. 11. Springer-Verlag; New York, USA: 2008. Symmetric log-domain diffeomorphic registration: a demons-based approach; pp. 754–761. [DOI] [PubMed] [Google Scholar]
- Vercauteren T, Pennec X, Perchant A, Ayache N. Diffeomorphic demons: efficient non-parametric image registration. Neuroimage. 2009 Mar;45(1 Suppl):S61–S72. doi: 10.1016/j.neuroimage.2008.10.040. http://www.dx.doi.org/10.1016/j.neuroimage.2008.10.040. [DOI] [PubMed]
- Wang L, Beg F, Ratnanather T, Ceritoglu C, Younes L, Morris JC, Csernansky JG, Miller MI. Large deformation diffeomorphism and momentum based hippocampal shape discrimination in dementia of the Alzheimer type. IEEE Trans Med Imaging. 2007;26(4):462–470. doi: 10.1109/TMI.2005.853923. ( http://www.dx.doi.org/10.1109/TMI.2005.853923) [DOI] [PMC free article] [PubMed]
- Winblad B, Palmer K, Kivipelto M, Jelic V, Fratiglioni L, Wahlund L-O, Nordberg A, Bäckman L, Albert M, Almkvist O, Arai H, Basun H, Blennow K, de Leon M, DeCarli C, Erkinjuntti T, Giacobini E, Graff C, Hardy J, Jack C, Jorm A, Ritchie K, van Duijn C, Visser P, Petersen RC. Mild cognitive impairment – beyond controversies, towards a consensus: report of the international working group on mild cognitive impairment. J Intern Med. 2004;256(3):240–246. doi: 10.1111/j.1365-2796.2004.01380.x. ( http://www.dx.doi.org/10.1111/j.1365-2796.2004.01380.x) [DOI] [PubMed]