Abstract
Rapid advances in neuroimaging techniques provide great potentials for study of Alzheimer’s disease (AD). Existing findings have shown that AD is closely related to alteration in the functional brain network, i.e., the functional connectivity between different brain regions. In this paper, we propose a method based on sparse inverse covariance estimation (SICE) to identify functional brain connectivity networks from PET data. Our method is able to identify both the connectivity network structure and strength for a large number o f brain regions with small sample sizes. We apply the proposed method to the PET data of AD, mild cognitive impairment (MCI), and normal control (NC) subjects. Compared with NC, AD shows decrease in the amount of inter-region functional connectivity within the temporal lobe especially between the area around hippocampus and other regions and increase in the amount of connectivity within the frontal lobe as well as between the parietal and occipital lobes. Also, AD shows weaker between-lobe connectivity than within-lobe connectivity and weaker between-hemisphere connectivity, compared with NC. In addition to being a method for knowledge discovery about AD, the proposed SICE method can also be used for classifying new subjects, which makes it a suitable approach for novel connectivity-based AD biomarker identification. Our experiments show that the best sensitivity and specificity our method can achieve in AD vs. NC classification are 88% and 88%, respectively.
Keywords: Brain connectivity, Sparse inverse covariance, Alzheimer’s, PET, Biomarker
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disorder characterized by progressive impairment of memory and other cognitive functions. It has been speculated by a number studies and accepted more widely recently that higher cognition results from different brain regions interacting with each other, rather than individual regions working independently (Horwitz, 2003; Delbeuck et al., 2003). This leads to the belief that AD, with major symptoms being dramatic global cognitive decline, may have abnormal functional brain connectivity patterns.
Functional connectivity refers to the coherence of the activities among distinct brain regions (Horwitz, 2003). Some past research in AD has shown that AD brains may have different connectivity patterns from normal brains. For example, functional connectivity is reduced between the hippocampus and other regions of AD brains (Supekar et al., 2008; Wang et al., 2007; Azari et al., 1992; Horwitz et al., 1987; Grady et al., 2001). A key pathological correlate, which may be implicated in hippocampal network disfunction relates to neurofibrillary tangles (NFTs), a hallmark of AD, which causes hippocampal neurodegeneration early in the course of AD pathology. This results from selective affects on specific cortical layers within the hippocampal formation (Hirano and Zimmerman, 1962), which raises the possibility that the functional interaction between the hippocampus and other related brain regions may be disrupted. In addition to changes in hippocampal networks, some studies of early AD and mild cognitive impairment (MCI) have found increased connectivity between the frontal lobe and other brain regions (Gould et al., 2006; Stern, 2006; Becker et al., 1996; Woodard et al., 1998; Saykin et al., 2004; Grady et al., 2003). This has been interpreted by some investigators as a compensatory reallocation or recruitment of cognitive resources (Gould et al., 2006). Since regions in the frontal lobe are typically affected later in the course of the disease, it is argued that an increase in frontal connectivity could help preserve some memory and attention ability in early AD patients (Stern, 2006).
Recent years have witnessed the rapid advancement of neuroimaging technologies, which provides an unprecedented opportunity for brain connectivity research. Based on the brain data acquired by functional neuroimaging techniques such as positron emission tomography (PET) and functional magnetic resonance imaging (fMRI), quite a number of analytic methodologies have been proposed to investigate functional brain connectivity.
Multivariate statistical methods have been used, such as principle component analysis (PCA) (Friston, 1994), PCA-based scaled subprofile model (Alexander and Moeller, 1994), independent component analysis (Calhoun et al., 2001; Calhoun et al., 2003), and partial least squares (McIntosh et al., 1996; Worsley et al., 1997). These methods tend to group brain regions into a few latent components. The brain regions within each component are believed to have strong connectivity, while the connectivity between components is weak. One limitation of these methods is that the latent components are mostly obtained based on statistical modeling consideration, so they may not necessarily correspond to biological entities, causing difficulty in interpretation.
A large body of functional connectivity modeling has been based on correlation analysis (Azari et al., 1992; Horwitz et al., 1987; Supekar et al., 2008; Stam et al., 2007). Correlation analysis captures pairwise information, which may not be able to effectively characterize the interactions of many brain regions working together. To overcome this limitation, partial correlation analysis has been adopted (Salvador et al., 2005a,b; Marrelec et al., 2006, 2007; Hampson et al., 2002). A partial correlation measures the association between two brain regions after factoring out the contribution to the pairwise correlation that might be due to global or third-party effects. Because partial correlations correspond to the off-diagonal entries of the inverse covariance (IC) matrix of the data, estimation of partial correlations is usually achieved by maximum likelihood estimation (MLE) of the IC matrix. A limitation of MLE is that reliable estimation requires the sample size of the data to be substantially larger than the number of brain regions modeled. This condition may be satisfied in studies based on fMRI data, in which the sample size corresponds to the length of the fMRI time series. However, in PET studies, because the samples size is the number of subjects which is usually very limited due to cost or availability constraints, existing partial correlation or IC based research has only been able to focus on a few (around ten) preselected brain regions.
We propose a new method for functional connectivity modeling, called sparse inverse covariance estimation (SICE), also known as Gaussian graphical models or graphical Lasso. This method imposes a “sparsity” constraint on the MLE of an IC matrix, which leads to reliable estimation of the IC with small sample sizes. Here, “small” means that the sample size can be close to or even less than the number of brain regions modeled. Using SICE to model brain connectivity is appropriate because many past studies based on anatomical brain databases have shown that the true brain network is sparse (Hilgetag et al., 2002; Kotter and Stephan, 2003; Sporns et al., 2004). Using sparse models of other kinds for brain connectivity modeling, such as multivariate or vector autoregressions, have been explored in the past, with a focus on time series data such as fMRI and EEG (Chiang et al., 2009; Thompson et al., 2009; Valdes-Sosa et al., 2005). In contrast, the proposal SICE can be used to model cross-sectional data such as PET.
Specifically in this paper, we apply one SICE method, developed by us in a previous paper (Huang et al. 2009), to identify brain connectivity models for AD, MCI, and normal control (NC) subjects based on FDG-PET data. SICE has been recognized as an effective tool for identifying the structure of an IC matrix, i.e., the zero and non-zero entries, but is not recommended to be used for estimating the magnitude of the non-zero entries. Therefore, we use SICE to identify brain connectivity model structures, i.e., existence and non-existence of functional connections between brain regions. Furthermore, we prove a monotone property of SICE, which enables us to develop a quasi-measure for the strength of functional connections. To our best knowledge, our work is among the first ones that utilize SICE and its associated property for functional brain connectivity structure and strength identification in AD studies. In addition, we show how to use the results of SICE to classify new subjects, which makes SICE a potential method for identifying connectivity-based biomarkers for AD. Another unique perspective of our work is that it utilizes PET data. While a majority of existing AD brain connectivity research has been based on fMRI data (Rajapakse and Zhou, 2007; Li et al., 2008; Zheng and Rajapkse, 2004; Zhuang et al., 2005; Chen and Herskovits, 2007), research based on PET data is still limited. Our work intends to bridge this gap.
Method
SICE for brain connectivity model structure identification
Suppose that there are p brain regions to be modeled, i.e., {X1, …, Xp}. The measurement data for each brain region is the regional cerebral metabolic rate for glucose by FDG-PET. The data can be reasonably assumed to follow a multivariate normality distribution, as this assumption has been adopted in a number of past publications. Statistical normality checks based on the data in our experiments also support this assumption (see Supplementary material). Given the measurement data of the brain regions from subjects, i.e., AD patients, SICE finds an estimate for the inverse covariance of the brain regions by solving the following optimization:
| (1) |
where θ and θ̂ denote the IC and its estimate, S is the sample covariance matrix, det (·) and tr (·) denote the determinant and trace of a matrix, ||·||1 denotes the sum of absolute values of all the entries in a matrix, and λ is a pre-selected so-called regularization parameter, λ>0. To help understand (1), (1) can be equivalently written as
| (2) |
where ∈ is reversely related to λ, ∈>0. It is easy to see from (2) that SICE aims to find a θ̂ that maximizes the likelihood function, under a constraint that the sum of the magnitudes of all entries in θ̂ is bounded by ∈ (or λ, equivalently). Furthermore, it can be seen from (2) that when ∈ is large enough (i.e., small λ), the constraint has little effect and SICE is just the usual MLE. However, when ∈ is small (i.e., big λ), SICE is able to produce an estimate for θ that is a shrunken version of the estimate by MLE. This is a advantage, because it has been found that the estimate for θ by MLE is likely to contain very few zero entries even when the θ is actually sparse, while the shrunken estimate for θ provided by SICE is able to recover those zero entries in θ. This advantage of SICE is especially significant under small sample sizes, which has been demonstrated by many papers (Yuan and Lin, 2007; Friedman et al., 2007; Schafer and Strimmer, 2005; Li and Gui, 2006).
Various methods have been developed to solve for the optimization in Eq. (1) and achieve the SICE in recent years (Yuan and Lin, 2007; Friedman et al., 2007; Levina et al., 2008; Li and Gui, 2006), including a method by us (Sun et al., 2009). A common characteristic of the SICE methods is that while they are good at discovering which entries in the IC matrix are zero and which are non-zero, they may not be good at estimating the magnitude of the non-zero entries due to the “shrinking” effect. Therefore, these methods may be more appropriate to be used for identifying the IC matrix structure, but not the parameters. As a result, once an estimate, θ̂, is obtained from SICE, we should use only the structural information (i.e., zero and non-zero entries) in to θ̂ build a brain connectivity model. Specifically, if we use a graph with nodes and undirected arcs to represent the brain connectivity model, we put an arc between nodes (i.e., brain regions) Xi and Xj if and only if θ̂ij ≠ 0, where θ̂ij is the entry at the ith row jth column of θ̂. An example of this is shown in Fig. 1.
Fig. 1.
(a) Estimated IC matrix by SICE θ̂ij ≠ 0. (b) Brain connectivity model built from the θ̂ in (a).
A monotone property of SICE for brain connectivity strength identification
The brain connectivity model obtained by SICE, such as the one in Fig. 1(b), may be interpreted in the following way. An arc between regions Xi and Xj may indicate that these two regions are directly connected in some functional process. This is because an arc represents a non-zero partial correlation which reflects the remaining association between two brain regions after the effect of other regions on their overall association has been factored out and the remaining association is likely to reflect the direct functional connection between the two regions. Furthermore, if two brain regions are not connected by an arc, but by a path consisting of more than one arc, they are not directed connected but may be reasonably considered as connected indirectly. For example, X1 and X3 in Fig. 1(b) may be considered as directly connected, while X1 and X5 may be considered as indirectly connected because there is not a single arc between them but two paths, X1–X3–X5 and X1–X4–X5.
The above discussion leads to the following definitions:
Definitions
Two brain regions are directly connected if there is an arc between them in the brain connectivity model. They are indirectly connected if there exists a path consisting of more than one arc between them. They are connected if they are either directly or indirectly connected.
The knowledge that two brain regions are connected is important for understanding the brain’s functional process. It is also important to find out the strength of the connection. However, a direct measure on the strength of connection is not possible, because the brain connectivity model obtained by SICE contains only structural information. A quasi-measure, on the other hand, may be possible due to the following considerations: The λ in the SICE formulation in Eq. (1) (or equivalently, the ∈ in Eq. (2)) has a similar effect to controlling the number of connections in the connectivity model estimated. A larger λ (smaller ∈) allows for a smaller number of connections. Therefore, as λ goes from λ1 to λ2, λ2 >λ1, some connections existing in the model corresponding to λ1 must drop. The connections that drop should be those such that the remaining connections maximize the likelihood (i.e., the objective functions in Eqs. (1) and (2)). To achieve this, obviously, the weakest connections should drop. In other words, as λ goes from λ1 to λ2, the connections that drop should be weaker than those that stay in. As a result, we can use λ2 as a quasi-measure for the strength of the connections that drop; in particular, the connections that drop at a bigger λ are stronger than those that drop at a smaller λ. Formally, the quasi-measure is defined as follows:
A quasi-measure for the strength of connection
A quasi-measure for the strength of connection between Xi and Xj is the critical λ value at which Xi and Xj change from being connected to being not connected.
For example, Fig. 2 shows the brain connectivity models estimated by SICE at four λ’s (λ1 < λ2 < λ3 < λ4). According to the above definition, the quasi-measure for the strength of connection between X6 and any of the other regions is λ2; that between X5 and X4 is λ3; that between any pair of regions in the cluster of X3, X2, and X1 is λ4.
Fig. 2.
Brain connectivity models at four λ’s (λ1 < λ2 < λ3 < λ4) showing the monotone property.
Note that in order for λ to be an appropriate quasi-measure for the strength of connections, we must be able to prove that if the connection between two brain regions drops at a certain λ, these two regions will never be connected again at larger λ’s. Otherwise, the strength of their connection cannot be uniquely determined. This is the so-called monotone property we discover, which is stated as follows (please see proof in the Appendix A):
Monotone property of SICE
If two brain regions are not connected in the connectivity model at a certain λ, they will never become connected as λ goes larger.
Discussion
Lastly in this section, we would like to provide some discussions on the use and potential benefits of the proposed quasi-measure:
With the aid of the quasi-measure, we can order the inter-region connections in terms of connection strength. For example, in Fig. 2, the connection between X6 and any of the other regions should be the weakest, that between X5 and X4 is the second weakest, and that between any pair of regions in the cluster of X3, X2, and X1 is the strongest.
Because λ is a quasi-measure, but not a direct one, for the strength of connections, there must be discrepancy between the measured strength by λ and the true strength even without sampling errors. Therefore, we would recommend using λ to order the connections, but not treating its value as a close estimate for the strength of a connection. Here is a simple example to illustrate this point: Even when two regions are not connected in the true brain network, the quasi-measure for their connection, i.e., the λ at which they change from being connected to being not connected in the estimated connectivity model by SICE, is still non-zero, because the λ in SICE is always positive.
λ is a quasi-measure for the strength of connections, but not for strength of the arcs in the brain connectivity model. Recall that arcs correspond to direct connections or partial correlations. In fact, we have found that the monotone property, although holds for connections, does not hold for direct connections, because an arc that drops at a certain λ may come back again as λ goes larger. Therefore, λ should not be used as a measure for the strength of the arcs. An example of this is shown in Fig. 2, in which the arc between X1 and X3 drops at λ2 but it comes back at λ3.
The potential benefits of the proposed quasi-measure are two-fold: First, it enables us to compare AD, MCI, and NC in terms of the order of inter-regions connections (results are shown in the next section). To our best knowledge, such a comparison has not been explored before, which provides new knowledge to AD studies. Second, in the literature of SICE, there has been a lack of a clear interpretation on λ, which is now being provided by our research. So, this research contributes to both the domain (AD) and the methodology (SICE).
Use of SICE for classifying new subjects
In this section, we will show how to use SICE to classify new subjects, which makes it possible to define connectivity-based AD biomarkers based on the connectivity models built from PET data. The basic idea is to apply SICE to the training data of AD and NC and estimate an IC matrix for each group. The estimated IC matrices will then be used to predict a new subject’s likelihood of being AD and NC, based on this subject’s PET measurement. The detailed procedure is illustrated as follows. Note that although the procedure is illustrated for AD vs. NC, it can be easily adapted for MCI vs. NC, or three-group (AD, MCI, and NC) classification.
Assume that there is a new subject with PET measurement x=[x1, …, xp], where xi is the PET measurement on brain region i, i =1, …, p, and p is the total number of brain regions. The objective is to determine whether to classify this subject as AD or NC. This may be achieved by comparing the likelihoods of x given that the subject is AD and NC, respectively. Because x follows a multivariate Gaussian distribution, its likelihood functions with respect to AD and NC can be written as,
respectively, where θAD and θNC are the inverse covariance matrices of AD and NC, respectively; the mean vector is considered to be zero without loss of generality. The estimates for θAD and θNC, θ̂AD and θ̂NC, can be obtained by SCIE based on training data. So, the classification rule can be: classify the new subject as AD if f(x|θ̂AD) > f(x|θ̂ NC) and NC otherwise. Note that we have mentioned previously that SCIE is good at identifying the IC matrix structure but not parameters. Therefore, to get better estimates for θ̂AD and θ̂NC, we can use the zero entries in the estimated IC matrices by SCIE as constraints and estimate the magnitudes of the non-zero entries by MLE, i.e., to solve a constrained optimization problem (Dempster, 1972).
Experiments and results
This section summarizes our experiments and findings for SICE-based brain connectivity modeling of AD, MCI, and NC using FDG-PET data. Purposes of our study focus on how aspects of the connectivity patterns exhibited in the models relate to existing findings in the literature, and on how other aspects may suggest further investigations in brain connectivity research.
Data acquisition and preprocessing
The data used in our study include FDG-PET images from 49 AD, 348 116 MCI, and 67 NC subjects downloaded from the Alzheimer’s 349 disease neuroimaging initiative (ADNI) database. Demographic information and MMSE 352 scores of the subjects are summarized in Table 1. The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. The initial goal of ADNI was to recruit 800 adults, ages 55 to 90, to participate in the research – approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years, and 200 people with early AD to be followed for 2 years.
Table 1.
Demographic information and MMSE scores.
| NC | MCI | AD | P-value | |
|---|---|---|---|---|
| Age (mean±SD) | 76.0±4.69 | 74.9±7.36 | 75.3±6.85 | 0.53 |
| Gender (male/female) | 43/24 | 76/40 | 27/22 | 0.77 |
| Years of education (mean±SD) | 15.9±3.24 | 16.0±2.86 | 14.7±3.02 | 0.01 |
| Baseline MMSE | 29.0±1.18 | 27.2±1.67 | 23.6±1.93 | <0.001 |
Preprocessing the images involves the following steps. Each subject’s FDG-PET image is spatially normalized to the MNI PET template, using the affine transformation and subsequent non-linear warping algorithm (Friston et al., 1995) implemented in SPM. The affine deformation adjusts each whole brain image for its position, orientation, size, and global shape based upon minimizing the mean residual variance (Zhilkin and Alexander, 2004). The non-linear warp, a linear combination of low spatial frequency basis discrete cosine functions, determines the optimal coefficients for each of the basis functions by minimizing the sum of squared differences between the raw MRI brain and the template image (Ashburner and Friston, 1999). Simultaneously, the smoothness of the transformation is maximized using maximum a posterior (MAP). Once in the MNI template, Automated Anatomical Labeling (AAL) (Tzourio-Mazoyer et al., 2002) is applied to extract data from each of the 116 anatomical volumes of interest (AVOI), and derived average of each AVOI for every subject based on the PET images.
Brain connectivity modeling by SICE and visualization techniques
42 AVOI are empirically chosen, which are brain regions known to be most affected by AD (Azari et al., 1992; Horwitz et al., 1987). These regions distribute in the frontal, parietal, occipital, and temporal lobes. Pease see Table 2 for names of the AVOI and the lobe each of them belong to.
Table 2.
Names of the AVOI for connectivity modeling (L=Left hemisphere, R=Right hemisphere).
| Frontal lobe | Parietal lobe | Occipital lobe | Temporal lobe | ||||
|---|---|---|---|---|---|---|---|
| 1 | Frontal_Sup_L | 13 | Parietal_Sup_L | 21 | Occipital_Sup_L | 27 | Temporal_Sup_L |
| 2 | Frontal_Sup_R | 14 | Parietal_Sup_R | 22 | Occipital_Sup_R | 28 | Temporal_Sup_R |
| 3 | Frontal_Mid_L | 15 | Parietal_Inf_L | 23 | Occipital_Mid_L | 29 | Temporal_Pole_Sup_L |
| 4 | Frontal_Mid_R | 16 | Parietal_Inf_R | 24 | Occipital_Mid_R | 30 | Temporal_Pole_Sup_R |
| 5 | Frontal_Sup_Medial_L | 17 | Precuneus_L | 25 | Occipital_Inf_L | 31 | Temporal_Mid_L |
| 6 | Frontal_Sup_Medial_R | 18 | Precuneus_R | 26 | Occipital_Inf_R | 32 | Temporal_Mid_R |
| 7 | Frontal_Mid_Orb_L | 19 | Cingulum_Post_L | 33 | Temporal_Pole_Mid_L | ||
| 8 | Frontal_Mid_Orb_R | 20 | Cingulum_Post_R | 34 | Temporal_Pole_Mid_R | ||
| 9 | Rectus_L | 35 | Temporal_Inf_L | ||||
| 10 | Rectus_R | 36 | Temporal_Inf_R | ||||
| 11 | Cingulum_Ant_L | 37 | Fusiform_L | ||||
| 12 | Cingulum_Ant_R | 38 | Fusiform_R | ||||
| 39 | Hippocampus_L | ||||||
| 40 | Hippocampus_R | ||||||
| 41 | ParaHippocampal_L | ||||||
| 42 | ParaHippocampal_R | ||||||
To build a brain connectivity model for AD, we first compute a sample covariance matrix, S, of the 42 AVOI, based on the measurement data of the 42 AVOI from 49 AD patients. Then, we apply SICE to solve the optimization problem in Eq. (1) based on the S and a pre-selected λ. The solution θ̂ is further converted to a graph consisting of nodes (AVOI) and arcs (non-zero entries in θ̂). Furthermore, considering that a graph of this kind may be too space-consuming for the paper, we adopt a matrix representation for the graph. Please see the first matrix in Fig. 3(a), for an example, which represents the brain connectivity model structure estimated by SICE at a certain λ. In the matrix, each row (column) corresponds to one of the 42 AVOI. A black cell corresponds to an arc. Because the matrix is symmetric, the total number of black cells is equal to twice the total number of arcs in the corresponding connectivity graph. Moreover, on each matrix, four red cubes are used to highlight the brain regions in each of the four lobes; that is, from top-left to bottom-right, the red cubes highlight the frontal, parietal, occipital, and temporal lobes, respectively.
Fig. 3.
(a) Brain connectivity models with total number of arcs equal to 60. (b) Brain connectivity models with total number of arcs equal to 120. (c) Brain connectivity models with total number of arcs equal to 180.
Furthermore, to facilitate the comparison between AD, MCI, and NC, connectivity models should also be developed for MCI and NC, respectively. The problem is how to select the λ value for each of three groups, so that the comparison between them will make sense. In this paper, we focus on comparing AD, MCI, and NC in terms of the distribution/organization of the connectivity, which has been less studied in the literature, but not in terms of the global scale of the connectivity, which has been studied substantially. To achieve this, we must factor out the connectivity difference between the three groups that is due to their difference at the global scale, so that the remaining difference will reflect their difference in the connectivity distribution/organization. A common strategy is to control the total number of arcs for each group to be the same, which has been adopted by a number of other studies (Supekar et al., 2008; Stam et al., 2007). We also adopt this strategy; specially, we adjust the λ in the estimation of the connectivity model of each group, such that the three models, corresponding to AD, MCI, and NC, respectively, will have the same total number of arcs. Also, by selecting different values for the total number of arcs, we can obtain models representing the brain connectivity at different strength levels. Specifically, given a small value for the total number of arcs, only strong arcs will show up in the resulting connectivity model, so the model is a model of strong brain connectivity; when increasing the total number of arcs, mild (or even weak) arcs will also show up in the resulting connectivity model, so the model is a model of mild-to-strong (or even weak-to-strong) brain connectivity. For example, Fig. 3 shows the connectivity models for AD, MCI, and NC with the total number of arcs equal to 60, 120, and 180.
Finally, we introduce some other ways to visualize the connectivity models to facilitate the comparison between AD, MCI, and NC, such as graphs of nodes and arcs (e.g., Fig. 4(i)) and brain images (e.g., Fig. 4(ii)):
Fig. 4.
(i) Hippocampus and parahippcampus sub-network connectivity for AD, i.e., connectivity within the network and connectivity between the network and other regions in temporal lobe; green, blue, red arcs represent connectivity from strong to weak. (ii) Four axial slices of AD brain, showing connectivity between ParaHippocampal_L (yellow) and other regions in the temporal lobe; green, blue, and red highlight regions connected with ParaHippocampal_L from strong to weak.
Fig. 4(i) displays a portion of the connectivity model for AD. Each node is an AVOI in Table 2 in the temporal lobe. This graph focuses on the connectivity regarding the sub-network consisting of Hippocampus_L & R ((X39 & X40)) and ParaHippocampal_L & R (X41 & X42), so it only displays the arcs between each region in the sub-network and other regions in the temporal lobe, as well as the arcs between the regions in the sub-network. Other arcs are omitted. Furthermore, green arcs are arcs (black cells) appearing in the matrix plot of AD in Fig. 3(a), so they represent strong connectivity. Blue arcs are arcs not appearing in the matrix plot of AD in Fig. 3(a), but appearing in that in Fig. 3(b), so they represent less strong connectivity. Red arcs are arcs not appearing in the matrix plots of AD in Fig. 3(a) or (b), but appearing in that in Fig. 3(c), so they represent even less strong connectivity.
Fig. 4(ii) shows four axial slices of an AD brain. This graph focuses on displaying the connectivity between region X41, ParaHippocampal_L, and other regions in the temporal lobe. ParaHippocampal_L is highlighted in yellow. Regions highlighted in green, blue, and red are those in the temporal lobe that have strong, less strong, even less strong connectivity with ParaHippocampal_L, respectively. In a similar way, Fig. 5(i) and (ii) are developed for NC. Note that similar graphs to Figs. 4 and 5 can be developed for other portions of the brain connectivity model, or even the whole brain, which are not shown here due to space limits.
Fig. 5.
(i) Hippocampus and parahippcampus sub-network connectivity for NC, i.e., connectivity within the network and connectivity between the network and other regions in temporal lobe; green, blue, red arcs represent connectivity from strong to weak. (ii) Four axial slices of NC brain, showing connectivity between ParaHippocampal_L (yellow) and other regions in the temporal lobe; green, blue, and red highlight regions connected with ParaHippocampal_L from strong to weak.
Comparison between AD, MCI, and NC in connectivity organization/distribution
The connectivity models estimated by SICE and various types of visualization techniques (matrix, graph, and brain slice) enable us to see the difference between AD, MCI, and NC in terms of connectivity organization/distribution. For example, Fig. 3(a) shows fewer black cells in the temporal lobe of AD than NC, but more black cells in the frontal lobe of AD than NC, where black cells correspond to direct connections between brain regions. While visual comparison is an important initial step to pick out the differences, it should be followed by rigorous statistical hypothesis testing to check if the observed differences are statistically significant. Therefore, we perform hypothesis testing to check if the number of black cells within each lobe, used to represent the amount of direct connections within that lobe, is significantly different between each pair of the study groups (i.e., AD, MCI, and NC). The hypothesis testing is also performed for the number of black cells between lobes. Here, we show the steps for testing if the number of black cells within the temporal lobe of AD, nAD_T, is significantly different from that of NC, nNC_T.
Draw samples of AD patients and samples of NC subjects, with replacement, from the original AD and NC datasets, respectively.
Apply the SICE method to learn one connectivity model for AD and one for NC, based on the samples drawn. During the learning of each connectivity model, adjust the λ such that the two models have the same total number of arcs.
Count the number of arcs (or equivalently, the number of black cells in the matrix representation) within the temporal lobe of the AD connectivity model. This number is a bootstrap sample for nAD_T; in a similar way, a bootstrap sample for nNC_T can be obtained.
Repeat (i)–(iii) N times and obtain N bootstrap samples for nAD_T and nNC_T, respectively.
Test the hypothesis that nAD_T and nNC_T are equal based on their respective bootstrap samples, and compute the P-value of the hypothesis test. Interpretation of the P-value is following: P-value <0.05 means strong evidence for nAD_T ≠ nAD_T; 0.05 < P <0.1 means some evidence (not strong though) for nAD_T ≠ nAD_T; P >0.1 means little evidence for nAD_T ≠ nAD_T, i.e., there is no significant difference between nAD_T and nNC_T.
Following similar steps to the above, we can also compare the amount of direct connections within other lobes as well as between lobes, for each pair of the study groups. The results are summarized in Table 3, which gives the P-value of the hypothesis testing. Specifically, a P-value is shown if it is smaller than 0.1 and replaced by a “–” otherwise. A P-value is highlighted if it is smaller than 0.05. The P-values presented here are those after correcting the effect of multiple testing using the standard False Discovery Rate (FDR) approach by Benjamini and Hochberg (1995). Note that because our multiple tests are not independent (the same datasets are used multiple times), the standard FDR approach might generate conservative results. This is a limitation of our current work and we will investigate how to overcome this limitation in future research. In addition to using P-value for comparison, we also develop box plots, as shown in Fig. 6 (only the box plots for AD vs. NC are shown due to the page limit).
Table 3.
P-value from the hypothesis test of connectivity difference between AD, MCI, and NC.
| (a) Total number of arcs=60 |
(b) Total number of arcs=120 |
(c) Total number of arcs=180 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AD vs. NC | Frontal | Parietal | Occipital | Temporal | AD vs. NC | Frontal | Parietal | Occipital | Temporal | AD vs. NC | Frontal | Parietal | Occipital | Temporal |
| Frontal | 0.013 | – | – | – | Frontal | 0.060 | – | – | – | Frontal | 0.091 | – | – | – |
| Parietal | – | 0.018 | – | Parietal | – | 0.007 | – | Parietal | – | 0.022 | 0.027 | |||
| Occipital | – | – | Occipital | – | 0.040 | Occipital | – | – | ||||||
| Temporal | 0.063 | Temporal | 0.001 | Temporal | 0.041 | |||||||||
| AD vs. MCI | Frontal | Parietal | Occipital | Temporal | AD vs. MCI | Frontal | Parietal | Occipital | Temporal | AD vs. MCI | Frontal | Parietal | Occipital | Temporal |
| Frontal | – | – | – | – | Frontal | – | – | – | – | Frontal | – | – | 0.036 | – |
| Parietal | – | 0.013 | – | Parietal | – | – | – | Parietal | – | – | – | |||
| Occipital | – | – | Occipital | – | 0.046 | Occipital | – | – | ||||||
| Temporal | 0.035 | Temporal | – | Temporal | – | |||||||||
| MCI vs. NC | Frontal | Parietal | Occipital | Temporal | MCI vs. NC | Frontal | Parietal | Occipital | Temporal | MCI vs. NC | Frontal | Parietal | Occipital | Temporal |
| Frontal | – | – | 0.020 | – | Frontal | – | – | 0.002 | – | Frontal | – | – | 0.007 | – |
| Parietal | – | – | – | Parietal | – | 0.078 | – | Parietal | – | – | 0.033 | |||
| Occipital | – | – | Occipital | – | – | Occipital | – | – | ||||||
| Temporal | – | Temporal | – | Temporal | – | |||||||||
Fig. 6.
Box plots for comparing AD (yellow) vs. NC (green) in terms of the amount of intra-and inter-lobe direct connections (i.e., black cells or arcs).
Inspection of the results from visualizations, hypothesis testing, and box plots reveals the following interesting observations:
Within-lobe connectivity
The temporal lobe of AD has a significantly lesser amount of direct connections than NC. This is true across the connectivity models at different strength levels (i.e., total arc number equal to 60, 120, and 180). In other words, even the direct connections between some strongly-connected brain regions in the temporal lobe may be disrupted by AD. In particular, it is clearly from Fig. 3(b) that the regions “Hippocampus” and “ParaHippocampal” (numbered by 39–42, located at the right-bottom corner of Fig. 3(b)) are much more separated from other regions in AD than in NC. The decrease in the amount of connections in the temporal lobe of AD, especially between the Hippocampus and other regions, has been extensively reported in the literature (Supekar et al., 2008; Wang et al., 2007; Azari et al., 1992; Horwitz et al., 1987; Grady et al., 2001). However, the temporal lobe of MCI does not show a significant decrease in the amount of direct connections, compared with NC. This may be because MCI does not disrupt the temporal lobe as severely as AD.
The frontal lobe of AD has a significantly more amount of direct connections than NC, which is true across the connectivity models at different strength levels. This is consistent with previous literature and has been interpreted as compensatory reallocation or recruitment of cognitive resources (Gould et al., 2006; Stern, 2006; Becker et al., 1996; Woodard et al., 1998; Saykin et al., 2004; Grady et al., 2003). Because the regions in the frontal lobe are typically affected later in the course of AD (our data are early AD), increase in the amount of connections in the frontal lobe may help preserve some cognitive functions in AD patients. Furthermore, the frontal lobe of MCI does not show a significant increase in the amount of direct connections, compared with NC. This indicates that the compensatory effect in MCI brain may not be as strong as that in AD brains.
There is no significant difference between AD, MCI, and NC in terms of the amount of direct connections within the parietal lobe and within the occipital lobe.
Between-lobe connectivity
In general, human brains tend to have a less amount of between-lobe connections than within-lobe connections. A majority of the strong connections occurs within lobes, but rarely between lobes. These can be clearly seen from Fig. 3 (especially Fig. 3(a)) in which there are a lot more black cells inside the red cubes than outside the red cubes, regardless of AD, MCI, and NC. Recall that the red cubes are used to highlight the four lobes.
AD has a significantly more amount of parietal-occipital direct connections than NC, which is true across the connectivity models at different strength levels. Increase in the amount of connections between the parietal and occipital lobes of AD has been previously reported in (Supekar, 2008). It may also be interpreted as a compensatory effect. Furthermore, MCI also shows increase in the amount of direct connections between the parietal and occipital lobes, compared with NC, but the increase is not as significant as AD.
While the amount of direct connections between the frontal and occipital lobes shows little difference between AD and NC, this amount for MCI shows a significant decrease. Also, AD has a less amount of temporal-occipital connections, a less amount of frontal-parietal connections, but a more amount of parietal-temporal connections than NC.
Between-hemisphere connectivity
We are also interested in knowing if there is a difference between AD, MCI, and NC, in terms of the amount of direct connections between hemispheres. To achieve this, we can count how many left–right pairs of the same regions have an arc (or black cell) between them in the connectivity models of AD, MCI, and NC, respectively. In addition to directly comparing the counts, we can also perform hypothesis testing (similar to the ones used for within-lobe and between-lobe comparisons). Results show that when the total number of arcs in the connectivity models is equal to 180 or 120, none of the tests is significant. However, when the total number of arcs is equal to 60, the P-value of the tests for “AD vs. NC”, “AD vs. MCI”, and “MCI vs. NC” are 0.038, 0.061, and 0.376, respectively. We further perform tests for the total number of arcs equal to 50 and find the P-value to be 0.026, 0.079, and 0.198 respectively. These results indicate that AD disrupts the strong connection between the same regions in the left and right hemispheres, whereas this disruption is not significant in MCI.
Comparison between AD, MCI, and NC in connection strength
We can use the quasi-measure developed in Section 2.2 to obtain an order for the inter-region connections in terms of the connection strength, for each of the three study groups. To present this order in a way that facilitates the comparison between the three groups, we propose a tree-like plot. As an illustrative example, Fig. 7 is a tree-like plot developed from the brain connectivity models in Fig. 2. One way to read off information from Fig. 7 is to look at it from right (small λ) to left (large λ). At a very small λ, i.e., λ=λ1, all regions are connected. As λ goes larger, i.e., λ1 < λ ≤λ2, X6, is the first region disconnected with other regions, so the connection between X6 and other regions is the weakest. As λ continues to go larger, i.e., λ2 − λ ≤ λ3, X4, X5, and the cluster of X1, X2, and X3 are disconnected, so the connection between them is the second weakest. Finally, with λ3 < λ, X1, X2, and X3 are disconnected, so the connectivity between them is the strongest.
Fig. 7.

A tree-like plot showing the order of connections between the brain regions in Fig. 2.
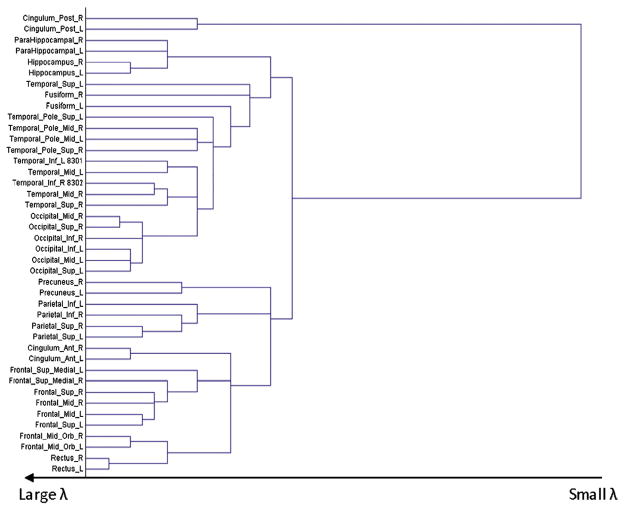
Following a similar manner, we develop a tree-like plot for AD, as shown in Fig. 8. Specially, the range of λ is determined such that the lower bound (λ=λL) corresponds to a “fully-connected” graph, i.e., every node has at least one arc attached, and the upper bound (λ=λU) corresponds to a “null” graph which has no arcs. Starting from the lower bound λ=λL, as λ goes larger, i.e., λL < λ ≤λ2, region “Tempora_Sup_L” is the first one disconnected with the rest of the brain, so “Tempora_Sup_L” may be the weakest connected region. As λ continues to go larger, i.e., λ2 < λ≤λ3, the rest of the brain further splits into three disconnected clusters, including the cluster of “Cingulum_Post_R” and “Cingulum_Post_L”, the cluster of “Fusiform_R” up to “Temporal_Sup_R”, and the cluster of the other regions. As λ continuously increases, each current cluster further splits into smaller clusters. Eventually, when λ reaches λU, all regions become disconnected. The sequence of the splitting gives an order for the inter-region connections in terms of the connection strength. Specifically, the earlier (i.e., smaller λ) a region or a cluster of regions becomes disconnected with the rest of the brain, the weaker it is connected with the rest of the brain. For example, in Fig. 8, it can be known that “Tempora_Sup_L” may be weakest connected with the rest of the brain in the brain network of AD; the second weakest ones are the cluster of “Cingulum_Post_R” and “Cingulum_Post_L”, and the cluster of “Fusiform_R” up to “Temporal_Sup_R”. It is very interesting to see that the weakest and second weakest connected brain regions in the brain network of AD include “Cingulum_Post_R” and “Cingulum_Post_L” as well as regions all in the temporal lobe, all of which have been found to be affected by AD early and severely (Supekar et al., 2008; Wang et al., 2007; Azari et al., 1992; Horwitz et al., 1987; Grady et al., 2001).
Fig. 8.
A tree-like plot for AD, showing the order of connections in terms of connection strength.
Next, to facilitate the comparison between AD and NC, a tree-like plot is also constructed for NC, as shown in Fig. 9. By comparing the plots for AD and NC, we can observe the following two distinct phenomena: First, in AD, between-lobe connections tend to be weaker than within-lobe connections. This can be seen from Fig. 8 which shows a clear pattern that the lobes become disconnected with each other before the regions within each lobe become disconnected with each other, as λ goes from small to large. This pattern does not show in Fig. 9 for NC. Second, the same brain regions in the left and right hemispheres are connected much weaker in AD than in NC. This can be seen from Fig. 9 for NC, in which the same brain regions in the left and right hemispheres are still connected even at a very large. However, this pattern does not show in Fig. 8 for AD.
Fig. 9.
A tree-like plot for NC, showing the order of connections in terms of connection strength.
Furthermore, a tree-like plot is also constructed for MCI (Fig. 10) and compared with the plots for AD and NC. In terms of the two phenomena discussed previously, MCI shows similar patterns to AD, but these patterns are not as distinct from NC as AD. Specifically, in terms of the first phenomenon, MCI also shows weaker between-lobe connections than within-lobe connections, which is similar to AD. However, this phenomenon is not as distinctive as AD. For example, a few regions in the temporal lobe of MCI, including “Temporal_Mid_R” and “Temporal_Sup_R”, appear to be more strongly connected with the occipital lobe than with other regions in the temporal lobe. In terms of the second phenomenon, MCI also shows weaker between-hemisphere connections in the same brain region than NC. However, this phenomenon is not as distinctive as AD. For example, several left–right pairs of the same brain regions are still connected even at a very large λ, such as “Rectus_R” and “Rectus_L”, “Frontal_Mid_Orb_R” and “Frontal_Mid_Orb _L”, “Parietal_Sup_R” and “Parietal_Sup_L”, as well as “Precuneus_R” and “Precuneus_L”. All above findings are consistent with the knowledge that MCI may be considered as a transition stage between normal aging and AD. Note that the tree-like plots in Figs. 8, 9 and 10 reveal the “observed” differences between AD, MCI, and NC. This serves as a starting point for comparing AD, MCI, and NC in terms of the connection strength. A challenging task following this may be to formulate an appropriate hypothesis testing to test the statistical significance of the observed difference, which will be investigated in future research.
Fig. 10.
A tree-like plot for MCI, showing the order of connections in terms of connection strength.
Use of SICE for classification of AD and NC
The purpose of this experiment is to assess the classification accuracy of the proposed method in Section 2.3. The experiment is performed on the PET dataset of 49 AD and 67 NC subjects. Leave-one-out cross-validation is applied. Specifically, we use each of the 116 (49 AD plus 67 NC) subjects as the “new” subject and the remaining subjects as the training data. Then, we apply the proposed method in Section 2.3 and obtain a “predicted” class (AD or NC) for the new subject. In this manner, we can obtain predicted classes for all 116 subjects. The predicted classes are compared with the true classes and classification accuracy is computed.
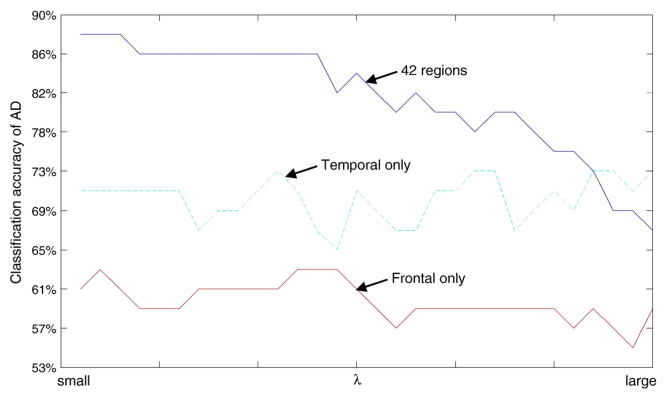
Results from the experiment are shown in Figs. 11 and 12. In particular, Fig. 11 shows the classification accuracy (vertical axis) of the 49 AD vs. λ values (horizontal axis), based on all 42 regions (blue curve), frontal regions only (red curve), and temporal regions only (green curve). Note that the classification accuracy varies with different λ’s, because different λ’s lead to different estimates for the inverse covariance matrices, θAD and θNC, which further affect performance of the classification based on them. In practice, we can choose a value for λ that achieves the desired classification accuracy for AD and NC, i.e., the desired sensitivity and specificity; and keep this λ for classifying future subjects.
Fig. 11.
Classification accuracy of AD.
Fig. 12.
Classification accuracy of NC.
Some observations can be made based on the results in Figs. 11 and 12. The best sensitivity and specificity the proposed method can achieve are 88% and 88%, respectively. However, they are not achieved at the same λ, because gain in sensitivity is associated with loss in specificity. Furthermore, performance of the classification based on all 42 regions is much better than that based on frontal or temporal regions alone. This may be because both frontal and temporal connectivity, as well as other connectivity patterns identified in the previous section of the paper (e.g., left-right hemisphere connectivity), have some discriminating power. Thus, using all 42 regions in the classification takes advantage of the combinatory effect of the discriminating powers of these local connectivity patterns.
Discussion
In this paper, we proposed SICE for identifying functional brain connectivity models from PET data. SICE was able to identify the connectivity model structures, and with the aid of a quasi-measure we developed, it can also identify the order of inter-region connections in terms of the connection strength. We applied the proposed method to the ADNI FDG-PET data of AD, MCI, and NC subjects. We compared these three groups in terms of the amounts of connections within lobes, between lobes, and between hemispheres, and in terms of the strength of connections. Note that “strength” of connections is not the same concept as “amount” of connections. For example, a lobe may have a large amount of connections between the regions in that lobe, but these regions may just be weakly connected to each other. Our findings showed that:
Decrease in the amount of connections
Comparing AD with NC, we found decrease in the amount of connections in the temporal lobe. Within the temporal lobe, we found that hippocampus, in conjunction with parahippocampus, is quite separated from other regions. These findings support that the temporal lobe, especially the area around hippocampus, is the first and most severely affected by AD and are consistent with previous findings in the literature (Supekar et al., 2008; Wang et al., 2007; Azari et al., 1992; Horwitz et al., 1987; Grady et al., 2001). Also, we found decrease in the amount of connections between the temporal and occipital lobes, and between the frontal and parietal lobes.
Increase in the amount of connections
We found increase in the amount of connections in the frontal lobe of AD brains. This may be interpreted as a compensatory effect or cognitive resource allocation (Gould et al., 2006; Stern, 2006; Becker et al., 1996; Woodard et al., 1998; Saykin et al., 2004; Grady et al., 2003; Supekar, 2008). In the literature, the compensatory effect has not only been found in early AD patients but also during memory tasks in healthy old adults compared with younger adults (Cabeza et al., 1997; Madden et al., 1999), suggesting that this might be a general response of human brains to functional loss resulting from various causes. Engagement of the frontal network is commonly found in studies of sustained attention (Fuster, 2000). Thus, significant increase in the amount of connections in the frontal lobe may reflect a greater engagement of the attentional resource to compensate for the decrease in the amount of connections in other part of the brain. Furthermore, most previous findings in the AD literature demonstrated the compensation effect during task performance. Our findings suggested that even during resting state, the increased recruitment of frontal resource still exists and may reflect a more general adaptation to the deficits of AD.
In addition, we also found increase in the amount of connections between the parietal and occipital lobes of AD brains. It is interesting to note that although there is significant increase in the amount of parietal-occipital connections, the amount of within-parietal and within-occipital connections of AD is not significant different from NC. This may indicate that the compensatory effect takes place between the parietal and occipital lobes but not within these lobes.
Strength of connections
The brain regions that are weakest connected to the rest of the brain in the brain network of AD include “Cingulum_Post_R” and “Cingulum_Post_L” as well as regions all in the temporal lobe, all of which have been found to be affected by AD early and severely (Supekar et al., 2008; Wang et al., 2007; Azari et al., 1992; Horwitz et al., 1987; Grady et al., 2001). Furthermore, between-lobe connections tend to be much weaker than within-lobe connectivity for AD, while this phenomenon is not significant for NC. Also, the same brain regions in the left and right hemispheres are connected much weaker in AD than in NC.
Findings about MCI
A unique perspective this paper provided is that we also studied MCI. While abundant literature exists in studying the brain connectivity difference between AD and NC, studies on MCI are limited. Our findings included that: MCI does not show as much decrease in the amount of connections in the temporal lobe as AD, nor does MCI show as much increase in the amount of connections in the frontal lobe and between the parietal and occipital lobes as AD. In addition, MCI does not seem to disrupt the strong connections between the same regions of the left and right hemispheres, as AD does. All these findings have supported the clinical observation that that MCI is a transition stage between normal aging and AD. One interesting finding about MCI is that it shows significant decrease in the amount of connections between the frontal and occipital lobes, compared with NC, while such decrease does not show for AD. This may suggest that MCI does have some uniqueness, or heterogeneity, of its own, rather than always being a clear pre-stage of AD.
On the other hand, our study on MCI does have limitations. First, the 116 MCI subjects used for brain connectivity modeling is a heterogeneous group. While they have all been diagnosed with MCI at the baseline-time checkup, some of them may convert to AD, some to NC, and others stay as MCI at later (e.g., 6th month or 12th month) checkups. However, during the time when this paper was developed, the diagnostic results of the 116 subjects at later checkups had not been available. Therefore, although the sample size of the MCI group is much larger than that of the AD and NC groups, the brain connectivity models of MCI may still be unreliable due to the data heterogeneity. In future work, we will split the current MCI group into MCI converters and non-converters according to the diagnostic results at later checkups, and build brain connectivity models for each subgroup. This will lead to more reliable models.
Clinical relevance of this research
First, this work may be used in clinical trials. Specifically, the modeling and analysis procedure on AD, MCI, and NC groups proposed in this paper can be readily applied to the groups given and not given a certain drug. Then, difference in the connectivity patterns of the two groups can be used to assess the drug efficacy. A significant advantage of applying this work to clinical trials is that SICE is able to produce reliable brain connectivity models with small sample sizes. This can help significantly lower the sample size requirement in clinical trials, increase the statistical power of the between-group comparison, and expedite drug efficacy assessment. Second, this work may be used for functional brain connectivity modeling based on fMRI data. The difference between PET and fMRI modeling is that the “samples” in PET modeling are subjects, while in fMRI modeling they are consecutive time points in the fMRI time series. As a result, the connectivity models built from PET data are models for each group (e.g., AD, MCI, or NC), while those from fMRI data are models for each subject. Once subject-level brain connectivity models are available, we can further identify connectivity-based biomarkers for AD and MCI. Note that most existing imaging biomarkers are based on individual brain regions, which may be greatly complemented by connectivity-based markers which characterize how the interactions between brain regions are affected by AD or MCI pathology. Third, it is also possible to define connectivity-based biomarkers based on the connectivity models built from PET data. To achieve this, we proposed a method in Section 2.3 and showed the experimental results using our data in Section 3.5.
Supplementary material and future work
Due the space limit, we put some additional results in the Supplementary Material: (1) Normality check of the data, in order to justify the multivariate normality assumption in SICE. The results showed that the data can be reasonably assumed to follow a multivariate normal distribution. (2) A whole-brain 116-region connectivity modeling. The results confirmed the major findings from the 42-region modeling.
Several future research directions are pointed out here. First, SICE provides a model for the linear interactions between brain regions, because it is based on the covariance matrix of the data. An interesting future direction is to explore the nonlinear interactions, which may be achieved by first discretizing the measurement of each brain region and then building a graphical model (e.g., a Bayesian network) of the brain regions based on the discretized measurements. Second, the current preprocessing procedure involves the use of the default SPM5 registration. We will explore the use of improved image registration algorithms in SPM5/8 (DARTEL).
Acknowledgments
Data collection and sharing for this project were funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI; Principal Investigator: Michael Weiner; NIH grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and through generous contributions from the following: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, Alzheimer’s Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging, with participation from the U.S. Food and Drug Administration. Industry partnerships are coordinated through the Foundation for the National Institutes of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory of Neuro Imaging at the University of California, Los Angeles.
Appendix A. Proof of the monotone property of the SICE method
A sufficient and necessary condition of the monotone property is as follow:
Theorem 1
Let and denote the clusters of nodes in the SICE-based brain connectivity models, with λ equal to λ1 and λ2 (λ1 < λ2), respectively. Here, a “cluster” of nodes means these nodes are all connected to one another either directly or indirectly. Then, for any , i ∈ {1, 2, …, L2}, there must exist a , such that .
This section proves the monotone property by proving that Theorem 1 is true.
(1) can be equivalently written as
| (A-1) |
It is known from (Banerjee et al., 2008) that the solution, Σ̂, is unique with a fixed positive λ, and Σ̂ must satisfy the equations in Eq. (A-2):
| (A-2) |
where (·)kl denotes the element at the kth row, lth column of a matrix.
When λ= λ1, denote the solution to Eq. (A-1) by Σ̂λ1. Furthermore, we can rearrange the rows and columns of Σ̂λ1, such that Σ̂λ1 becomes a block diagonal matrix and each sub-matrix along the main diagonal of the rearranged Σ̂λ1 correspond to a cluster of nodes in the SICE-based graphical model. Denote the sub-matrices by , j =1, …, L1. Recall that is the jth cluster of nodes in the graphical model. As a result, Σ̂λ1 can be written as:
| (A-3) |
A sufficient condition for Theorem 1 being true is that the solution to Eq. (A-1) when λ= λ2, denoted by Σ̂λ2, must share the same structure as Eq. (A-3), i.e., Σ̂λ2 can be written as:
| (A-4) |
To prove this sufficient condition, our strategy will include two steps: step one aims to find a matrix having the same structure as Σ̂λ1; step two aims to prove that this matrix is a solution to Eq. (A-2) with λ=λ2.
Step One
The rows and columns of the sample covariance matrix, S, can be rearranged in the same way as Σ̂λ1, i.e.,
| (A-5) |
Next, one optimization problem can be formulated corresponding to one sub-matrix , j=1,…, L1, i.e.,
| (A-6) |
Furthermore, the solutions to (A-6), i.e., , j=1,…, L1, can be put together and form a big matrix Yλ2, i.e.,
| (A-7) |
It is obvious that Ŷλ2 has the same structure as Ŷλ1.
Step Two
This step aims to prove that the Ŷλ2 in Eq. (A-7) satisfies Eq. (A-2) with λ=λ2. To prove this, we need to prove that (i) the elements in , j=1,…, L1, satisfy Eq. (A-2) and that (ii) the elements not in , all of which are equal to zero, also satisfy Eq. (A-2).
(i) Suppose that (Ŷλ2)kl is an element in , j ∈ {1, …, L1}; more specifically, suppose that (Ŷλ2)kl is the element at the hth row, sth column of , i.e.,
| (A-8) |
Because Yλ2 is the solution to the optimization in Eq. (A-6), it must satisfy Eq. (A-9):
| (A-9) |
It is easy to know that is in fact the element at the kth row, lth column of S, i.e.,
| (A-10) |
and is the element at the kth row, lth column of (Ŷλ2)−1, i.e.,
| (A-11) |
Inserting Eqs. (A-8), (A-10), and (A-11) into Eq. (A-9) results in Eq. (A-2) with λ=λ2.
(ii) Suppose that (Ŷλ2)kl is an element not in , j = 1, …, L1, i.e., (Ŷλ2)kl = 0. Furthermore, it can be known that ((Ŷλ2)−1)kl = 0 because Ŷλ2 is a block diagonal matrix. Since ((Ŷλ2)−1)kl = 0, to prove that (Ŷλ2)kl satisfies Eq. (A-2) with λ= λ2 is to prove that | (S)kl − (Ŷλ2)kl | ≤ λ2. It can be derive that | (S)kl − (Ŷλ2)kl | = |(S)kl| = | (S)kl − (Ŷλ1)kl | ≤ λ1, where the second equality holds because (Σ̂λ1)kl = 0, and the “≤” holds due to the last equation in Eq. (A-1) with λ= λ1. Also, it has been known that λ1 = λ2. Therefore, | (S)kl − (Ŷλ2)kl | ≤ λ1 ≤ λ2.
Appendix B. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2009.12.120.
References
- Alexander G, Moeller J. application of the scaled subprofile model: a statistical approach to the analysis of functional patterns in neuropsychiatric disorders: a principal component approach to modeling regional patterns of brain function in disease. Hum Brain Mapp. 1994:79–94. [Google Scholar]
- Ashburner J, Friston KJ. Nonlinear spatial normalization using basis functions. Hum Brain Mapp. 1999;7:254–266. doi: 10.1002/(SICI)1097-0193(1999)7:4<254::AID-HBM4>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azari NP, Rapoport SI, Grady CL, Schapiro MB, Salerno JA, Gonzales-Aviles A. Patterns of interregional correlations of cerebral glucose metabolic rates in patients with dementia of the Alzheimer type. Neurodegeneration. 1992;1:101–111. [Google Scholar]
- Banerjee O, Ghaoui LE, d’Aspremont A. Model selection through sparse maximum likelihood estimation for multivariate gaussian or binary data. J Mach Learn Res. 2008;9:485–516. [Google Scholar]
- Becker JT, Mintun MA, Aleva K, Wiseman MB, Nichols T, DeKosky ST. Compensatory reallocation of brain resources supporting verbal episodic memory in Alzheimer’s disease. Neurology. 1996;46:692–700. doi: 10.1212/wnl.46.3.692. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Cabeza R, Grady CL, Nyberg L, McIntosh AR, Tulving E, Kapur S, Jennings JM, Houle S, Craik FIM. Age-related differences in neural activity during memory encoding and retrieval: a positron emission tomography study. J Neurosci. 1997;17:391–400. doi: 10.1523/JNEUROSCI.17-01-00391.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. Spatial and temporal independent component analysis of functional MRI data containing a pair of task-related waveforms. Hum Brain Mapp. 2001;13:43–53. doi: 10.1002/hbm.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pekar JJ, Pearlson GD. Latency (in)sensitive ICA. Group independent component analysis of fMRI data in the temporal frequency domain. Neuro Image. 2003;20:1661–1669. doi: 10.1016/s1053-8119(03)00411-7. [DOI] [PubMed] [Google Scholar]
- Chen R, Herskovits EH. Graphical-model-based multivariate analysis of functional magnetic-resonance data. Neuro Image. 2007;35 (2):635–647. doi: 10.1016/j.neuroimage.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang J, Wang ZJ, McKeown MJ. Sparse Multivariate Autoregressive (mAR)-based Partial Directed Coherence (PDC) for Electroencephalogram (EEG) Analysis. Proceedings of the 2009 IEEE International Conference on Acoustics, Speech and Signal Processing; 2009.2009. pp. 457–460. [Google Scholar]
- Delbeuck X, Van der Linden M, Collette F. Alzheimer’s disease as a disconnection syndrome? Neuropsychol Rev. 2003;13 (2):79–92. doi: 10.1023/a:1023832305702. [DOI] [PubMed] [Google Scholar]
- Dempster AP. Covariance selection. Biometrics. 1972;28 (1):157–175. [Google Scholar]
- Friedman J, Tastie, Tibsirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2007;8 (1):1–10. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ. Functional and effective connectivity: a synthesis. Hum Brain Mapp. 1994;2:56–78. [Google Scholar]
- Friston KJ, Ashburner J, Frith CD, Pline JB, Heather JD, Frachowiak RSJ. Spatial registration and normalization of images. Hum Brain Mapp. 1995;2:89–165. [Google Scholar]
- Fuster JM. Executive frontal functions. Exp Brain Res. 2000;133:66–70. doi: 10.1007/s002210000401. [DOI] [PubMed] [Google Scholar]
- Gould RL, Arroyo B, Brown RG, Owen AM, Bullmore ET, Howard RJ. Brain mechanisms of successful compensation during learning in Alzheimer disease. Neurology. 2006;67:1011–1017. doi: 10.1212/01.wnl.0000237534.31734.1b. [DOI] [PubMed] [Google Scholar]
- Grady CL, Furey ML, Pietrini P, Horwitz B, Rapoport SI. Altered brain functional connectivity and impaired short-term memory in Alzheimer’s disease. Brain. 2001;124:739–756. doi: 10.1093/brain/124.4.739. [DOI] [PubMed] [Google Scholar]
- Grady CL, McIntosh AR, Beig S, Keightley ML, Burian H, Black SE. Evidence from functional neuroimaging of a compensatory prefrontal network in Alzheimer’s disease. J Neurosci. 2003;23:986–993. doi: 10.1523/JNEUROSCI.23-03-00986.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson M, Peterson BJ, Skudlarski P, Gatenby JC, Gore JC. Detection of functional connectivity using temporal correlations in MR images. Hum Brain Mapp. 2002;15:247–262. doi: 10.1002/hbm.10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgetag C, Kotter R, Stephan KE. Computational methods for the analysis of brain connectivity. In: Ascoli GA, editor. Computational Neuroanatomy. Humana Press; Totowa, NJ: 2002. [Google Scholar]
- Hirano A, Zimmerman HM. Alzheimer’s neurofibrillary changes. A topographic study. Arch Neurol. 1962;7:227–242. doi: 10.1001/archneur.1962.04210030065009. [DOI] [PubMed] [Google Scholar]
- Horwitz B. The elusive concept of brain connectivity. Neuro Image. 2003;19:466–470. doi: 10.1016/s1053-8119(03)00112-5. [DOI] [PubMed] [Google Scholar]
- Horwitz B, Grady CL, Sclageter NL, Duara R, Rapoport SI. intercorrelations of regional glucose metabolic rates in Alzheimer’s disease. Brain Res. 1987;407:294–306. doi: 10.1016/0006-8993(87)91107-3. [DOI] [PubMed] [Google Scholar]
- Huang S, Li J, Sun Li, Liu J, Wu T, Chen K, Fleisher A, Reiman E, Ye J. Learning Brain Connectivity of Azheimer’s Disease from Neuroimaging Data; Proceedings of Neural Information Processing Systems Conference (NIPS) 2009 (acceptance rate 8%); Vancouver, B.C., Canada. September 7–9, 2009.2009. [Google Scholar]
- Kotter R, Stephan ME. Network participation indices: characterizing component roles for information processing in neural networks. Neural Netw. 2003;16:1261–1275. doi: 10.1016/j.neunet.2003.06.002. [DOI] [PubMed] [Google Scholar]
- Levina E, Rothman AJ, Zhu J. Sparse Estimation of Large CovarianceMatrices via a Nested Lasso Penalty. Ann Appl Stat. 2008;2:245–263. [Google Scholar]
- Li H, Gui J. Gradient directed regularization for sparse gaussian concentration graphs, with applications to inference of genetic networks. Biostatistics. 2006;7:302–317. doi: 10.1093/biostatistics/kxj008. [DOI] [PubMed] [Google Scholar]
- Li JN, Wang ZJ, Palmer SJ, McKeown MJ. Dynamic Bayesian network modeling of fMRI: a comparison of group-analysis methods. Neuro Image. 2008;41:398–407. doi: 10.1016/j.neuroimage.2008.01.068. [DOI] [PubMed] [Google Scholar]
- Madden DJ, Turkington TG, Provenzale JM, Denny LL, Hawk TC, Gottlob LR, Coleman RE. Adult age differences in the functional neuroanatomy of verbal recognition memory. Hum Brain Mapp. 1999;7:115–135. doi: 10.1002/(SICI)1097-0193(1999)7:2<115::AID-HBM5>3.0.CO;2-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrelec G, Krainik A, Duffau H, Pélégrini-Issac M, Lehéricy S, Doyon J, et al. Partial correlation for functional brain interactivity investigation in functional MRI. Neuro Image. 2006;32:228–237. doi: 10.1016/j.neuroimage.2005.12.057. [DOI] [PubMed] [Google Scholar]
- Marrelec, et al. Using partial correlation to enhance structural equation modeling of functional MRI data. Magn Reson Imaging. 2007;25:1181–1189. doi: 10.1016/j.mri.2007.02.012. [DOI] [PubMed] [Google Scholar]
- McIntosh AR, Bookstein FL, Haxby JV, Grady CL. Spatial pattern analysis of functional brain images using partial least squares. Neuro Image. 1996;3:143–157. doi: 10.1006/nimg.1996.0016. [DOI] [PubMed] [Google Scholar]
- Rajapakse JC, Zhou J. Learning effective brain connectivity with dynamic Bayesian networks. Neuro Image. 2007;37:749–760. doi: 10.1016/j.neuroimage.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Salvador R, Suckling J, Coleman M, Pickard JD, Menon D, Bullmore E. Neurophysiological architecture of functional magnetic resonanceimages of human brain. Cereb Cortex. 2005a;34:387–413. doi: 10.1093/cercor/bhi016. [DOI] [PubMed] [Google Scholar]
- Salvador R, Suckling J, Schwarzbauer C, Bullmore E. Undirected graphs of frequency dependent functional connectivity in whole brain networks. Philos Trans R Soc Lond B Biol Sci. 2005b;360:937–946. doi: 10.1098/rstb.2005.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ, Wishart HA, Rabin LA, et al. Cholinergic enhancement of frontal lobe activity in mild cognitive impairment. Brain. 2004;127:1574–1583. doi: 10.1093/brain/awh177. [DOI] [PubMed] [Google Scholar]
- Schafer J, Strimmer KA. Shrinkage approach to large-scale covariance matrix estimation and implications for functional genomics. Stat Appl Genet Mol Biol. 2005;4(1) doi: 10.2202/1544-6115.1175. Article 32. [DOI] [PubMed] [Google Scholar]
- Sporns O, Chialvo DR, Kaiser M, Hilgetag CC. Organization, development and function of complex brain networks. Trends Cogn Sci. 2004;8:418–425. doi: 10.1016/j.tics.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Stam CJ, Jones BF, Nolte G, Breakspear M, Scheltens P. Small-world networks and functional connectivity in Alzheimer’s disease. Cereb Cortex. 2007;17:92–99. doi: 10.1093/cercor/bhj127. [DOI] [PubMed] [Google Scholar]
- Stern Y. Cognitive reserve and Alzheimer disease. Alzheimer Disease Associated Disorder. 2006;20:69–74. doi: 10.1097/01.wad.0000213815.20177.19. [DOI] [PubMed] [Google Scholar]
- Sun L, Patel R, Liu J, Chen K, Wu T, Li J, Reiman E, Ye J. Mining Brain Region Connectivity for Alzheimer’s Disease Study via Sparse Inverse Covariance Estimation. Proceedings of Knowledge Discovery and Data Mining Conference KDD; 2009.2009. [Google Scholar]
- Supekar K, Menon V, Rubin D, Musen M, Greicius MD. Network analysis of intrinsic functional brain connectivity in Alzheimer’s disease. PLoS Comput Biol. 2008;4 (6):1–11. doi: 10.1371/journal.pcbi.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WK, Barber A, Siegle GJA. Bayesian sparse vector autoregressive model for resting state connectivity analyses. Neuro Image. 2009;47 (Supplement 1):S39–S41. [Google Scholar]
- Tzourio-Mazoyer N, et al. Automated anatomical labelling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single subject brain. Neuro Image. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Valdés-Sosa PA, Sanchez-Bornot JM, Lage-Castellanos A, Vega-Hernandez M, Bosch-Bayard J, Melie-García L, Canales-Rodriguez E. Estimating brain functional connectivity with sparse multivariate autoregression. Phil Trans R Soc B. 2005:969–981. doi: 10.1098/rstb.2005.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Liang M, Wang L, Tian L, Zhang X, Li K, Jiang T. Altered functional connectivity in early Alzheimer’s disease: a resting-state fMRI study. Hum Brain Mapp. 2007;28:967–978. doi: 10.1002/hbm.20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodard JL, Grafton ST, Votaw JR, Green RC, Dobraski ME, Hoffman JM. Compensatory recruitment of neural resources during overt rehearsal of word lists in Alzheimer’s disease. Neuropsychology. 1998;12:491–504. doi: 10.1037//0894-4105.12.4.491. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Poline JB, Friston KJ, Evans AC. Characterizing the response of PET and fMRI data using multivariate linear models. Neuro Image. 1997;6:305–319. doi: 10.1006/nimg.1997.0294. [DOI] [PubMed] [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in the Gaussian graphical model. Biometrika. 2007;94 (1):19–35. [Google Scholar]
- Zheng X, Rajapakse JC. Graphical models for brain connectivity from functional imaging data. Proceedings of 2004 IEEE International Joint Conference on Neural Networks.2004. [Google Scholar]
- Zhilkin P, Alexander ME. Affine registration: a comparison of several programs. Magn Reson Imaging. 2004;22:55–66. doi: 10.1016/j.mri.2003.05.004. [DOI] [PubMed] [Google Scholar]
- Zhuang JC, Laconte S, Peltier S, Zhang K, Hu XP. Connectivity exploration with structural equation modeling: an fMRI study of bimanual motor coordination. Neuro Image. 2005;25:462–470. doi: 10.1016/j.neuroimage.2004.11.007. [DOI] [PubMed] [Google Scholar]