Abstract
Streptococcus pneumoniae expressing serogroup 6 capsules frequently causes pneumococcal infections and the evolutionary origins of the serogroup 6 strains have been extensively studied. However, these studies were performed when serogroup 6 had only two known members (serotypes 6A and 6B) and before the two new members (serotypes 6C and 6D) expressing wciNβ were found. We have therefore reinvestigated the evolutionary origins of serogroup 6 by examining the profiles of the capsule gene loci and the multilocus sequence types (MLSTs) of many serogroup 6 isolates from several continents. We confirmed that there are two classes of cps locus sequences for serogroup 6 isolates. In our study, class 2 cps sequences were limited to a few serotype 6B isolates. Neighbour-joining analysis of cps sequence profiles showed a distinct clade for 6C and moderately distinct clades for class 1 6A and 6B sequences. The serotype 6D cps profile was found within the class 1 6B clade, suggesting that it was created by recombination between 6C and 6B cps loci. Interestingly, all 6C isolates also had a unique wzy allele with a 6 bp deletion. This suggests that serotype switching to 6C involves the transfer of a large (>4 kb) gene segment that includes both the wciNβ allele and the ‘short’ wzy allele. The MLST studies of serotype 6C isolates suggest that the 6C cps locus is incorporated into many different pneumococcal genomic backgrounds but that, interestingly, 6C cps may have preferentially entered strains of the same genomic backgrounds as those of serotype 6A.
INTRODUCTION
Streptococcus pneumoniae (pneumococcus) is a common colonizer of the human nasopharynx, yet it is an important human pathogen responsible for several diseases, mainly in children, the elderly and the immune-compromised (Lynch & Zhanel, 2009). Using a polysaccharide (PS) capsule, of which there are at least 93 structurally distinct types (Bratcher et al., 2010; Calix & Nahm, 2010; Henrichsen, 1995; Park et al., 2007b), this bacterium is able to shield its surface from recognition by the host innate immune system, thereby making the capsule a potent colonization/virulence factor (Avery & Dubos, 1931; Bogaert et al., 2004). Some capsule types (serotypes) are more prevalent in disease than others, with the serogroup 6 strains, which include serotypes 6A, 6B, 6C and 6D, being more commonly isolated from infections than the majority of other serotypes. Vaccination strategies in use today target the pneumococcal capsule from the most prevalent serotypes, and most pneumococcal vaccines, including the widely used 7-valent conjugate vaccine (PCV-7), contain the serotype 6B PS.
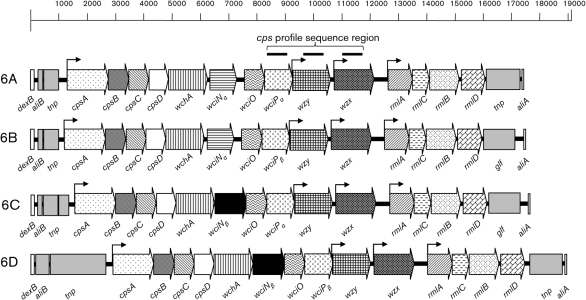
Because of its clinical importance, the evolution of the serogroup 6 strains has been previously studied in detail (Mavroidi et al., 2004; Robinson et al., 2002). Serogroup 6 capsular PS synthesis loci (cps) encode 14 ORFs and range in size from ∼17 to ∼19.1 kb due to variations in the non-coding regions found at either end (Fig. 1) (Mavroidi et al., 2004, 2007; Park et al., 2007a). Like most serotypes, the central, serotype-specific cps region of all serogroup 6 strains has a G+C content lower than the pneumococcal background, and three of these genes (wciP, wzy and wzx) are highly specific to serogroup 6 (Mavroidi et al., 2004). Using DNA sequences of selected parts of these serogroup 6-specific genes to create a ‘cps profile’, Mavroidi et al. (2004) showed that the only consistent genetic difference between serotypes 6A and 6B was a single nonsynonymous polymorphism in the wciP gene: the wciPα gene of serotype 6A has a G at nucleotide 584 while wciPβ of 6B has an A (Mavroidi et al., 2004). They also showed that the serogroup 6 cps loci can be divided into two distinct classes based on the presence of an INDEL sequence (Mavroidi et al., 2004). The majority of 6A and 6B strains do not have an INDEL and fall into class 1. In contrast, some 6B isolates and very few 6A isolates have an INDEL and fall into class 2. The capsule loci from the different classes have a 5.4 % sequence divergence whereas those of class 1 differ by only 1–2 % (Mavroidi et al., 2004).
Fig. 1.
Capsule gene loci of 6A (GenBank accession no. CR931638), 6B (GenBank accession no. CR931639), 6C (GenBank accession no. EF538714) and 6D (GenBank accession no. HM171374) strains. All ORFs involved in capsule synthesis are shown as horizontal arrows, and their direction indicates the transcriptional orientation. The three regions of the wciP (645 bases), wzy (492 bases) and wzx (477 bases) genes used for cps profiling are shown as three black bars above the 6A cps diagram. The wciN and wciP alleles are indicated by α and β. The size of a gene fragment from the beginning of wciNβ to the end of wzx is about 4130 bases. Bent arrows indicate potential transcription sites.
Since these studies were published, two new serogroup 6 members have been discovered. One new serotype, 6C, discovered in 2007 is serologically similar to 6A, but has a glucose residue replacing the galactose residue in the 6A PS (Park et al., 2007b). The 6C cps is 98 % similar to the 6A cps except that it contains a unique wciN gene (referred to as wciNβ) which does not have significant sequence homology with other pneumococcal genes, including the wciNα gene of 6A and 6B (Fig. 1) (Park et al., 2007a). While PCV-7, which contains 6B PS, has been shown to reduce the occurrence of both carriage and invasive disease resulting from vaccine-related serotype 6A, serotype 6C seems to be able to evade this cross-reactive protection afforded by the vaccine and is, therefore, increasing in prevalence (Carvalho Mda et al., 2009; Leach et al., 2009; Nahm et al., 2009; Park et al., 2008; Tocheva et al., 2010). Also, serotype 6D was recently described (Bratcher et al., 2010; Jin et al., 2009) and can be genetically distinguished from serotype 6C by the wciP gene (Bratcher et al., 2009). Specifically, serotype 6C contains a wciPα gene whereas serotype 6D has a wciPβ gene. In view of these recent discoveries in serogroup 6, we have reinvestigated the genetic evolution of serogroup 6 strains by studying the origins of these new serotypes as well as the mechanism of expansion of serotype 6C.
METHODS
PCR, sequencing and sequence analysis.
Genomic DNA was purified from pneumococci using phenol/chloroform extraction and was amplified by PCR as described previously (Mavroidi et al., 2004) using appropriate primers and PCR mixture. Primer sequences are shown in Table 1. The PCR mix contained 37.5 μl sterile water, 1 μl genomic DNA, 2 μl each primer (5 pmol), 2 μl 10 mM dNTP, 5 μl 10× LA Taq buffer solution and 0.5 μl LA Taq polymerase (2.5 U μl−1; Takara). The DNA sequence of the PCR product was determined by the Genomics Core Facility at University of Alabama at Birmingham.
Table 1.
List of primers used in the study
| Primer name (direction)* | Location | Sequence | Source or reference |
|---|---|---|---|
| 5117 (F) | wchA | 5′-ATCAAGTGGTATTGGAAGCGGG | This study |
| 5386 (F) | wciNβ | 5′-CTGCTTTCCAAAGAGTTCG | This study |
| 5106 (F) | wchA | 5′-TACCATGCAGGGTGGAATGT | Park et al. (2007a) |
| 5108 (F) | wciP | 5′-ATGGTGAGAGATATTTGTCAC | Mavroidi et al. (2004) |
| 5140 (F) | wzy | 5′-CCTAAAGTGGAGGGAATTTCG | Mavroidi et al. (2004) |
| 5141 (F) | wzx | 5′-TTCGAATGGGAATTCAATGG | Mavroidi et al. (2004) |
| 3386 (R) | wciNβ | 5′-TAATATACCTATCAACTCCACCGC | This study |
| 3102 (R) | wciP | 5′-CTGGCATGTCATCTTTAGAAAA | This study |
| 3101 (R) | wciO | 5′-CCATCCTTCGAGTATTGC | Park et al. (2007a) |
| 3107 (R) | wciP | 5′-AGCATGATGGTATATAAGCC | Mavroidi et al. (2004) |
| 3143 (R) | wzy | 5′-CCTCCCATATAACGAGTGATG | Mavroidi et al. (2004) |
| 3144 (R) | wzx | 5′-GCGAGCCAAATCGGTAAGTA | Mavroidi et al. (2004) |
*F, Forward; R, reverse.
DNA sequences of selected parts of the wciP, wzy and wzx genes were subjected to cps profiling studies using previously described approaches (Mavroidi et al., 2004). Alleles were assigned according to the designations previously used, and new alleles were given arbitrarily numbered designations. The sequences of wciP, wzy and wzx were concatenated, and the concatenated sequences were subjected to neighbour-joining analysis (Saitou & Nei, 1987) to investigate the evolutionary relationship among the cps loci of different serogroup 6 isolates. All evolutionary trees were drawn using mega4 (Tamura et al., 2007). Pairwise/evolutionary differences were computed using the maximum composite likelihood method in mega4 (Tamura et al., 2004, 2007), and in both analyses, positions containing gaps were eliminated from the dataset (complete deletion option). The percentages of replicate trees in which the associated sequences clustered together in the bootstrap test (500 replicates) are reported as the bootstrap values.
wciN and flanking regions from serotype 6A (CR931638), a class 2 6B (AF246897), 6C (EF538714), 33F (CR931702) and 4 (CR931635) sequences were analysed for possible recombination events. Recombinational analysis was performed by the RDP (recombination detection program) method (Martin & Rybicki, 2000) using rdp3 (Martin et al., 2005). For the recombination analysis, two sequences were created by randomly inserting the wciN gene or the wciN with its flanking regions into one insertion site of the serotype 4 cps locus in order to create a ‘mock foreign source’ for the wciN gene.
PCR amplicons were sequenced and the sequences were subjected to multilocus sequence typing (MLST) analysis as described previously (Enright & Spratt, 1998). Known alleles were then identified using the pneumococcal MLST website (http://spneumoniae.mlst.net), and numbers were assigned to new alleles by the database curator. All the MLST data listed in Table 1 have been submitted to the online pneumococcal MLST database. Evolutionary relations among MLST types were determined with the eBURST algorithm of the Department of Infectious Disease Epidemiology at Imperial College, London, as described by Enright & Spratt (1998).
Pneumococcal isolates.
Fifty-seven isolates were collected between 1999 and 2008 from four different continents (Table 2). Twenty-four of the isolates were 6A, 25 were 6C, 6 were 6B and 2 were 6D. These 57 isolates were subjected to cps profiling as well as MLST studies. To supplement the cps profiling studies, we studied an additional 12 6B isolates that were already in our collection. The resulting panel of 69 isolates included 18 isolates from Asia, 18 from Europe, 14 from North America, and 19 from South America. Full information on the additional 12 6B isolates is provided in Supplementary Table S1 (available with the online version of this paper).
Table 2.
Pneumococcal isolates used for the study
All isolates are from this study except MNZ595 and MNZ604, which are from the study by Bratcher et al. (2009).
| Strain | Location | Year isolated | Serotype | Clonal complex | ST | cps profile data | ||
|---|---|---|---|---|---|---|---|---|
| wciP | wzy | wzx | ||||||
| MNZ680 | S. America | 2001 | 6A | CC176 | 4598 | 2 | 8 | 1 |
| MNZ616 | S. America | 2004 | 6A | CC176 | 4598 | 2 | 8 | 1 |
| MNZ631 | S. America | 2003 | 6A | CC176 | 4598 | 2 | 1 | 1 |
| MNZ632 | S. America | 2002 | 6A | CC176 | 4598 | 2 | 1 | 1 |
| MNZ681 | S. America | 2001 | 6A | CC176 | 4600 | 2 | 1 | 1 |
| MNZ664 | S. America | 1999 | 6A | CC176 | 4622 | 2 | 1 | 1 |
| MNZ208 | Europe | 1999 | 6A | Singleton | 2611 | 2 | 6 | 1 |
| MNZ446 | N. America | 2007 | 6A | CC2090 | 376 | 2 | 6 | 1 |
| MNZ428 | N. America | 2007 | 6A | CC2090 | 1538 | 2 | 6 | 1 |
| MNZ677 | S. America | 1999 | 6A | CC315 | 1093 | 2 | 6 | 1 |
| MNZ218 | Europe | 2001 | 6A | CC395 | 327 | 15 | 6 | 1 |
| MNZ459 | N. America | 2007 | 6A | CC460 | 460 | 2 | 6 | 1 |
| MNZ497 | N. America | 2004 | 6A | CC460 | 460 | 1 | 1 | 1 |
| MNZ479 | N. America | 2004 | 6A | CC460 | 460 | 1 | 13 | 1 |
| MNZ239 | Europe | 2008 | 6A | CC473 | 813 | 2 | 1 | 1 |
| MNZ471 | N. America | 2007 | 6A | CC473 | 1876 | 14 | 1 | 1 |
| MNZ462 | N. America | 2007 | 6A | CC473 | 1876 | 14 | 1 | 1 |
| MNZ226 | Europe | 2003 | 6A | CC473 | 4595 | 2 | 1 | 3 |
| MNZ212 | Europe | 1999 | 6A | CC490 | 4363 | 2 | 1 | 1 |
| MNZ222 | Europe | 2002 | 6A | CC690 | 690 | 14 | 1 | 1 |
| MNZ28 | Asia | 2008 | 6A | CC81 | 81 | 2 | 1 | 1 |
| MNZ13 | Asia | 2008 | 6A | CC81 | 81 | 2 | 1 | 1 |
| MNZ19 | Asia | 2008 | 6A | CC81 | 282 | 2 | 1 | 1 |
| MNZ17 | Asia | 2008 | 6A | Singleton | 4620 | 2 | 1 | 1 |
| MNZ31 | Asia | 2008 | 6B | CC176 | 4604 | 16 | 15 | 1 |
| MNZ59 | Asia | 2008 | 6B | CC176 | 4605 | 16 | 15 | 1 |
| MNZ02 | Asia | 2008 | 6B INDEL | CC180 | 1624 | 8 | 14 | 7 |
| MNZ39 | Asia | 2008 | 6B INDEL | CC90 | 90 | 8 | 7 | 7 |
| MNZ62 | Asia | 2008 | 6B INDEL | CC90 | 90 | 8 | 14 | 7 |
| MNZ60 | Asia | 2008 | 6B INDEL | CC90 | 4606 | 8 | 14 | 7 |
| MNZ440 | N. America | 2007 | 6C | CC1379 | 1379 | 13 | 10 | 1 |
| MNZ613 | S. America | 2002 | 6C | CC1379 | 1379 | 9 | 10 | 1 |
| MNZ472 | N. America | 2007 | 6C | CC1379 | 1379 | 13 | 10 | 1 |
| MNZ595 | S. America | 2001 | 6C | CC1379 | 4599 | 13 | 10 | 1 |
| MNZ480 | N. America | 2004 | 6C | CC1390 | 1390 | 13 | 12 | 1 |
| MNZ488 | N. America | 2004 | 6C | CC1390 | 1390 | 13 | 10 | 1 |
| MNZ435 | N. America | 2007 | 6C | CC1390 | 1390 | 13 | 10 | 1 |
| MNZ458 | N. America | 2007 | 6C | CC1439 | 4602 | 13 | 10 | 1 |
| MNZ219 | Europe | 2002 | 6C | CC1715 | 1715 | 9 | 10 | 1 |
| MNZ240 | Europe | 2008 | 6C | CC176 | 2689 | 9 | 10 | 1 |
| MNZ23 | Asia | 2008 | 6C | CC176 | 4596 | 9 | 10 | 12 |
| MNZ34 | Asia | 2008 | 6C | CC176 | 4596 | 9 | 10 | 12 |
| MNZ38 | Asia | 2008 | 6C | CC176 | 4597 | 9 | 10 | 12 |
| MNZ604 | S. America | 1999 | 6C | CC3854 | 4601 | 9 | 10 | 1 |
| MNZ597 | S. America | 2002 | 6C | CC3854 | 4601 | 9 | 10 | 1 |
| MNZ683 | S. America | 2001 | 6C | CC3854 | 4601 | 9 | 10 | 1 |
| MNZ592 | S. America | 2003 | 6C | CC3854 | 4601 | 9 | 10 | 1 |
| MNZ678 | S. America | 1999 | 6C | CC3854 | 4621 | 9 | 10 | 1 |
| MNZ221 | Europe | 2002 | 6C | CC395 | 395 | 9 | 10 | 1 |
| MNZ213 | Europe | 1999 | 6C | CC395 | 395 | 9 | 10 | 1 |
| MNZ432 | N. America | 2007 | 6C | CC473 | 473 | 9 | 10 | 11 |
| MNZ16 | Asia | 2008 | 6C | Singleton | 855 | 9 | 10 | 11 |
| MNZ18 | Asia | 2008 | 6C | Singleton | 855 | 9 | 10 | 11 |
| MNZ227 | Europe | 2004 | 6C | Singleton | 3531 | 9 | 11 | 1 |
| MNZ209 | Europe | 1999 | 6C | Singleton | 4603 | 9 | 11 | 1 |
| MNZ21 | Asia | 2008 | 6D | CC81 | 282 | 5 | 1 | 1 |
| MNZ22 | Asia | 2008 | 6D | CC81 | 282 | 5 | 1 | 1 |
RESULTS
wciN and flanking region sequence variations among 6C and 6D isolates
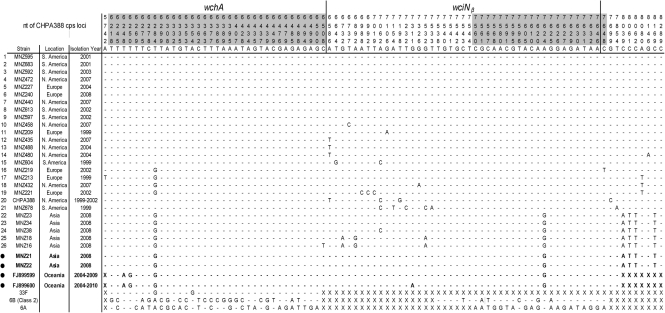
The wciNβ gene was sequenced from 25 new 6C isolates (Fig. 2, Table 2), which were obtained from four different continents over a 10 year period. In addition, we included data from the 6C strain (CHPA388) (Fig. 2) for which the entire cps locus has been sequenced and published (GenBank accession no. EF538714) for comparison. By comparing the sequences, all the variations were found at 17 (1.51 %) of the 1125 bases in wciNβ of 6C serotype. The total sequence variation is also very small: 0.11 %, 31 bases differed out of a total of 29 250 (=26×1125) bases. This difference is consistent with the heterogeneity observed for the cps sequences (GenBank accession nos AF246898, AY078347 and CR931638) of three 6A isolates (0.1–0.2 %), which presumably represent randomly chosen isolates. Furthermore, nine clinical isolates collected from three different continents (Europe, North America and South America) over an 8 year period have exactly the same nucleotide sequences for 2782 bases including wciNβ and flanking regions (Fig. 2, top 9 rows). Thus, it is likely that wciNβ was introduced to the serogroup 6 cps locus from a foreign, probably non-pneumococcal, source only once.
Fig. 2.
Sequence diversity of the wciNβ and its flanking regions for serotype 6C and 6D strains. The DNA sequence was determined from base 5671 to base 8452. The consensus sequence (top line) and the numbering system (numbers are given vertically) are based on the 6C cps sequence (GenBank accession no. EF538714). Heavy vertical bars at the top indicate the two ends of the wciNβ ORF. The flanking regions of the ‘foreign gene’ that produced wciNβ are shaded (between bases 6209 and 6508 and bases 7545 and 7655). Most sequences were from serotype 6C isolates except for the four sequences from 6D isolates (marked by •). One of 26 6C sequences is a published sequence (CHPA388, GenBank accession no. EF538714). Two Oceania 6D sequences are from GenBank (accession nos FJ899599 and FJ899600). X, No corresponding sequences. Sequences from 33F (GenBank accession no. CR931697) and a class 2 6B strain (GenBank accession no. AF246897) are included to show the similarity in the 5′ and 3′ flanking regions, respectively. The sequence for 6A (GenBank accession no. CR931638) is shown for comparison at the bottom.
Our previous study (Park et al., 2007a) suggested that the insertion of wciNβ may have been facilitated by two clearly identifiable flanking regions (grey shading in Fig. 2), which are about 300 and 110 bases long in the 5′ and 3′ regions, respectively. The flanking regions were defined by an intermediate level of genetic similarity (80–90 %) when comparing 6A and 6C cps loci, while the central region (i.e. the foreign gene) has no homology (29 % similarity) and outside of the flanking regions is highly similar (>98 %) (Park et al., 2007a). These flanking regions and their margins would vary among 6C isolates if the foreign gene was introduced multiple times to form serotype 6C cps. We have therefore determined the sequence of the wciNβ flanking regions from the 26 6C isolates and the two 6D isolates from Korea (Fig. 2). We also included the sequences of two 6D isolates from Fiji available from GenBank in the analysis (Fig. 2) (Jin et al., 2009). When the 410 bases in the two flanking regions from the 30 isolates were compared, only five bases varied, and all of these are located in the middle of the flanking regions, suggesting no variation at the margin. Thus, analysis of flanking regions also supports a single incorporation of the wciNβ gene in the serogroup 6 cps.
Interestingly, the 5′ flanking region is very similar to the corresponding region of pneumococcal serotype 33F cps and the 3′ flanking region is very close to the corresponding region of a class 2 6B strain cps, as shown at the bottom of Fig. 2. Potential recombination events that could have produced the 6C wciN and its flanking regions were investigated with the RDP method. The analysis suggests that generation of the 6C cps locus would involve complex recombination events requiring sources for the wciNβ and the ‘short’ wzy in addition to 33F and class 2 serotype 6B cps.
Analysis of the capsule gene loci of 6C and 6D isolates
Recently, isolates expressing serotype 6D were discovered in nature. To examine the evolutionary relationship of serotype 6D with the other three serotypes of serogroup 6, we determined the sequence of the entire cps locus of a serotype 6D isolate (GenBank accession no. HM171374). As illustrated in Fig. 1, the 6D cps is bound by dexB and aliA, as are the other capsule gene loci (García et al., 2000), and has transposase-like regions at each end. Between these transposase-like regions, the 6D cps has the same 14 functional genes found in the other serogroup 6 cps loci. In addition, the sequence of the 6D cps locus is almost identical (98.6 % identity in the 14 933 bp that include all the ORFs) to that of 6C (GenBank accession no. EF538714) except for the known difference in the wciP gene and differences in the non-coding regions flanking the cps locus. Similar to other members of serogroup 6, the G+C content of the region from wciNβ to wzy for 6D is only 31 %, which is lower than the rest of the pneumococcal genome (39 %) (Tettelin et al., 2001).
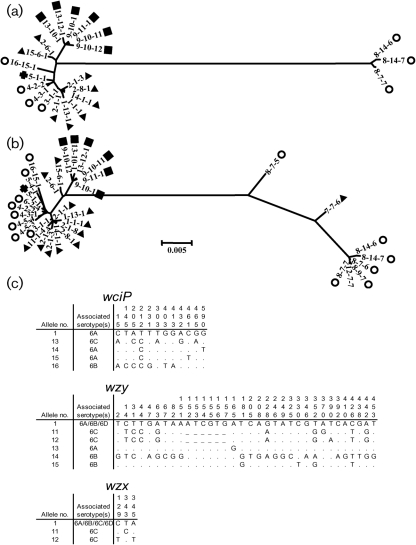
To study the evolutionary relationship between serotype 6C and serotypes 6A and 6B, we determined the cps profiles of the 57 isolates listed in Table 2 along with 12 archived 6B isolates. The cps profile was determined as previously described by sequencing a portion of three genes (wciP, wzy and wzx) that are common in all serogroup 6 capsule gene loci (Mavroidi et al., 2004). Our study identified 11 new alleles (shown in Fig. 3c): four for wciP, five for wzy and two for wzx. Interestingly, wzy alleles 10 (Mavroidi et al., 2004), 11 and 12 are used only by serotype 6C isolates and have a very distinct 6 bp deletion (Fig. 3c).
Fig. 3.
Neighbour-joining trees of unique cps profiles. (a) Tree constructed with 22 unique cps profiles from our study, which included 23 6A (▴), 18 6B (○), 25 6C (▪) and 2 6D (+) isolates. Table 2 shows the cps profiles of all the strains included in our study. Note that three class 2 6B isolates are shown on the right of the tree and all class 1 strains are shown on the left. (b) Tree constructed with cps profile data from both our current study and a published study (Mavroidi et al., 2004). Class 2 isolates are shown on the right of the tree and two cross-over strains (profiles 8-7-5 and 7-7-6) are shown in the middle. The bar represents a genetic distance of 0.5 %. (c) Polymorphic nucleotides for all new alleles of the cps genes (wciP, wzy and wzx). Allele 1 is shown at the top for comparison. Note that wzy alleles 11 and 12, which are associated with serotype 6C, have 6 base deletions (positions 152–157). –, No 6 bp deletions; ., identical bases.
When the three (wciP, wzy and wzx) sequences of each isolate were concatenated and the evolutionary tree was determined for the 69 isolates using the neighbour-joining method (Fig. 3a), the class 2 isolates (i.e. isolates with an INDEL) could be clearly separated from the class 1 isolates (isolates without an INDEL), as described previously (Mavroidi et al., 2004), with 99 % bootstrap support and the genetic distance between them being greater than 5.4 %. When class 1 isolates were examined, the 6C strains formed a distinct clade (99 % bootstrap support) suggesting a single origin (Fig. 3a). The class 1 6A and 6B strains formed moderately distinct clades (79 % bootstrap support) and a common ancestor for the two clades could not be excluded (Mavroidi et al., 2004). The genetic distance between class 1 6A and 6B isolates was >0.26 %, whereas the genetic distance of the 6C cluster from the 6A and 6B clusters was >0.78 % by pairwise differences computed using mega4 (Fig. 3a). Nevertheless, these clades are largely serotype-specific, and so we refer to the two clades by the dominant serotype in that clade. The two 6D isolates were nestled within the class 1 6B clade. The clustering pattern of the serotypes within clades did not change when additional published cps profile data (Mavroidi et al., 2004) were merged with our data (Fig. 3b). Also, analysis of the clustering patterns of individual gene segments did not provide additional information (see Supplementary Fig. S1, available with the online version of this paper). The cps profile analysis provides additional support that all the 6C cps loci shared a distinct origin but the class 1 6A and 6B clades are less distinct.
While most isolates in one clade are of a single serotype, there are some exceptions. Two obvious exceptions are the serotype 6D isolates expressing profile 5-1-1, which is within the 6B cluster. While more serotype 6D isolates should be examined, the cps loci of these two 6D isolates were most likely generated when 6C and 6B cps recombined at a location between wciN and wciP. The other exception is a 6B strain expressing 3-1-1, which is located within the 6A cluster. A previous study suggested that this profile may have arisen from the 6A serotype by a mutation of the wciP gene (Mavroidi et al., 2004). Additional outliers are the 6A strains expressing cps profiles 2-6-1 and 15-6-1. They have a relatively distinct wzy sequence containing four single nucleotide polymorphisms separating them from the class 1 6A and 6B sequences. These single nucleotide polymorphisms are shared by the 6C sequences, suggesting these outlier 6A strains may have been the source of the wzy sequence in the 6C strains.
The 6C cluster is associated with three very similar wzy alleles that have a 6 nt in-frame ‘ATCGTG’ deletion (Fig. 3c). This association was observed for the 25 6C isolates studied here and for 14 additional 6C isolates that were collected for our previous study (Park et al., 2007a; unpublished data). In contrast, all 42 isolates expressing serotypes 6A and 6B have normal wzy alleles. Consequently, the 6C cps locus has two genetic markers (wciNβ and the ‘short’ wzy) that are separated by about 4130 bp, which spans almost the entire stretch of serotype-specific genes in the cps locus. This finding suggests that the serotype switching event that results in expressing the 6C capsule involves a genetic transfer of not only wciNβ but also a large piece of DNA including wciNβ and the ‘short’ wzy.
Analysis of the genomic background of 6C and 6D clinical isolates
Since the introduction of conjugate pneumococcal vaccines, the prevalence of serotype 6C has greatly increased in several parts of the world. To investigate whether all or selected clones of serotype 6C were increasing in prevalence, we performed an MLST analysis on a global set of 6C isolates. Table 2 shows the sequence type (ST) and clonal complex of all of the pneumococcal isolates used in this study. The clonal complex of each isolate was identified by performing eBURST analysis using resources available at http://www.mlst.net. Our results show that some 6A isolates possess genetic backgrounds of ST81, ST376 and ST1538, which are associated with well-known antibiotic resistant clones (Spain23F-1 and North Carolina6A-23). Interestingly, ST282 expressed by serotype 6D is a single-locus variant of the Spain23F-1 clone. The cps gene of serotype 6C is associated with STs in multiple clonal complexes, as others have also found (Carvalho Mda et al., 2009; Jacobs et al., 2009; Nunes et al., 2009), but no 6C STs in this list could be linked with antibiotic resistant clones. However, it was shown that serotype 6C can have an antibiotic resistant ST (e.g. ST376) (Carvalho Mda et al., 2009).
DISCUSSION
A previous study of cps profiles of serogroup 6 isolates revealed two classes of cps profiles (named classes 1 and 2) and only a small evolutionary separation between class 1 6A and 6B clades. Since that study had been performed before serotypes 6C and 6D were discovered, a fresh look at the evolutionary relationships amongst the serogroup 6 serotypes was warranted. We have now studied the cps profiles of many additional isolates, including those of serotypes 6C and 6D. In addition to confirming the two classes of profiles found earlier, our cps profile studies show three clades within class 1, with two moderately distinct clades for 6A and 6B, as seen previously, and a new clade associated with serotype 6C.
A surprising and unexpected finding is that all 25 6C isolates included in the current study have ‘short’ wzy alleles lacking ATCGTG. In contrast, all 42 isolates expressing serotypes 6A and 6B that were included in this study have the normal-sized wzy. When we studied an additional 14 6C isolates (Park et al., 2007a), we found that all of them have the ‘short’ wzy alleles (unpublished data). Furthermore, Mavroidi et al. (2004) studied 102 serogroup 6 isolates before serotype 6C was discovered and described two serogroup 6 isolates as expressing a ‘short’ wzy allele. We were able to test one of the two isolates and found that it expressed serotype 6C (unpublished observation). Thus, the ‘short’ wzy allele is very strongly associated with serotype 6C cps. However, the ‘short’ wzy is not essential for expressing the serotype 6C capsule because we were previously able to convert a 6A serotype strain to a 6C strain by replacing wciNβ alone, without replacing wzy (Park et al., 2007a).
Our studies shed new insights into the origins of serotype 6C cps. Previously, we proposed that a wciNβ gene of an unknown origin (about 1500 bases) was inserted into the 6A cps with help from the two flanking regions (Park et al., 2007a). Although a recombination event involving the serotype 33F cps and class 2 serotype 6B cps was also considered, a more likely possibility is that 6C cps was created by transfer of a large foreign gene segment spanning wciNβ and the ‘short’ wzy to pneumococcus. The source of the foreign gene is unclear at the moment but it is likely to have come from bacteria in the nasopharynx, where pneumococci normally reside. Oral streptococcal species are logical candidates since they commonly coexist with pneumococci in the nasopharynx, often have capsules like pneumococci (Mavroidi et al., 2007) and may have served as the source for antibiotic-resistance genes for pneumococci (Guerin et al., 2000; Hakenbeck et al., 1998). As we find limited heterogeneity in the sequences of serotype 6C cps, serotype 6C was probably created once. Thus, we may be able to deduce the cps of the 6C founder. We believe that the founder may have had a cps profile of 9-10-1 based on the frequency and diversity of the cps alleles (Table 2). While the knowledge of the 6C founder sequence with two genetic markers may help its identification, the bacterial gene pool in the nasopharynx is very large, indicating that the identification of the exact source of 6C cps would not be simple.
While determining the origin of serotype 6C cps requires additional studies, the cps of the two Korean 6D isolates appears to have been created by a genetic recombination between serotypes 6B and 6C: the cps of these two serotypes may have recombined between wciN and wciP. However, more 6D isolates should be studied before we can conclude that all 6D isolates arose in this manner. Recently, 6D isolates were found in Fiji (Jin et al., 2009) and Finland (unpublished information). The cps profile of the Finnish 6D is 5-1-4. This is almost identical to the Korean 6D cps profile since alleles 4 and 1 of wzx differ in only 1 nt. The Fijian 6D isolates also have wciP allele 5 (Jin et al., 2009) as do the Korean 6D isolates. This suggests that serotype 6D cps may have been created once and spread throughout the world. Given its potentially wide distribution, it is interesting to note that reports of serotype 6D are currently very rare and that it was not found in several large screens of clinical isolates (Bratcher et al., 2009; Carvalho Mda et al., 2009; Hermans et al., 2008; Jacobs et al., 2010). One of the reasons for the rarity of serotype 6D may be difficulties in distinguishing between serotypes 6C and 6D. The presence or absence of ‘short’ wzy may be useful for distinguishing them because, unlike 6C, both Korean and Finnish 6D have normal-sized wzy.
While serotype 6C cps profiles have remained quite distinct from those of other strains of serogroup 6 (Fig. 3), our data as well as those from others (Carvalho Mda et al., 2009; Jacobs et al., 2009) show that serotype 6C is often associated with STs associated with other members of serogroup 6. For instance, many 6C isolates possess STs of 395, 473, 1379 and 1390, which are the most common STs for serotype 6A. While there are multiple potential explanations for this situation, we favour the explanation that the entire cps can easily move among different pneumococcal backgrounds as already shown for serotype 19A (Brueggemann et al., 2007). This movement may be easy because both ends of the cps have transposase-like regions whose exact function is still not well understood but may facilitate cps exchanges. Another explanation may be that two penicillin-resistance genes found at two extremes of the cps locus may facilitate the retention of transfers involving the entire cps, as was shown experimentally (Trzciński et al., 2004). However, serotype 6C appears to preferentially share its STs with those of serotype 6A but not 6B or other serotypes (e.g. serotype 19A). More studies are needed to explain how pneumococcal cps can so easily yet selectively move among different genetic backgrounds.
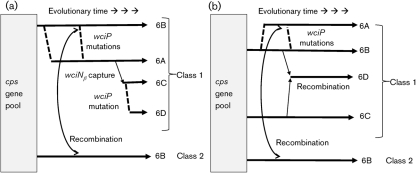
Our studies significantly clarify the evolutionary origins of serogroup 6 cps loci (Fig. 4). Previous studies suggested two independent origins of serogroup 6 cps: cps of class 1 and class 2 isolates (Fig. 4a). Although class 1 6A and 6B isolates could be grouped into two clades, the two clades were not so distinct, and class 1 6A and 6B isolates were presumed to have a single origin since the two serotypes could interconvert with a single mutation (Fig. 4a) (Mavroidi et al., 2004). Following the discovery of serotypes 6C and 6D, we then presumed that 6C cps arose from 6A cps by capturing the wciNβ gene (Park et al., 2007a) and that 6C cps became 6D cps by a somatic mutation (Fig. 4a). However, our data show that the clade for serotype 6C cps is distinct and it was probably produced by capturing a DNA fragment spanning the entire low G+C region from a currently ill-defined gene pool in the nasopharynx (Fig. 4b). Serotype 6D cps appears to have resulted from a recombination between 6B cps and 6C cps (Fig. 4b). These findings show that serogroup 6 cps loci, despite their high degree of sequence similarity, have at least three independent origins: for 6C cps, class 1 6B cps and class 2 6B cps. Perhaps these multiple independent origins are possible because the nasopharyngeal microbiome provides a large gene pool for pneumococcal cps, which allows S. pneumoniae to express a great variety of capsular structures.
Fig. 4.
Two models for the evolution of the serogroup 6 strains. (a) The model previously proposed by Mavroidi et al. (2004) modified to include the new serotypes 6C and 6D. (b) Proposed new model based on the new data. The nature of the ‘cps gene pool’ from which the capsule genes have originated is undefined at the moment. Genetic recombination events are shown with thin arrows. Mutations are shown with dashed lines.
Acknowledgments
The work was supported with funding from the National Institutes of Health (AI-31473) to M. H. N. The University of Alabama at Birmingham has applied for the Intellectual Property Rights for serotypes 6C and 6D. M. H. N. and I. H. P. are employed by the University of Alabama at Birmingham, which may represent a potential conflict of interests. We thank Drs A. Robinson and W. Benjamin for careful reading and comments.
Abbreviations
MLST, multilocus sequence typing
PCV-7, 7-valent conjugate vaccine
PS, polysaccharide
ST, sequence type
Footnotes
The GenBank/EMBL/DDBJ accession number for the entire cps locus of a serotype 6D isolate is HM171374.
A supplementary table and a supplementary figure are available with the online version of this paper.
References
- Avery, O. T. & Dubos, R. (1931). The protective action of a specific enzyme against type III pneumococcus infection in mice. J Exp Med 54, 73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaert, D., De Groot, R. & Hermans, P. W. (2004). Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis 4, 144–154. [DOI] [PubMed] [Google Scholar]
- Bratcher, P. E., Park, I. H., Hollingshead, S. K. & Nahm, M. H. (2009). Production of a unique pneumococcal capsule serotype belonging to serogroup 6. Microbiology 155, 576–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratcher, P. E., Kim, K. H., Kang, J. H., Hong, J. Y. & Nahm, M. H. (2010). Identification of natural pneumococcal isolates expressing serotype 6D by genetic, biochemical, and serological characterization. Microbiology 156, 555–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueggemann, A. B., Pai, R., Crook, D. W. & Beall, B. (2007). Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog 3, e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calix, J. J. & Nahm, M. H. (2010). A new pneumococcal serotype, 11E, has variably inactivated wcjE gene. J Infect Dis 202, 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho Mda, G., Pimenta, F. C., Gertz, R. E., Jr, Joshi, H. H., Trujillo, A. A., Keys, L. E., Findley, J., Moura, I. S. & other authors (2009). PCR-based quantitation and clonal diversity of the current prevalent invasive serogroup 6 pneumococcal serotype, 6C, in the United States in 1999 and 2006 to 2007. J Clin Microbiol 47, 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright, M. C. & Spratt, B. G. (1998). A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144, 3049–3060. [DOI] [PubMed] [Google Scholar]
- García, E., Llull, D., Munoz, R., Mollerach, M. & Lopez, R. (2000). Current trends in capsular polysaccharide biosynthesis of Streptococcus pneumoniae. Res Microbiol 151, 429–435. [DOI] [PubMed] [Google Scholar]
- Guerin, F., Varon, E., Hoi, A. B., Gutmann, L. & Podglajen, I. (2000). Fluoroquinolone resistance associated with target mutations and active efflux in oropharyngeal colonizing isolates of viridans group streptococci. Antimicrob Agents Chemother 44, 2197–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakenbeck, R., Konig, A., Kern, I., van der Linden, M., Keck, W., Billot-Klein, D., Legrand, R., Schoot, B. & Gutmann, L. (1998). Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level beta-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J Bacteriol 180, 1831–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichsen, J. (1995). Six newly recognized types of Streptococcus pneumoniae. J Clin Microbiol 33, 2759–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans, P. W., Blommaart, M., Park, I. H., Nahm, M. H. & Bogaert, D. (2008). Low prevalence of recently discovered pneumococcal serotype 6C isolates among healthy Dutch children in the pre-vaccination era. Vaccine 26, 449–450. [DOI] [PubMed] [Google Scholar]
- Jacobs, M. R., Bajaksouzian, S., Bonomo, R. A., Good, C. E., Windau, A. R., Hujer, A. M., Massire, C., Melton, R., Blyn, L. B. & other authors (2009). Occurrence, distribution, and origins of Streptococcus pneumoniae Serotype 6C, a recently recognized serotype. J Clin Microbiol 47, 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, M. R., Dagan, R., Bajaksouzian, S., Windau, A. R. & Porat, N. (2010). Validation of factor antiserum 6d for serotyping Streptococcus pneumoniae serotype 6C. J Clin Microbiol 48, 1456–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, P., Kong, F., Xiao, M., Oftadeh, S., Zhou, F., Liu, C., Russell, F. & Gilbert, G. L. (2009). First report of putative Streptococcus pneumoniae serotype 6D among nasopharyngeal isolates from Fijian children. J Infect Dis 200, 1375–1380. [DOI] [PubMed] [Google Scholar]
- Leach, A. J., Morris, P. S., McCallum, G. B., Wilson, C. A., Stubbs, L., Beissbarth, J., Jacups, S., Hare, K. & Smith-Vaughan, H. C. (2009). Emerging pneumococcal carriage serotypes in a high-risk population receiving universal 7-valent pneumococcal conjugate vaccine and 23-valent polysaccharide vaccine since 2001. BMC Infect Dis 9, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, J. P., III & Zhanel, G. G. (2009). Streptococcus pneumoniae: epidemiology, risk factors, and strategies for prevention. Semin Respir Crit Care Med 30, 189–209. [DOI] [PubMed] [Google Scholar]
- Martin, D. & Rybicki, E. (2000). rdp: detection of recombination amongst aligned sequences. Bioinformatics 16, 562–563. [DOI] [PubMed] [Google Scholar]
- Martin, D. P., Williamson, C. & Posada, D. (2005). rdp2: recombination detection and analysis from sequence alignments. Bioinformatics 21, 260–262. [DOI] [PubMed] [Google Scholar]
- Mavroidi, A., Godoy, D., Aanensen, D. M., Robinson, D. A., Hollingshead, S. K. & Spratt, B. G. (2004). Evolutionary genetics of the capsular locus of serogroup 6 pneumococci. J Bacteriol 186, 8181–8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavroidi, A., Aanensen, D. M., Godoy, D., Skovsted, I. C., Kaltoft, M. S., Reeves, P. R., Bentley, S. D. & Spratt, B. G. (2007). Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci. J Bacteriol 189, 7841–7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahm, M. H., Lin, J., Finkelstein, J. A. & Pelton, S. I. (2009). Increase in the prevalence of the newly discovered pneumococcal serotype 6C in the nasopharynx after introduction of pneumococcal conjugate vaccine. J Infect Dis 199, 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes, S., Valente, C., Sa-Leao, R. & de Lencastre, H. (2009). Temporal trends and molecular epidemiology of recently described serotype 6C of Streptococcus pneumoniae. J Clin Microbiol 47, 472–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, I. H., Park, S., Hollingshead, S. K. & Nahm, M. H. (2007a). Genetic basis for the new pneumococcal serotype, 6C. Infect Immun 75, 4482–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, I. H., Pritchard, D. G., Cartee, R., Brandao, A., Brandileone, M. C. & Nahm, M. H. (2007b). Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J Clin Microbiol 45, 1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, I. H., Moore, M. R., Treanor, J. J., Pelton, S. I., Pilishvili, T., Beall, B., Shelly, M. A., Mahon, B. E. & Nahm, M. H. (2008). Differential effects of pneumococcal vaccines against serotypes 6A and 6C. J Infect Dis 198, 1818–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, D. A., Briles, D. E., Crain, M. J. & Hollingshead, S. K. (2002). Evolution and virulence of serogroup 6 pneumococci on a global scale. J Bacteriol 184, 6367–6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N. & Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Nei, M. & Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101, 11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Nei, M. & Kumar, S. (2007). mega4: Molecular Evolutionary Genetics Analysis (mega) software version 4.0. Mol Biol Evol 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tettelin, H., Nelson, K. E., Paulsen, I. T., Eisen, J. A., Read, T. D., Peterson, S., Heidelberg, J., DeBoy, R. T., Haft, D. H. & other authors (2001). Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293, 498–506. [DOI] [PubMed] [Google Scholar]
- Tocheva, A. S., Jefferies, J. M., Christodoulides, M., Faust, S. N. & Clarke, S. C. (2010). Increase in serotype 6C pneumococcal carriage, United Kingdom. Emerg Infect Dis 16, 154–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzciński, K., Thompson, C. M. & Lipsitch, M. (2004). Single-step capsular transformation and acquisition of penicillin resistance in Streptococcus pneumoniae. J Bacteriol 186, 3447–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]